
Improved Accuracy of Stomata Micrograph Classification Using
YOLOv11 with Gamma Correction and CLAHE
Yusri Simang
1
, Indrabayu
1
a
and Siti Andini Utiarahman
2
1
Department of Informatics Engineering, Hasanuddin University, Makassar, Indonesia
2
Department of Electrical Engineering, Hasanuddin University, Makassar, Indonesia
Keywords: Micrograph, Stomata, YOLOv11, Gamma Correction, CLAHE.
Abstract: This study aims to improve the accuracy of stomata classification in micrograph images through the You Only
Look Once version 11 (YOLOv11) algorithm combined with Gamma Correction and Contrast Limited
Adaptive Histogram Equalization (CLAHE) as preprocessing techniques. Microscopic images often suffer
from uneven illumination and weak contrast, which hinder automatic stomata detection. Gamma Correction
adjusts image brightness non-linearly, while CLAHE enhances local contrast without amplifying noise.
Stomata images were captured using a binocular microscope, annotated based on morphology, and divided
into training, validation, and test sets for model development. Experimental results show that preprocessing
improves YOLOv11 performance, with precision increasing from 0.93 to 0.94, recall from 0.91 to 0.92, and
mean Average Precision (mAP) at 50% intersection over union from 0.95 to 0.96. For the Graminoid class,
precision increased from 0.84 to 0.86, recall from 0.88 to 0.89, and mAP@50 from 0.82 to 0.83. The Peak
Signal-to-Noise Ratio (PSNR) also improved from 27.9–28.35 dB to 28.44–29.40 dB, indicating better image
quality. These results demonstrate that the integration of Gamma Correction and CLAHE effectively enhances
image clarity and improves stomata detection performance, supporting more reliable and efficient automation
in botanical analysis.
1 INTRODUCTION
Stomata are microscopic pores on the surface of plant
leaves and stems that play a vital role in gas exchange
and the regulation of water loss through transpiration.
Their number, size, and morphology are important
parameters in plant physiology, ecological
adaptation, and taxonomic classification (Haworth et
al., 2023). Traditionally, stomata are identified
manually using light microscopy, a process that is
time-consuming, subjective, and highly dependent on
individual expertise. The challenge becomes greater
when micrograph images suffer from blurriness,
uneven illumination, or low contrast, which
significantly hampers accurate morphological
analysis (Gibbs & Burgess, 2024). To overcome these
limitations, computer vision and deep learning
approaches are increasingly applied for automatic
stomata classification.
You Only Look Once (YOLO) is a family of one-
stage object detection algorithms that perform
a
https://orcid.org/0000-0003-2026-1809
classification and localization simultaneously,
enabling real-time analysis. The latest version,
YOLOv11, introduces modules such as C2f, attention
mechanisms, and Spatial Pyramid Pooling Fast
(SPPF), which improve small-object detection and
computational efficiency. Several studies have
demonstrated its effectiveness for stomatal analysis
Zhang et al. (2021) combined YOLO with superpixel
segmentation for stomata recognition, Zhang et al.
(2023) proposed the DeepRSD method for randomly
oriented stomata, while Yang et al. (2025) developed
StomaYOLO for maize stomata detection. Beyond
stomata, YOLO has also been applied successfully in
plant disease classification (Alhwaiti et al., 2025).
These studies confirm YOLO’s robustness,
supporting its adoption in this research.
However, detection performance with YOLO
strongly depends on input image quality. Microscopic
images often suffer from uneven lighting, subtle
noise, and low contrast that obscure stomatal
structures. To address this, image enhancement
68
Simang, Y., Indrabayu, and Utiarahman, S. A.
Improved Accuracy of Stomata Micrograph Classification Using YOLOv11 with Gamma Correction and CLAHE.
DOI: 10.5220/0014275100004928
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Innovations in Information and Engineering Technology (RITECH 2025), pages 68-75
ISBN: 978-989-758-784-9
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
methods were employed before YOLOv11 detection.
Gamma Correction was used to normalize
illumination by amplifying darker regions without
overexposing bright areas, as shown effective in low-
light enhancement (Jeon et al., 2024; Wang et al.,
2023; Mim et al., 2025). Meanwhile, Contrast
Limited Adaptive Histogram Equalization (CLAHE)
was applied to enhance local contrast while
controlling noise, a technique widely validated in
biomedical and plant imaging (Liu et al., 2021; Gibbs
et al., 2021; Narla et al., 2024; Buriboev et al., 2024).
The integration of Gamma Correction and
CLAHE has also proven beneficial. For example,
Chang et al. (2018) reported that dual gamma
correction with CLAHE improved visual quality in
low-light images, while Benchabane & Charif (2025)
achieved a 6–9% accuracy gain in COVID-19 X-ray
classification using this combination. Mim et al.
(2025) further confirmed the effectiveness of
integrating these techniques for structural clarity and
classification performance. Despite these advances,
no studies have explicitly incorporated Gamma
Correction and CLAHE into the YOLOv11 pipeline
for stomatal classification. This research therefore
proposes an automatic stomata classification system
based on YOLOv11 with integrated Gamma
Correction and CLAHE preprocessing, aimed at
improving accuracy in tropical leaf micrographs and
supporting the automation of botanical analysis and
precision agriculture.
2 METHODOLOGY
This study focuses on the development of a stomata
classification and detection system using a deep
learning algorithm under uneven lighting conditions
in stomata micrograph images. The dataset used in
this study consisted of 312 micrograph images of
stomata from herbal plant leaves, captured using a
biological binocular microscope. These images were
distributed across four stomata classes: Anomocytic
(76 images), Diacytic (84 images), Graminoid (80
images), and Paracytic (72 images). The dataset was
divided into training (218 images, 70%), validation
(62 images, 20%), and testing (31 images, 10%).
To enhance the training set and reduce overfitting,
data augmentation was applied. For each training
image, three additional augmented images were
generated by applying 90° clockwise rotation, 90°
counter-clockwise rotation, and a flipped orientation.
This process expanded the training set from 218 to
588 images. By introducing these geometric
variations, the model was exposed to stomata with
different orientations, ensuring better class balance
and improving robustness in recognizing
morphological structures under diverse conditions.
Following the annotation process, bounding boxes
and class labels were assigned to each stomata
instance. Since a single micrograph may contain
multiple stomata, the number of annotated instances
exceeds the total number of images. Table 1
summarizes the distribution of labels across the four
stomata classes.
After dataset preparation and annotation, the
workflow of the proposed system, including
preprocessing, model training, and evaluation, is
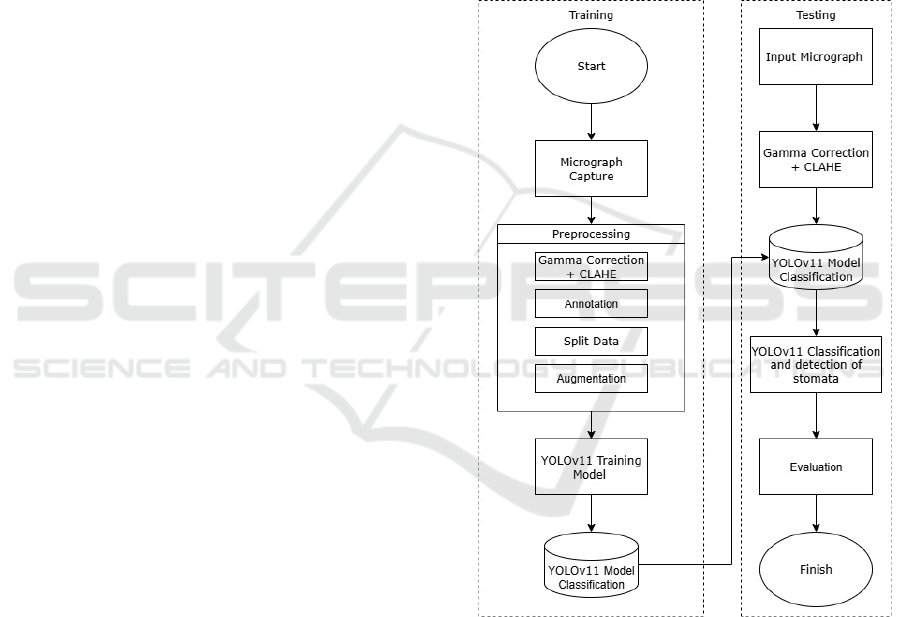
illustrated in Figure 1.
Figure 1: System Design Flow.
2.1 Training Phase
During the training phase, the YOLOv11 model was
fine-tuned using the Ultralytics framework. The pre-
processed dataset (Gamma Correction and CLAHE
applied) consisted of 588 augmented training images,
62 validation images, and 31 test images. Training
was performed for 100 epochs with a batch size of 4
and an input resolution of 640 × 640 pixels. The
learning rate was initialized at 0.001 with the default
Improved Accuracy of Stomata Micrograph Classification Using YOLOv11 with Gamma Correction and CLAHE
69
optimizer provided by Ultralytics. All experiments
were executed on a workstation equipped with an
NVIDIA RTX 4070 GPU (8 GB VRAM). The model
checkpoint with the highest validation mAP@50 was
saved for subsequent evaluation.
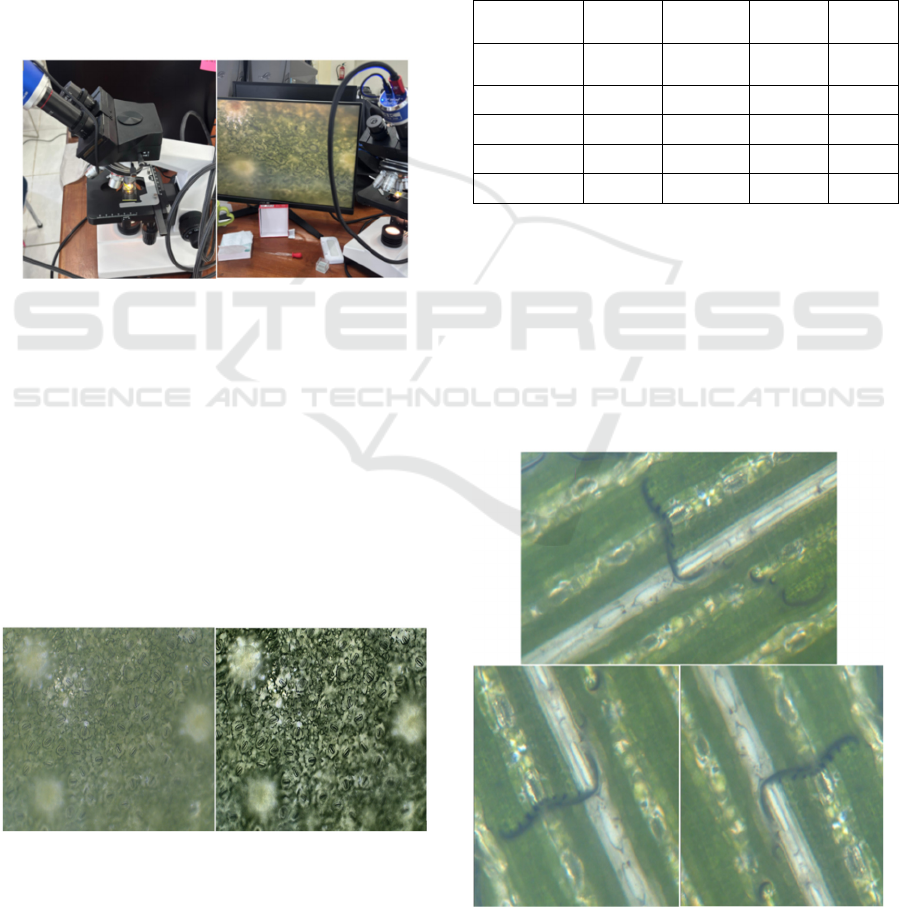
2.1.1 Data Acquisition
Stomata images were acquired using a biological
binocular microscope at 3× and 4× magnification,
covering the four stomata types analyzed in this
study. The samples were obtained from four different
herbal plants, with one representative species used for
each stomata type. The process of data acquisition as
shown in Figure 2.
Figure 2: Stomata micrograph data acquisition.
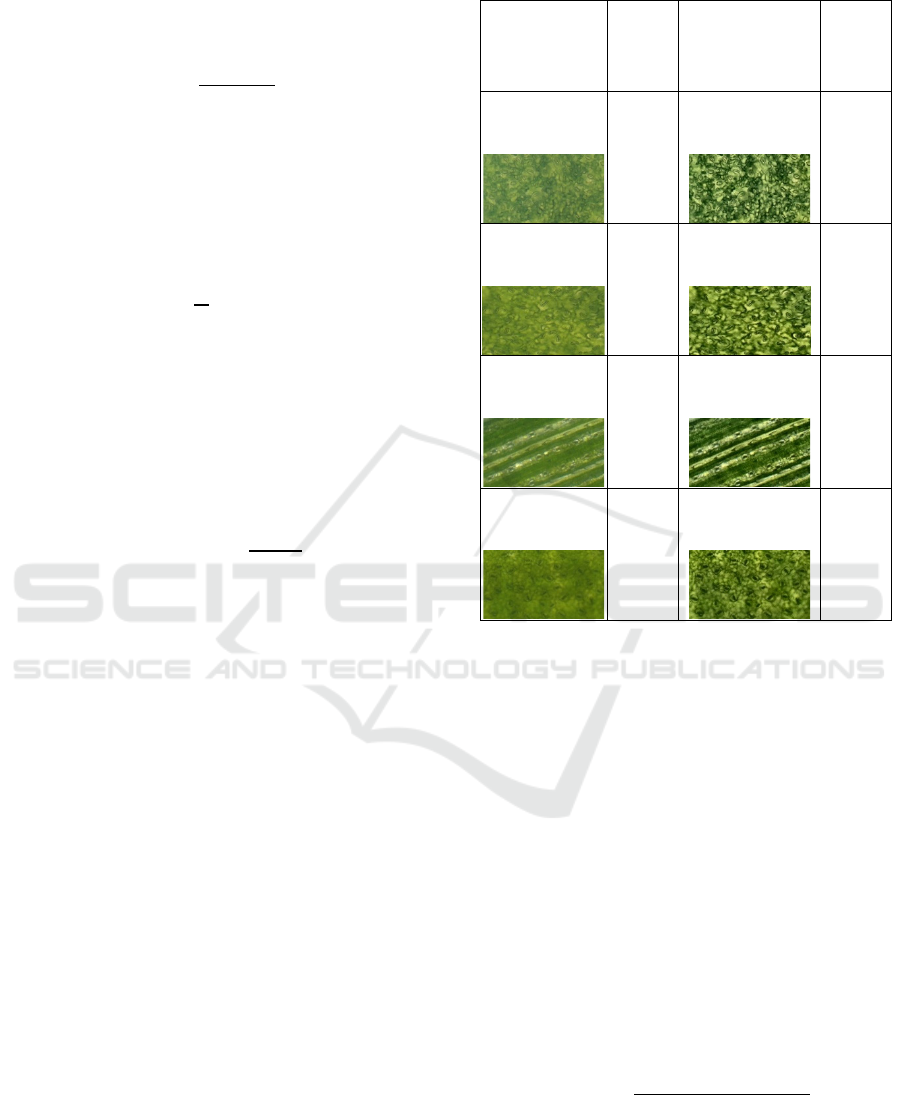
2.1.2 Preprocessing
The image was pre-processed using Gamma
Correction (γ = 0.9) to non-linearly adjust brightness,
making fine stomatal structures more visible in darker
regions. In addition, CLAHE was applied with a clip
limit of 3.0 and a tile grid size of 8×8 to enhance local
contrast while preventing excessive noise. These
enhancements ensured that the input images
preserved morphological details critical for stomata
recognition. Figure 3 illustrates one of preprocessing
result.
Figure 3: Preprocessing results.
2.1.3 Annotations
Stomata are given bounding boxes and morphological
labels to train the YOLOv11 model to recognize and
distinguish stomata types.
2.1.4 Split Data
The dataset was split into training, validation, and
testing subsets for model training and evaluation as
shown in Table 1.
Table 1: Distribution of stomata labels.
Class
Labels
Train
Labels
Validation
Labels
Testin
g
Label
count
Anomocyti
c
3679 339 230 4248
Diacytic 4925 770 141 5836
Graminoid 3532 520 217 4269
Paracytic 4618 266 355 5239
Total 16754 1895 943 19592
2.1.5 Data Augmentation
In the Figure 4, each original stomata image was
augmented into three variations: one rotated 90°
clockwise, one 90° counterclockwise, and one kept
unchanged. This increased the dataset from 219 to
590 images, helping the YOLOv11 model generalize
better by simulating different microscope
orientations.
Figure 4: Data augmentation results.
RITECH 2025 - The International Conference on Research and Innovations in Information and Engineering Technology
70
2.1.6 Training Model YOLOv11
The pre-processed and annotated images were then
used to train YOLOv11. This integration allowed the
model to learn more discriminative visual features
from images with improved lighting and contrast,
leading to better stomata classification performance.
2.1.7 Build Model
The model is stored after validation and is ready to be
used for classification and detection of stomata in test
data.
2.2 Testing Phase
In the testing phase, the trained YOLOv11 model was
evaluated on the held-out test set of 31 images, which
had been preprocessed with Gamma Correction and
CLAHE. No augmentation or annotation was applied
during testing to ensure fairness. The model
predictions were compared with ground-truth labels
using precision, recall, mAP@50, mAP@50–95, and
F1-score as evaluation metrics.
2.2.1 Input Data
The images tested are images that have never been
used before and are entered into the system to
evaluate the final performance of the model. The
results of this stage provide an overview of the extent
to which the model is capable of working on
completely new data.
2.2.2 Preprocessing (Testing)
In the testing phase, only Gamma Correction and
CLAHE were applied to the images. Unlike the
training phase, no annotation or augmentation was
performed, as the goal was to evaluate the model on
unseen images with enhanced visibility and contrast,
ensuring a fair and unbiased evaluation.
2.2.3 Yolov11 Stomata Detection &
Classification
The trained YOLOv11 model directly received the
pre-processed test images and performed stomata
detection and classification. By using enhanced
images, the model was able to achieve higher
precision and recall in identifying small stomatal
structures.
2.2.4 Evaluation
To assess the performance of the proposed YOLOv11
model, detection results were compared against the
ground truth annotations using widely adopted
evaluation metrics, including Accuracy, Precision,
Recall, mean Average Precision (mAP), and Peak
Signal-to-Noise Ratio (PSNR).
To evaluate the performance of the YOLOv11
model in detecting various types of stomata under
different image processing scenarios (without
preprocessing, with Gamma Correction, and with
CLAHE), a confusion matrix was employed as the
primary evaluation technique (Figure 5). The
confusion matrix provides a detailed summary of
classification outcomes by categorizing predictions
into four main groups: True Positive (TP),
representing stomata objects that were correctly
detected and classified by the model; False Negative
(FN), referring to stomata objects that were missed
and not detected by the model; False Positive (FP),
representing non-stomata regions that were
incorrectly classified as stomata; and True Negative
(TN), indicating non-stomata regions that were
correctly identified as not belonging to the stomata
class.
Figure 5: Confusion Matrix.
Where TP (True Positive) denotes the number of
correctly detected stomata, TN (True Negative)
denotes the number of correctly rejected non-stomata
regions, FP (False Positive) represents the number of
incorrectly detected regions, and FN (False Negative)
represents the number of missed stomata.
Precision (1) measures the proportion of
correctly predicted stomata among all detected
regions.
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 =
𝑇𝑃
𝑇𝑃
+
𝐹𝑃
(1
)
Improved Accuracy of Stomata Micrograph Classification Using YOLOv11 with Gamma Correction and CLAHE
71
Recall (2) measures the proportion of correctly
detected stomata with respect to all actual stomata in
the ground truth.
𝑅𝑒𝑐𝑎𝑙𝑙 =
𝑇𝑃
𝑇𝑃
+
𝐹𝑁
(2)
Where AP
i
represents the Average Precision for
class i, and N is the total number of classes. The mAP
(3) is widely used in object detection to evaluate the
overall detection performance across all classes.
𝑚𝐴𝑃 =
1
𝑁
𝐴𝑃
(3)
Where MAX
I
is the maximum possible pixel
value (255 for 8-bit images), and MSE is the Mean
Squared Error between the original image and the
processed image. PSNR (4) quantifies the quality of
image enhancement during preprocessing and
detection.
𝑃𝑆𝑁𝑅 = 10 ∙ 𝑙𝑜𝑔
(
𝑀𝐴𝑋
𝑀
𝑆
𝐸
)
(4)
3 RESULTS AND DISCUSSION
3.1 Peak Signal-to-Noise Ratio (PSNR)
Evaluation
The PSNR values in all three images showed an
increase after preprocessing using Gamma Correction
and CLAHE. In Table 2, the initial PSNR was in the
range of 27.9–28.35 dB and increased to 28.44–29.40
dB after preprocessing. This increase indicates an
improvement in the visual quality of the image,
especially in terms of lighting and local contrast,
without compromising important details. This proves
that the preprocessing technique used is effective in
clarifying the structure of the stomata, thus
supporting the automatic detection process by the
YOLOv11 model more accurately.
Table 2: PSNR evaluation.
Gamma
PSNR
(dB)
Gamma +
CLAHE
PSNR
(dB)
27.96
28.33
28.01
28.69
27.88
28.63
28.43
29.32
3.2 YOLOv11 Training Results with
Gamma Correction + CLAHE
The training was conducted on an NVIDIA RTX
4070 GPU (8 GB VRAM) using the Ultralytics
YOLO framework. Each epoch required ~2.5
minutes, with a total training time of 4.2 hours for 100
epochs, and inference reached 45 FPS (≈22 ms per
image), showing near real-time suitability. As shown
in Table 3, preprocessing improved YOLOv11
performance compared to the baseline (P = 0.93, R =
0.91, mAP@50 = 0.95, mAP@50–95 = 0.54).
Gamma Correction slightly raised recall and
mAP@50–95, CLAHE increased precision, and their
combination gave the best results (P = 0.94, R = 0.92,
mAP@50 = 0.96, mAP@50–95 = 0.55). Accuracy
was calculated as.
𝐴
𝑐𝑐𝑢𝑟𝑎𝑐𝑦 =
𝑇𝑃 + 𝑇𝑁
𝑇𝑃+𝑇𝑁+𝐹𝑃+𝐹𝑁
(5
)
The proposed method achieved 95% accuracy,
higher than Zhang et al. (2021) with 91.6%, Gibbs et
al. (2021) with 90.2%, Zhang et al. (2023) with
RITECH 2025 - The International Conference on Research and Innovations in Information and Engineering Technology
72
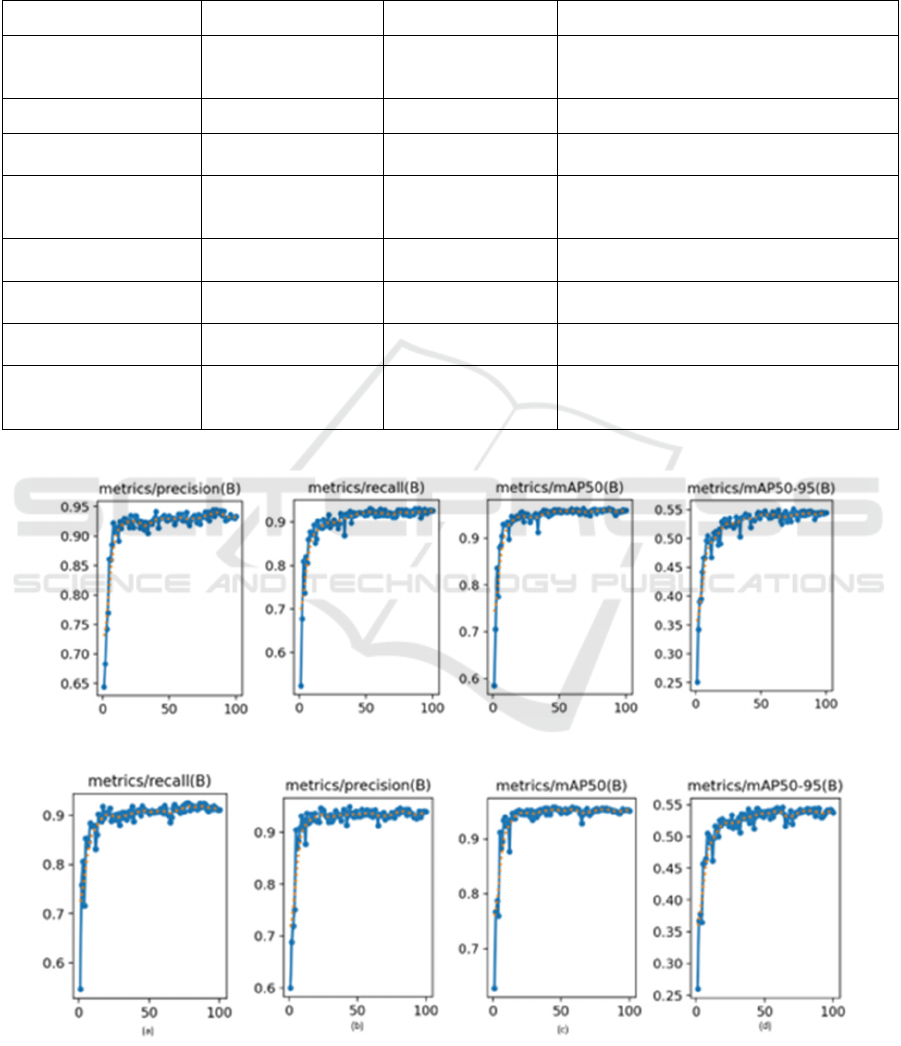
94.3%, and Utiarahman (2025) with 93.5%. These
results confirm that Gamma + CLAHE preprocessing
enhances YOLOv11 as shown in Figure 7, providing
the better balance of precision, recall, and accuracy
rather than without Gamma + CLAHE (Figure 6)..
Table 3: Results of YOLOv11 Original and YOLOv11 Training with Gamma + CLAHE.
Study / Method Dataset / Target Approach Reported Performance
Zhang et al., 2021
Stomata micrographs
(various plants)
YOLO + entropy rate
superpixel
segmentation
Accuracy = 93%
Zhang et al., 2023 Corn leaves DeepRSD Accuracy= 94.3%
Gibbs et al., 2021
Wheat stomata
micrographs
CNN-based Accuracy > 90%
Utiarahman, 2025 Herbal plants
Benchmark
YOLOv8–v11
YOLOv11: Precision = 0.933, Recall =
0.886, mAP@50 = 0.951, mAP@50–95 =
0.515
This study (YOLOv11
b
aseline)
Herbal plant stomata (4
types)
YOLOv11 without
p
reprocessing
Accuracy = 94% Precision = 0.93, Recall =
0.91, mAP@50 = 0.95, mAP@50
–
95 = 0.54
This study (YOLOv11 +
Gamma
)
Herbal plant stomata (4
t
yp
es
)
YOLOv11 with
Gamma
Accuracy = 95% Precision = 0.93, Recall =
0.92, mAP
@
50 = 0.95, mAP
@
50
–
95 = 0.55
This study (YOLOv11 +
CLAHE
)
Herbal plant stomata (4
t
yp
es
)
YOLOv11 with
CLAHE
Accuracy = 94% Precision = 0.94, Recall =
0.91, mAP
@
50 = 0.94, mAP
@
50
–
95 = 0.54
This study (YOLOv11 +
Gamma + CLAHE)
Herbal plant stomata (4
types)
YOLOv11 with
Gamma Correction +
CLAHE
Accuracy = 95% Precision = 0.94, Recall =
0.92, mAP@50 = 0.96, mAP@50–95 = 0.55
Figure 6: Graph of the results of the original YOLOv11 training (mAP 50, mAP 50-95, precision, and recall).
Figure 7: Graph of YOLOv11 training results with Gamma Correction + CLAHE (mAP 50, mAP 50-95, precision, and
recall).
Improved Accuracy of Stomata Micrograph Classification Using YOLOv11 with Gamma Correction and CLAHE
73
3.3 YOLOv11 Testing Results with
Gamma + CLAHE
The test results in Table 4 showed that the YOLOv11
model with Gamma Correction and CLAHE
preprocessing outperformed the original model. In the
Anomocytic and Paracytic (AD) classes, there was an
increase in precision, mAP, and accuracy. For the
Graminoid class, recall increased from 0.88 to 0.89
and precision from 0.84 to 0.86, indicating improved
detection. Meanwhile, the Diacytic class achieved
consistently high results on both models (precision
0.95–0.96; accuracy 0.99), indicating optimal
detection even without preprocessing.
Table 4: YOLOv11 Testing Results with Gamma + CLAHE.
Metho
d
Class TP FP FN TN Precision Recall Accurac
y
mAP 50 mAP 50-95
YOLOv11
Original
Anomocytic 202 28 32 787 0.91 0.83 0.94 0.93 0.52
Diac
y
tic 138 3 7 901 0.95 0.97 0.99 0.98 0.57
Graminoi
d
200 17 44 788 0.84 0.88 0.93 0.82 0.43
Parac
y
tic 352 3 23 671 0.95 0.98 0.98 0.99 0.59
YOLOv11 +
Gamma
Correction
Anomocytic 201 29 30 797 0.90 0.83 0.94 0.93 0.53
Diacytic 138 3 9 907 0.95 0.97 0.99 0.99 0.58
Graminoi
d
200 17 50 790 0.85 0.90 0.94 0.83 0.44
Parac
y
tic 350 5 26 676 0.95 0.98 0.97 0.99 0.60
YOLOv11 +
CLAHE
Anomoc
y
tic 199 31 33 795 0.92 0.83 0.94 0.92 0.52
Diacytic 138 18 47 794 0.96 0.98 0.99 0.99 0.57
Graminoi
d
199 18 47 794 0.86 0.87 0.94 0.82 0.43
Paracytic 351 4 27 676 0.95 0.98 0.97 0.99 0.59
YOLOv11 +
Gamma
Correction +
CLAHE
Anomoc
y
tic 204 26 32 793 0.91 0.85 0.95 0.94 0.52
Diac
y
tic 138 3 10 904 0.96 0.98 0.99 0.99 0.58
Graminoi
d
201 16 10 790 0.86 0.89 0.94 0.83 0.43
Paracytic 351 4 22 678 0.95 0.98 0.98 0.99 0.60
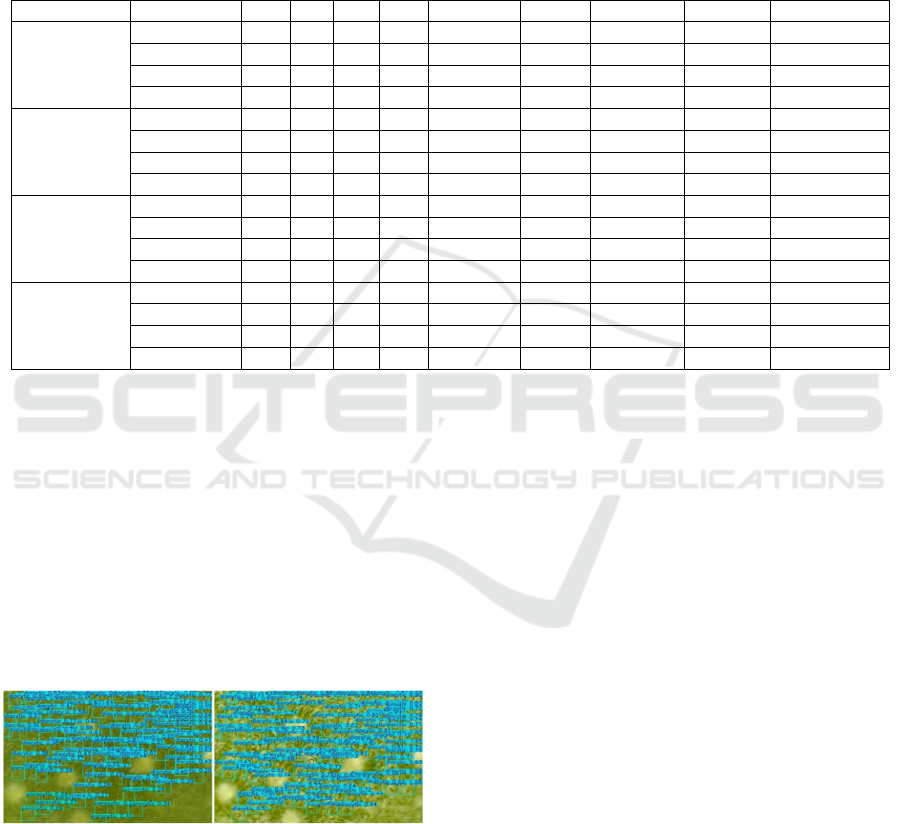
3.4 Results of Gamma+CLAHE
Application
Figure 8 presents the results of stomata detection on
micrograph images processed with Gamma
Correction and CLAHE. The YOLOv11 model
successfully detects stomata, as indicated by the
bounding boxes labeled "diacytic" and "paracytic" in
the relevant regions. CLAHE enhances local contrast,
making stomatal structures clearer even in previously
blurry or dim areas, while Gamma Correction
balances overall illumination across the image.
Figure 8: Comparison of the application of Gamma
Correction + CLAHE.
4 CONCLUSIONS
This study developed a YOLOv11-based automatic
stomata classification system with Gamma
Correction and CLAHE preprocessing. The results
showed improved image quality (higher PSNR) and
better detection performance in terms of precision,
recall, and mAP. The model provided more stable and
accurate detection of different stomata types,
confirming the benefit of combining image
enhancement with deep learning for botanical
analysis.
However, the study has limitations, including a
relatively small dataset restricted to four stomata
types and moderate performance gains from
preprocessing. Although YOLOv11 achieved
acceptable inference speed, further optimization is
needed for large-scale or real-time use.
Future work should involve real-time
experiments, the integration of attention mechanisms
to enhance feature extraction, and expansion of the
dataset with additional plant species to improve
robustness and generalizability. These efforts are
expected to increase both the efficiency and accuracy
of automatic stomata detection for broader
applications in plant science and precision
agriculture.
RITECH 2025 - The International Conference on Research and Innovations in Information and Engineering Technology
74

ACKNOWLEDGEMENTS
The authors express their gratitude to colleagues and
collaborators for their valuable support, and to the
laboratory staff for assistance during data collection.
Special thanks are also due to the reviewers for their
constructive feedback, which improved the quality of
this manuscript. The use of generative AI tools is
acknowledged for enhancing the language and
readability of this paper under human oversight.
REFERENCES
Alhwaiti, Y., Khan, M., Asim, M., Siddiqi, M. H., Ishaq,
M., & Alruwaili, M. (2025). Leveraging YOLO deep
learning models to enhance plant disease identification.
Scientific Reports, 15(1).
https://doi.org/10.1038/s41598-025-92143-0
Benchabane, A., & Charif, F. (2025). Enhanced COVID-19
Detection Through Combined Image Enhancement and
Deep Learning Techniques. Informatica (Slovenia),
49(16), 67–76.
https://doi.org/10.31449/inf.v49i16.5869
Buriboev, A. S., Khashimov, A., Abduvaitov, A., & Jeon,
H. S. (2024). CNN-Based Kidney Segmentation Using
a Modified CLAHE Algorithm. Sensors, 24(23).
https://doi.org/10.3390/s24237703
Gibbs, J. A., & Burgess, A. J. (2024). Application of deep
learning for the analysis of stomata: a review of current
methods and future directions. In Journal of
Experimental Botany (Vol. 75, Issue 21, pp. 6704–
6718). Oxford University Press.
https://doi.org/10.1093/jxb/erae207
Gibbs, J. A., Mcausland, L., Robles-Zazueta, C. A.,
Murchie, E. H., & Burgess, A. J. (2021). A Deep
Learning Method for Fully Automatic Stomatal
Morphometry and Maximal Conductance Estimation.
Frontiers in Plant Science, 12.
https://doi.org/10.3389/fpls.2021.780180
Haworth, M., Marino, G., Materassi, A., Raschi, A., Scutt,
C. P., & Centritto, M. (2023). The functional
significance of the stomatal size to density relationship:
Interaction with atmospheric [CO2] and role in plant
physiological behaviour. In Science of the Total
Environment (Vol. 863). Elsevier B.V.
https://doi.org/10.1016/j.scitotenv.2022.160908
Jeon, J. J., Park, J. Y., & Eom, I. K. (2024). Low-light
image enhancement using gamma correction prior in
mixed color spaces. Pattern Recognition, 146.
https://doi.org/10.1016/j.patcog.2023.110001
Liu, H., Zhao, Z., & She, Q. (2021). Self-supervised ECG
pre-training. Biomedical Signal Processing and
Control, 70.
https://doi.org/10.1016/j.bspc.2021.103010
Mim, S. A., Cheng, J., & Zhang, J. (2025). Fusion-based
framework for low-light image enhancement using
BM3D filter and adaptive gamma correction. Optics
and Laser Technology, 192.
https://doi.org/10.1016/j.optlastec.2025.113662
Narla, V. L., Suresh, G., Rao, C. S., Awadh, M. Al, &
Hasan, N. (2024). A multimodal approach with firefly
based CLAHE and multiscale fusion for enhancing
underwater images. Scientific Reports, 14(1).
https://doi.org/10.1038/s41598-024-76468-w
Utiarahman, S. A., Indrabayu, & Nurtanio, I. (2025).
Performance Evaluation of YOLOv8 to YOLOv11 for
Accurate Detection and Classification of Stomata in
Microscopic Images of Herbal Plants. Proceedings of
the NMITCON 2025 International Conference. IEEE.
https://doi.org/10.1109/NMITCON65824.2025.11188
393
Wang, Y., Liu, Z., Liu, J., Xu, S., & Liu, S. (2023). Low-
Light Image Enhancement with Illumination-Aware
Gamma Correction and Complete Image Modelling
Network. http://arxiv.org/abs/2308.08220
Yang, Z., Liao, Y., Chen, Z., Lin, Z., Huang, W., Liu, Y.,
Liu, Y., Fan, Y., Xu, J., Xu, L., & Mu, J. (2025).
StomaYOLO: A Lightweight Maize Phenotypic
Stomatal Cell Detector Based on Multi-Task Training.
Plants, 14(13). https://doi.org/10.3390/plants14132070
Zhang, F., Ren, F., & Li, J. (2021). Automatic stomata
recognition and measurement based on improved
YOLO deep learning model and entropy rate superpixel
algorithm. Ecological Informatics, 68, 101521.
https://doi.org/10.1016/j.ecoinf.2021.101521
Zhang, F., Wang, B., Lu, F., & Zhang, X. (2023). Rotating
Stomata Measurement Based on Anchor-Free Object
Detection and Stomata Conductance Calculation. Plant
Phenomics, 5.
https://doi.org/10.34133/plantphenomics.0106
Improved Accuracy of Stomata Micrograph Classification Using YOLOv11 with Gamma Correction and CLAHE
75
