
Identification of Thalassemia Using Support Vector Machine
Gurbinder Kaur and Vijay Kumar Garg
School of Computer Science and Engineering, Lovely Professional University, Phagwara, Punjab, India
Keywords: Thalassemia, Support Vector Machine (SVM), Machine Learning, Hematological Parameters.
Abstract: Thalassemia is an inherited blood disorder where a person does not have normal hemoglobin production and
is due to abnormal genes present in the DNA of a person. Consanguineous marriages bring about an increase
in the occurrence of such disorders especially in the Thai regions of the world thus, making it a serious public
health issue. An early and precise diagnosis for this type of blood disorder is crucial for better patient
management as well as providing genetic counseling effectively. This study focuses on the application of
Support Vector Machine to identify thalassemia from CBC data. We aim to develop a reliable and robust
model by integrating various hematological parameters including hemoglobin concentration, red blood cell
indices and other Thalassemia relevant biomarkers. The concerned dataset was pre-treated to take care of the
missing values and maximization of SVM efficacy was achieved by data normalization. It was found that the
SVM model is quite efficient in terms of accuracy of classification, hence it can be considered as a good tool
for diagnosis for Thalassemia. The SVM technique has the potential to make the identification, treatment
expeditious and litheness and the scope of Thalassemia detection relatively large.
1 INTRODUCTION
The genetic disorder ‘Thalassemia’ is
heterogeneously grouped disease with low
production of hemoglobin (Hb) alpha and beta chains.
Hemoglobin is responsible for carrying oxygen to
various parts of body, the component of red blood
cells. It is formed by alpha and beta proteins. When
the human body does not produce any of these
proteins, red blood cells are grown enough to supply
sufficient oxygen that causes anemia in early
childhood stage and remain for life-long (Weatherall
et al. 2001). Thalassemia is genetically transfer from
one of the parents to their child. The reason of disease
can be mutation or deletion of certain genes. Alpha
thalassemia patients have no alpha-globin gene due to
which there is either decrease or completely absence
of alpha globin chain. Four alleles are present in the
alpha globin gen, and depending on the quantity of
alleles deleted, alpha thalassemia varies from mild to
severe. The most severe form involves the deletion of
all four alleles, resulting in no production of alpha
globin’s, while excess gamma chains, which are
typically present during fetal development, form
tetramers. This condition is not compatible with life
and leads to hydrops fetalis. In contrast, the deletion
of just one allele represents the mildest form and is
often clinically silent. In beta thalassemia, body is not
able to produce enough beta-globin genes, one of the
essential components of hemoglobin, which are
crucial for RBCs. The shortage of RBCs occurs due
to decease in beta globin that cause anemia (Origa R.
(2017)). The severity and type of beta thalassemia
depend on the degree of anemic patients. Sometimes
it is mild anemia, and this type of the disease is
commonly called as minor beta thalassemia or having
beta thalassemia trait. This is starting type of beta
thalassemia and does not usually require medical
treatment. The patient with moderate anemia has
intermedia type of Beta thalassemia requires blood
transfusion Tung. Patient suffering from beta
thalassemia major need continuous medical care
along with therapy of blood transfusion.
Thalassemia is diagnosed through blood tests and
genetic analysis in secure lab conditions traditionally
(Weatherall et al. 2001). These diagnosed methods
are accurate but slow and expensive. In some regions
it is not possible to diagnose thalassemia due to
limited resources. Presently with the machine
learning techniques, disease diagnosis become less
costly, scalable and faster. This research paper’s
objective is to create SVM model for classification
between normal and thalassemia patients using CBC
parameters.
600
Kaur, G. and Garg, V. K.
Identification of Thalassemia Using Support Vector Machine.
DOI: 10.5220/0013917500004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 4, pages
600-604
ISBN: 978-989-758-777-1
Proceedings Copyright © 2026 by SCITEPRESS – Science and Technology Publications, Lda.

2 RELATED WORK
Study (Lachover-Roth et al. 2024) diagnose α
thalassemia carriers based on RBC indices and
analysis of hemoglobin. The SVM formula developed
and then validated using a dataset of 1334 suspect
carrier women for detection, compared to molecular
analysis. Shine and Lal, as well as Support Vector
Machine formulas were effective in the detection of
thought-to-be α-thalassemia carriers with high
sensitivity and NPV(Negative predictive value). SVM
formula in detecting α thalassemia carriers: The
sensitivity of the SVM by 99.33%, NPV - 99.93%
Shine and Lal formula - Sensitivity (85.54%), NPV
(98.93%).
Study (Fu et al. 2021) construct the machine-
learning classifier by combining existing indices and
novel ones using machine learning algorithm. A total
of 350 patients suspected to have thalassemia
contributed a dataset for the training and validation of
our classifier, compared in performance with thirteen
indices. This method first created a classifier through
adopting SVM model and cross-validation (10-fold)
was then employed for selecting optimal parameters
for SVM model. The average AUC and error rate of
this classifier were 0.76 (the highest) and 0.26,
respectively.
Authors (Sa'Id, A. A et al. 2021) used Twin
Support Vector Machines (TSVM) in this study as one
of the machines learning developed techniques
inspired by Support Vector Machines (SVM), as this
technique purposed to find the nonparallel
hyperplanes to solve binary classification problem.
This was done by three popular kernels based on many
studies such as Linear, Polynomial, and Radial Basis
Function (RBF). The model TSVM gives best
performance using kernel function ‘Rbf’ having
average accuracy 99.32%, precision 99.75% and F1
score 99.24%. the model performance on
‘Polynomial’ kernel is 99.79% average recall.
Article (Wirasati et al. 2021) compare the
performance of SVM model with ‘Linear’,
‘Polynomial’ and ‘Gaussian Radial Basis Function’ to
classify thalassemia dataset. The training and testing
ratio used was 90% and 10% respectively. The
accuracy of kernel function ‘Rbf’ was 99.63%. The
performance of model on ‘Linear’ and ‘Polynomial’
was with accuracy of 98.23% and 97.9% respectively.
Authors (Laengsri et al. 2019) create a machine
learning model classify thalassemia and iron-
deficiency anemia using hemoglobin parameters
derived from CBC analysis. In this paper, five ML
techniques – random forest, support vector machine,
decision tree, artificial neural network and k-NN was
used to create a model. The dataset of 186 patients
with 13 indices and discriminate formulas was used to
train the model. The accuracy of this ThalPred model
was 95.59% with performance value AUC of 0.98 and
MCC of 0.87. SVM model performs better than other
models to discriminate IDA and TT.
Research (Laeli et al. 2020) objective was to
analyze thalassemia dataset with SVM model by
doing hyperparameters tuning using Grid Search. The
regularization parameter C and gamma parameter with
kernel function ’rbf’ was used in experiment. The
proposed model gives accuracy of 100% with
parameters value of 428.13 and 0.0000183 for C and
gamma respectively. The training data of 90% was
trained on holdout validation technique. The 10-fold
cross validation technique also shows accuracy of
100% with value of 4832.93 and 0.0000183 for C and
gamma parameters. It values gives better performance
with C and gamma parameters than using their default
values in ‘rbf’ function. The accuracy of 73.33% on
holdout and accuracy of 57.14% for on 10-fold cross
validation was shown by model.
Study (Roth et al. 2018) published formulas on a
database of more than 22,000 samples, and created a
new formula based on an SVM to identify β-
thalassemia carriers. The formulas were judged
according to their sensitivity, specificity and negative
predictive value. The study found that the SVM
formula and Shine's formula both showed sensitivity
of >98% and value of negative predictive value (NPV)
> 99.77% in detecting β-thalassemia carriers. All other
published formulas gave inferior results.
Paper (Amendolia et al. 2003) uses ML models for
thalassemia screening using SVM, MLP and KNN.
The analysis proposes a dual-classifier framework
predicated on SVM: the initial layer distinguishes
between cases of pathological and non-pathological
patients, and the next layer differentiates between two
distinct pathologies. The findings indicate that the
MLP classifier yields marginally superior outcomes in
comparison to SVM, achieving a sensitivity of 92%
and a specificity of 95%. However, SVM
demonstrates a sensitivity of 83% coupled with a
specificity of 95%. Although both classifiers perform
admirably, this study highlights the nuanced
differences in their effectiveness.
3 SUPPORT VECTOR MACHINE
(SVM)
Support Vector Machines are indeed employed in the
medical field for various tasks: disease classification,
Identification of Thalassemia Using Support Vector Machine
601
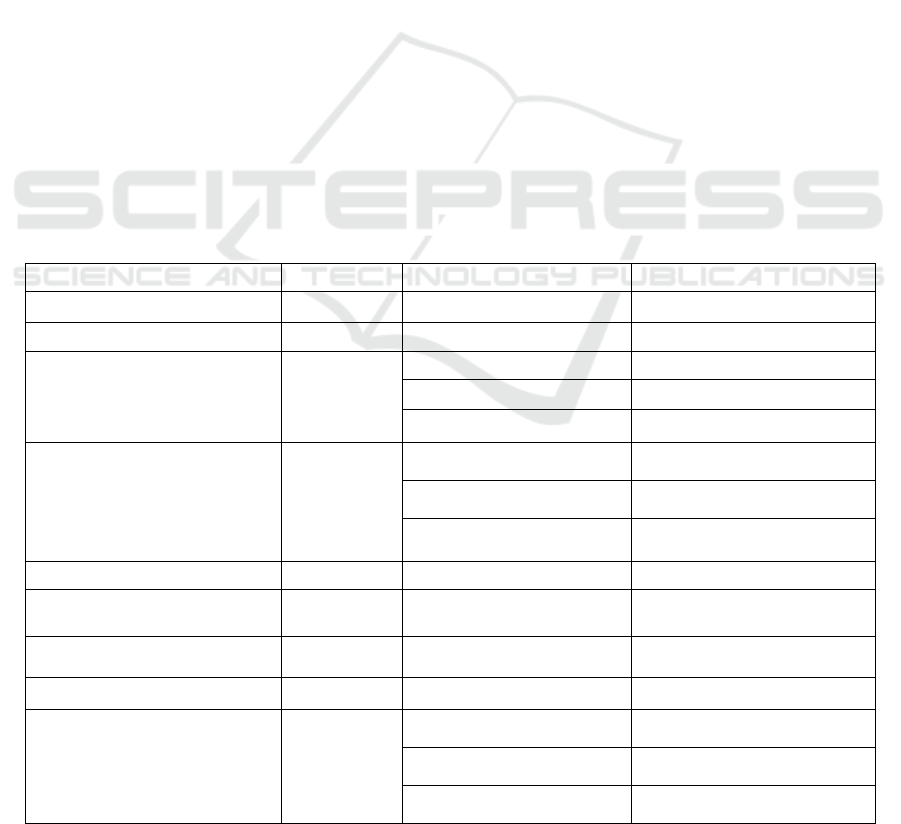
image analysis and outcome prediction. However,
there are circumstances in which alternative machine
learning algorithms like deep learning models might
be preferred for certain medical applications because
they can handle complex and data of high-dimensions
(Rustam et al. 2022). In this realm of SVM, a
hyperplane functions as the decision boundary that
separates data points into distinct categories. In a two-
dimensional space, this appears as a line; in higher
dimensions, it transforms into a plane or
hypersurface. SVM is particularly well-known for its
use of "kernels," which enable it to perform
exceptionally well even when confronted with data
that is not linearly separable (Widodo et al. 2007).
Support Vector Machines (SVMs) have demonstrated
exceptional proficiency in managing intricate
medical data, primarily because of their capacity to
navigate nonlinear relationships among no. of
different features along with classes. The primary
advantage of SVM classifier is its prowess of
pinpointing an optimized decision boundary, which
epitomizes the maximum margin to separate different
classes. This optimal hyperplane is formed using
training samples having a limited subset of total
samples, known as support vectors (SVs) that are
important data structures of SVM. These support
vectors discard the irrelevant training samples
keeping the integrity of the SVM's decision function,
called the ‘optimal hyperplane’ (Guido R et al 2024).
4 METHODOLOGY
The modules are designed for identification of
thalassemia patients into ‘Normal’ and ‘Beta-
Thalassemia’. The modules are as under (Figure 1):
• Dataset and its preprocessing: The CBC
parameters (Table-1) of patients have been
gathered from local labs and hospitals for to
distinguish individuals who may, however, be
at risk for thalassemia. The target class of
dataset is ‘Normal’ and ‘Beta Thalassemia’.
The dataset has 1 parameter including target
class.
• Data Splitting, outlier detection and data
balancing: Train-Test split is done before
outlier handing and balancing of dataset using
Smote Tomek is performed on train data to
ensure the integrity of the data.
• Supervised Support Vector Classifier on train
data is initialized, hyperparameter tuning and
Stratified fold using 5-fold is used for cross
validation of train data.
Table 1: SVM parameters.
CBC Paramete
r
Abb.use
d
Age Group Normal Range
Mean Corpuscular Volume MCV All ages 80-100 fL
Mean Corpuscular Hemoglobin MCH All ages 27-31 pg
Red Blood Cell Count RBC
Adults (Men) 4.5-5.9 x 10^12/L
Adults (Women) 4.1-5.1 x 10^12/L
Children (1-12 years) 4.0-5.5 x 10^12/L
Hemoglobin HB
Adults (Men) 13.8-17.2 g/dL
Adults (Women) 12.1-15.1 g/dL
Children (1-12 years) 11.0-13.5 g/dL
Red Cell Distribution Width RDW All ages 11.5-14.5%
Mean Corpuscular Hemoglobin
Concentration
MCHC All ages 0.5-2.5%
Platelet Count PLT All ages 32-36 g/dL
White Blood Cell Count WBC All ages 150,000-400,000 per µL
Hematocrit HCT
Adults (Men) 4,000-11,000 per µL
Adults (Women) 38.3-48.6(0.383-0.486)
Children (1-12 years) 35.5-44.9% (0.355-0.449)
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
602
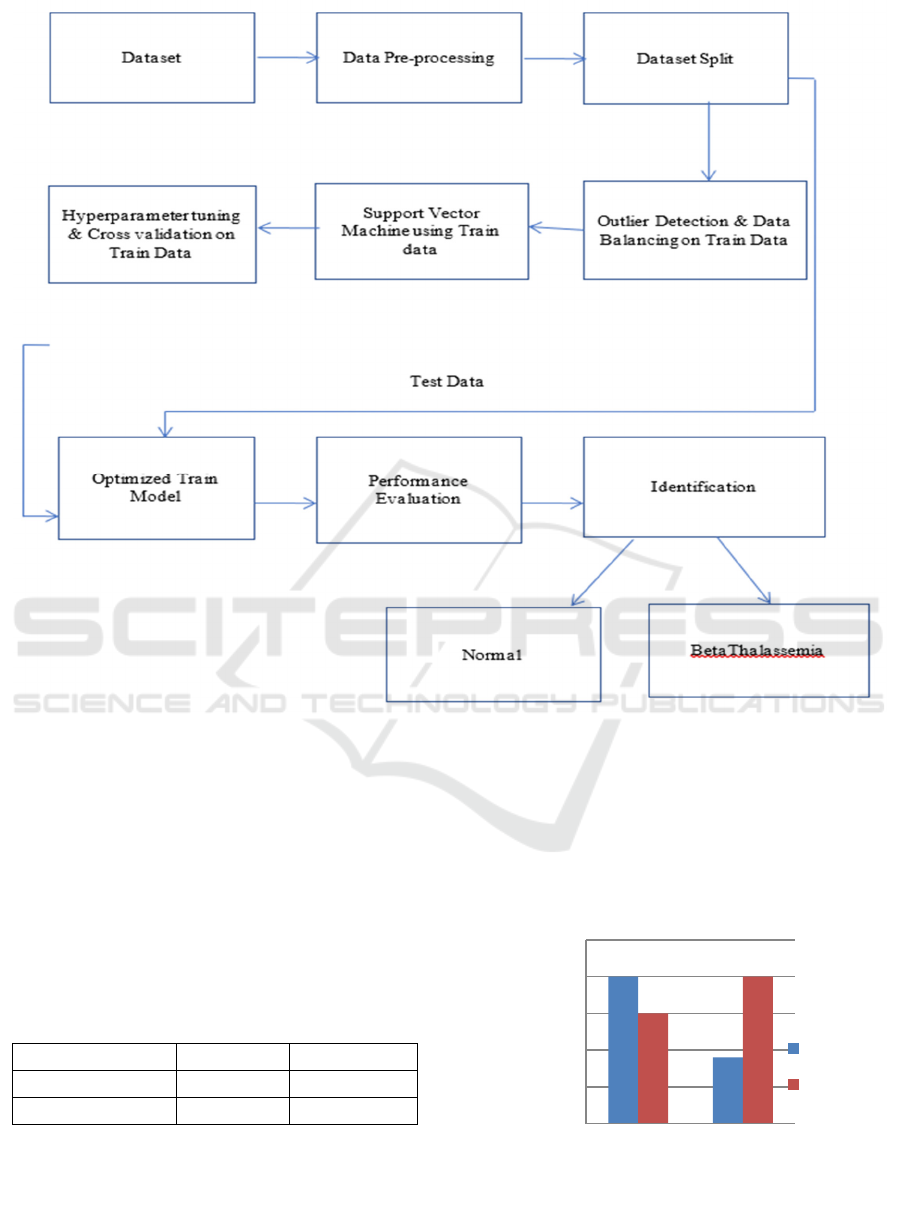
Figure 1: Proposed methodology. Source: Made by author.
• Optimized train model is tested by test data
and Performance metrics Accuracy, Precision,
Recall and F1 score is calculated. Finally, the
model identified
• The patient with ‘Normal’ or ‘Beta
Thalassemia’.
5 RESULT ANALYSIS
Table 2: Precision and recall on test data.
Class Label Precision Recall
0 1.00 0.89
1 0.95 1.00
The real-world thalassemia dataset is used in
classifying ‘Normal’ and ‘Thalassemia’ patients.
CBC indices including demographic features ‘Age’,
‘Gender’ and target classes are used to create model
with SVM technique. The following table 2 shows the
performance metrics on test data. The accuracy score
of SVM model is 96%.
The graphical representation of Precision and
Recall is shown in figure 2:
Figure 2: Precision and recall graph.
0.8
0.85
0.9
0.95
1
1.05
Precision Recall
Class-0
Class-1
Identification of Thalassemia Using Support Vector Machine
603
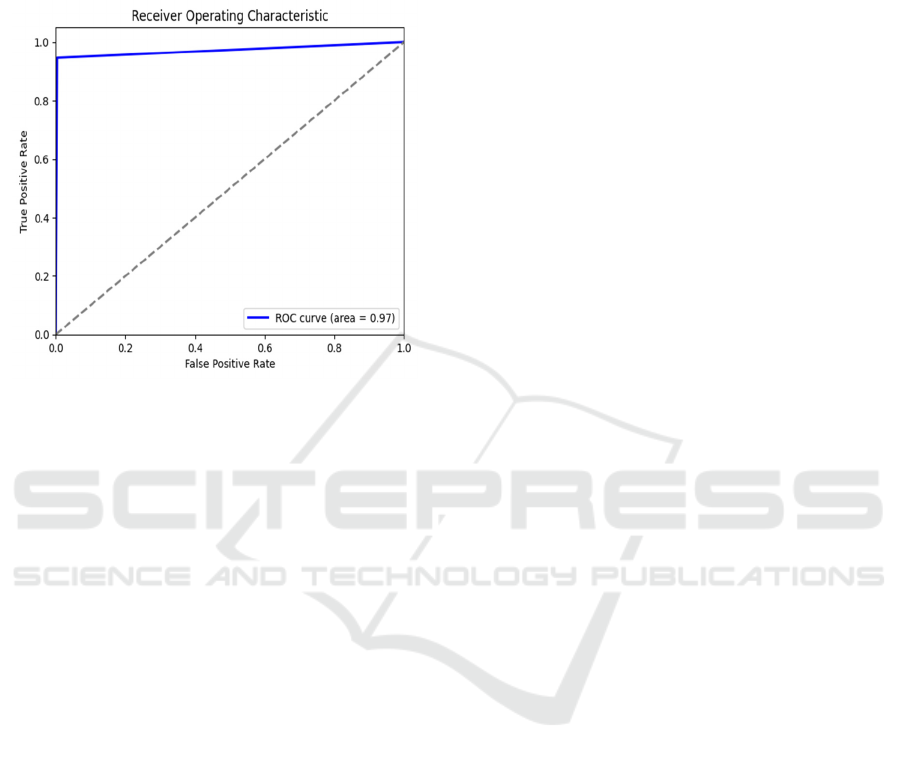
Receiver Operating Curve is graphical
representation of how the model will classify data. At
different threshold levels, true positive rate is plotted
against false positive rate. The AUC-ROC is 0.97%
for this SVM model is shown in figure 3.
Figure 3: ROC curve. Source: Made by author.
6 CONCLUSIONS
The methodology to classify thalassemia with real-
world dataset using SVM technique can assist
medical professional using analysis of CBC indices.
It is clear from accuracy of the model that
practitioners can relies on this model for predicting
thalassemia condition. In future other techniques will
be applied on same dataset for better performance.
REFERENCES
Amendolia, S. R., Cossu, G., Ganadu, M. L., Golosio, B.,
Masala, G. L., & Mura, G. M. (2003). A comparative
study of k-nearest neighbour, support vector machine
and multi-layer perceptron for thalassemia
screening. Chemometrics and Intelligent Laboratory
Systems, 69(1-2), 13-20.
Ferih, K., Elsayed, B., Elshoeibi, A. M., Elsabagh, A. A.,
Elhadary, M., Soliman, A., Abdalgayoom, M., &
Yassin, M. (2023). Applications of Artificial
Intelligence in Thalassemia: A Comprehensive
Review. Diagnostics (Basel, Switzerland), 13(9), 1551.
https://doi.org/10.3390/diagnostics13091551
Fu, Y. K., Liu, H. M., Lee, L. H., Chen, Y. J., Chien, S. H.,
Lin, J. S., ... & Lai, J. Y. (2021). The TVGH-NYCU
Thal-Classifier: Development of a Machine-Learning
Classifier for Differentiating Thalassemia and Non-
Thalassemia Patients. Diagnostics (Basel) 2021; 11 (9):
1725. DOI: https://doi.
org/10.3390/diagnostics11091725. PMID:
https://www. ncbi. nlm. nih. gov/pubmed/34574066.
Guido R, Ferrisi S, Lofaro D, Conforti D. (2024). An
Overview on the Advancements of Support Vector
Machine Models in Healthcare Applications: A
Review. Information, 15(4):235.
https://doi.org/10.3390 /info15040235
Lachover-Roth, I., Peretz, S., Zoabi, H., Harel, E., Livshits,
L., Filon, D., ... & Koren, A. (2024). Support Vector
Machine-Based Formula for Detecting Suspected α
Thalassemia Carriers: A Path toward Universal
Screening. International Journal of Molecular
Sciences, 25(12), 6446.
Laeli, A. R., Rustam, Z., Hartini, S., Maulidina, F., &
Aurelia, J. E. (2020, November). Hyperparameter
optimization on support vector machine using grid
search for classifying thalassemia data. In 2020
International Conference on Decision Aid Sciences and
Application (DASA) (pp. 817-821). IEEE.
Laengsri, V., Shoombuatong, W., Adirojananon, W.,
Nantasenamat, C., Prachayasittikul, V., & Nuchnoi, P.
(2019). ThalPred: a web-based prediction tool for
discriminating thalassemia trait and iron deficiency
anemia. BMC medical informatics and decision
making, 19, 1-14.
Origa R. (2017). β-Thalassemia. Genetics in medicine:
official journal of the American College of Medical
Genetics, 19(6), 609–619.
https://doi.org/10.1038/gim.2016.173
Roth, I. L., Lachover, B., Koren, G., Levin, C., Zalman, L.,
& Koren, A. (2018). Detection of β-thalassemia carriers
by red cell parameters obtained from automatic
counters using mathematical formulas. Mediterranean
journal of hematology and infectious diseases, 10(1).
Rustam, F., Ashraf, I., Jabbar, S. et al. Prediction of β-
Thalassemia carriers using complete blood count
features. Sci Rep 12, 1999(2022). https:// doi.org/
10.1038/s41598-022-22011-8.
Sa'Id, A. A., Rustam, Z., Novkaniza, F., Setiawan, Q. S.,
Maulidina, F., & Wibowo, V. V. P. (2021, September).
Twin support vector machines for thalassemia
classification. In 2021 International Conference on
Innovation and Intelligence for Informatics,
Computing, and Technologies (3ICT) (pp. 160-164).
IEEE.
Weatherall, D. J., & Clegg, J. B. (2001). The Thalassaemia
Syndromes.
Widodo, A., & Yang, B. S. (2007). Support vector machine
in machine condition monitoring and fault
diagnosis. Mechanical systems and signal
processing, 21(6), 2560-2574.
Wirasati, I., Rustam, Z., Aurelia, J. E., Hartini, S., &
Saragih, G. S. (2021). Comparison some of kernel
functions with support vector machines classifier for
thalassemia dataset. Int J Artif Intell ISSN, 2252(8938),
8938.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
604
