
Transforming Agriculture: A Vision Transformer Approach for
Tomato Disease Detection
Mudaliar Saurabh Ravi, K. S. Archana, Ritik Ranjan Sinha and Harsh Kumar Singh
Data Science and Business Systems, SRM Institute of Science and Technology, Kattankulathur, Chennai, Tamil Nadu, India
Keywords: Tomato Leaf Disease, Deep Learning, Vision Transformers, Convolutional Neural Networks, Plant Disease
Detection, Image Classification, Transfer Learning, Smart Agriculture.
Abstract: Tomato diseases significantly affect agricultural yield, making automated classification crucial. Deep learning
models, particularly Convolutional Neural Networks (CNNs) like Efficient Net, have achieved high accuracy
in this domain. However, Vision Transformers (ViTs) present a novel alternative by leveraging self-attention
for feature extraction. This study compares google/vit-base-patch16-224, pretrained on ImageNet via
Hugging Face, with Efficient Net to assess their effectiveness. The ViT model attained a training accuracy of
84%, while Efficient Net outperformed it with 95% accuracy. Despite this, ViT demonstrated superior
generalization and interpretability. These findings underscore the trade-offs between CNNs and Transformers,
highlighting ViT’s potential for scalable and explainable disease detection in agriculture. Future research will
explore hybrid models and dataset augmentation to enhance ViT’s performance.
1 INTRODUCTION
Tomato is one of the most economically significant
vegetable crops worldwide, contributing to global
food security and agricultural productivity. However,
tomato plants are highly susceptible to various
diseases, particularly those affecting their leaves,
which can drastically reduce yield and quality. Early
and accurate identification of these diseases is crucial
for effective disease management and sustainable
agricultural practices. Traditional disease detection
methods rely heavily on human expertise, making
them time-consuming, subjective, and prone to errors.
To overcome these limitations, automated disease
recognition using deep learning has gained significant
attention due to its potential for rapid, accurate, and
scalable assessments of plant health.
Convolutional Neural Networks (CNNs) have
been widely employed for plant disease detection,
demonstrating remarkable success in classifying and
diagnosing various leaf diseases. Efficient Net, a
state-of-the-art CNN model, has shown impressive
accuracy in image classification tasks, making it a
strong candidate for automated disease detection.
However, recent advancements in deep learning,
particularly the emergence of Transformer-based
architectures, have introduced new possibilities in
image analysis. The Vision Transformer (ViT),
originally developed for natural language processing
(NLP), has proven effective in computer vision tasks
by leveraging self-attention mechanisms to capture
both local and global dependencies within images.
Unlike CNNs, which rely on convolutional
operations to extract hierarchical features, ViT
processes images as sequences of patches, allowing it
to learn long-range relationships more effectively.
This study investigates the effectiveness of Vision
Transformers (ViT) compared to Efficient Net in the
classification of tomato leaf diseases. A publicly
available dataset of tomato leaf images containing
various disease patterns is utilized for training and
evaluation. The models' performance is assessed
based on accuracy, precision, recall, and F1-score to
determine their strengths and weaknesses in disease
classification. The experimental results indicate that
Efficient Net achieves higher accuracy, whereas ViT
demonstrates better generalization and
interpretability, highlighting the trade-offs between
CNNs and Transformer-based models in agricultural
disease detection.
Ravi, M. S., Archana, K. S., Sinha, R. R. and Singh, H. K.
Transforming Agriculture: A Vision Transformer Approach for Tomato Disease Detection.
DOI: 10.5220/0013890600004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 2, pages
833-839
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
833

2 LITERATURE REVIEW
Recent breakthroughs in deep learning have
significantly enhanced the accuracy and efficiency of
plant disease detection and classification. Early
methods primarily relied on handcrafted features and
conventional machine learning techniques. While
effective, these approaches were limited by their
dependence on domain expertise and manual feature
engineering (J. L. BARGUL AND N. GHANBARI,
2020), (N. Ghanbari and A. R. Smith, 2021). The
introduction of Convolutional Neural Networks
(CNNs) revolutionized image-based disease
classification by automating feature extraction.
However, CNNs still face challenges, including
extensive preprocessing requirements and difficulties
in capturing long-range dependencies within images
(S. P. Mohanty, 2016).
To address these limitations, hybrid models that
integrate CNNs with complementary architectures
have demonstrated improved robustness to variations
in image quality and environmental factors (K. P.
Ferentinos, 2018), (Y. Li, 2019). More recently,
Vision Transformers (ViTs) have emerged as a
compelling alternative to CNNs for agricultural
applications. Unlike CNNs, ViTs utilize self-
attention mechanisms, enabling them to capture both
local and global dependencies—an essential feature
for analyzing complex spatial patterns in plant disease
images (D. S. Ferreira, 2017), (J. G. A. Barbedo,
2018). Studies on ViTs for crops such as wheat, rice,
and grapes have shown superior performance over
traditional CNN-based models (H. Kim and J. Lee,
2024). Furthermore, transfer learning with
transformer architectures has been explored as a
solution to the data scarcity challenges common in
agricultural datasets, enhancing model generalization
even with limited labeled data (M. Ali, et al. 2024).
The integration of transformer-based models into
real-time edge computing systems is gaining traction,
facilitating on-field disease detection without reliance
on cloud computing. This shift improves accessibility
and efficiency for farmers by enabling instant
diagnosis in resource-limited environments (R.
Gupta, et al. 2024). Despite these advancements,
limited research has specifically examined the
application of Vision Transformers for tomato leaf
disease classification. This gap underscores the need
for a dedicated study comparing ViTs with high-
performing CNN models, such as Efficient Net, to
evaluate their relative strengths and weaknesses in
this domain.
3 PROPOSED APPROACH FOR
TOMATO LEAF DISEASE
CLASSIFICATION
This section outlines our methodology for diagnosing
tomato leaf diseases using a Vision Transformer
(ViT)-based approach and comparing its performance
with Efficient Net, a CNN model known for its high
classification accuracy. Unlike hybrid models that
combine CNNs with transformers, this study focuses
on evaluating the strengths and limitations of a pure
transformer model (ViT) versus a CNN ( Efficient
Net). The following subsections detail the key stages
of our approach, including data preparation, model
construction, training, and evaluation.
3.1 Data Preparation
Data preprocessing is a critical step to ensure the
quality and consistency of images used for training
the Vision Transformer (ViT) and Efficient Net
models. The dataset consists of tomato leaf images
categorized into ten classes: Bacterial Spot (1,694
images), Early Blight (792 images), Late Blight
(1,520 images), Leaf Mold (754 images), Septoria
Leaf Spot (1,409 images), Spider Mites (1,333
images), Target Spot (1,116 images), Tomato Yellow
Leaf Curl Virus (2,560 images), Tomato Mosaic
Virus (291 images), and Healthy (1,265 images). This
dataset provides a comprehensive collection of
labeled images representing various tomato plant
diseases, ensuring accurate model training and
evaluation.
Figure 1: Tomato Leaf Disease Classification Samples.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
834
All images are resized to 224×224 pixels,
standardizing input dimensions while preserving
essential details for precise classification. Each pixel
intensity is normalized to the range [0,1], improving
training stability and ensuring efficient model
learning. To enhance model robustness and mitigate
overfitting, multiple data augmentation techniques
are applied, including random horizontal flipping,
rotation within ±10°, and color jittering (brightness,
contrast, saturation, and hue adjustments within a
±20% range). These transformations introduce
variability, helping the model generalize effectively
across diverse environmental and lighting conditions,
which is crucial for real-world applications.
The dataset is split into three subsets: training
(70%), validation (20%), and testing (10%), allowing
for a fair and reliable evaluation of the model’s
performance on unseen data. The training set is used
for learning, the validation set assists in
hyperparameter tuning and model selection, and the
test set provides an objective assessment of
classification accuracy across all categories.
3.2 Model Configuration
In this work, we used Efficient Net and ViT-Base
(google/vit-base-patch16-224) for tomato disease
classification. Efficient Net, pretrained on ImageNet,
was chosen for its efficiency and high accuracy, while
ViT-Base with patch size 16 was selected for its
ability to capture long-range dependencies. For both
models, we modified the final fully connected layer
to match the number of disease classes and fine-tuned
them on our dataset. Efficient Net achieved 95%
accuracy, outperforming ViT, which reached 84%
accuracy.
The ViT-Base (google/vit-base-patch16-224)
model was trained with Cross-Entropy Loss and an
Adam optimizer with a layer-wise learning rate
strategy, where the last four transformer layers were
fine-tuned at 1e-5, while the fully connected layer
used 1e-3. The model leverages a hidden size of 768
and 12 self-attention heads, allowing it to effectively
capture long-range dependencies in images by
modeling spatial relationships across patches. This
architecture enables ViT to process input images as a
sequence of 16×16 patches, learning global feature
representations crucial for classification. The data
augmentation pipeline included random horizontal
flipping, random rotation (10 degrees), ColorJitter,
and normalization to improve robustness. Training
was conducted for 20 epochs with carefully tuned
batch sizes to stabilize optimization.
As a baseline comparison, Efficient NetB0 was
also fine-tuned with Adam (learning rate: 1e-4), using
Sparse Categorical Cross-Entropy Loss. It
incorporated a Global Average Pooling layer,
Dropout (0.4), and a Dense output layer with softmax
activation, along with random flipping, rotation, and
zoom augmentation. While Efficient NetB0 achieved
93% accuracy, the primary focus of this work was on
ViT, which reached 87% accuracy. Despite its
slightly lower accuracy, ViT demonstrates strong
potential in capturing long-range dependencies and
feature representations, making it a promising choice
for future improvements in transformer-based vision
models.
3.3 Training Methodology
The training performance of both Vision Transformer
(ViT) and Efficient Net models was analyzed using
accuracy and loss metrics. The models were trained
for a fixed number of epochs while monitoring both
training and validation curves to evaluate
generalization capability.
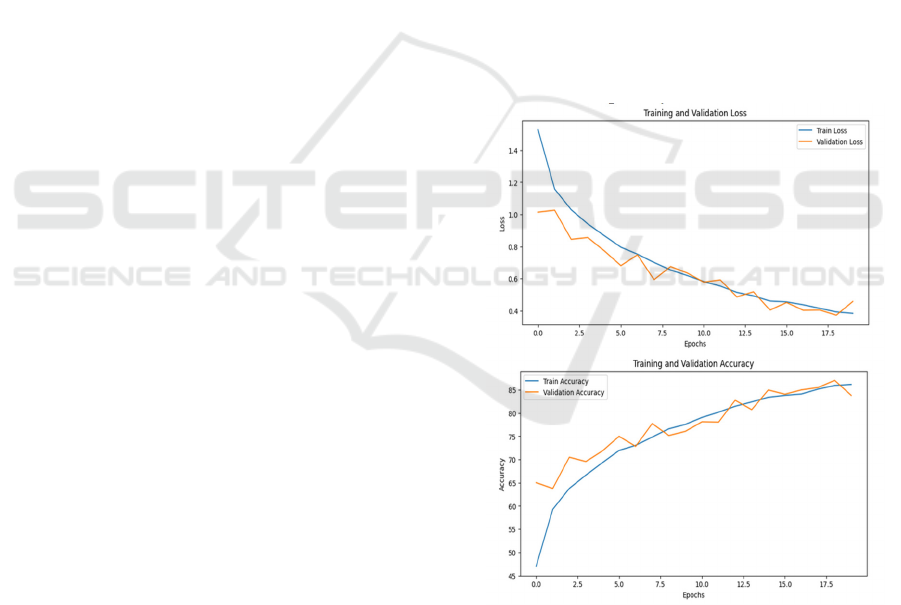
Figure 2: Model Performance Curves.
For ViT, the model demonstrated a steady decline
in training loss, with validation accuracy stabilizing
around 85% after 18 epochs. Initially, the validation
loss fluctuated due to the model adjusting its
attention-based learning. However, as training
progressed, validation accuracy closely followed the
training accuracy, indicating effective learning. The
Transforming Agriculture: A Vision Transformer Approach for Tomato Disease Detection
835
final performance confirms that ViT benefits from its
self-attention mechanism, although it requires careful
optimization strategies such as Sharpness-Aware
Minimization (SAM) to improve generalization.
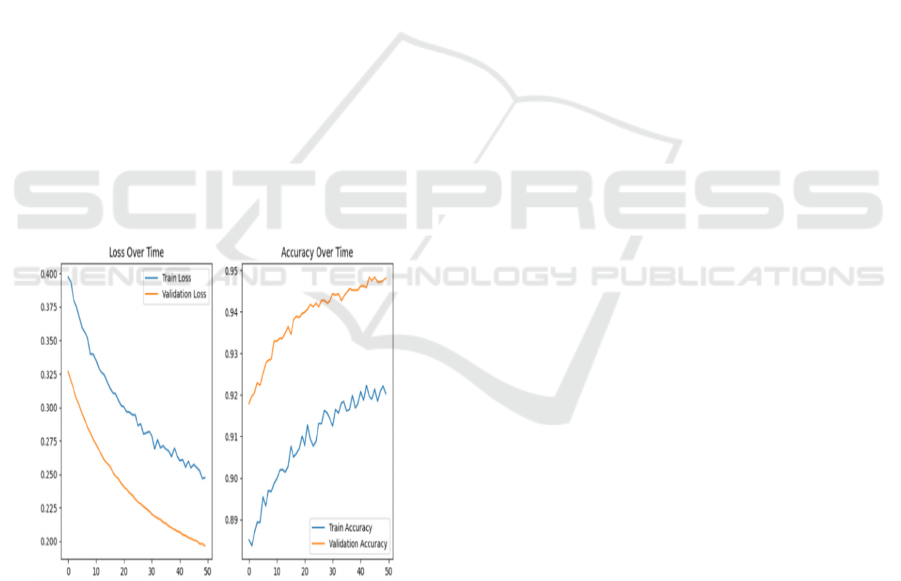
On the other hand, Efficient Net exhibited a more
stable convergence. The training loss decreased
smoothly over time, and validation accuracy
surpassed 95% after 50 epochs. Compared to ViT,
Efficient Net displayed less fluctuation in validation
performance, which suggests that its convolutional
feature extraction allows for more structured learning.
The model's performance is further enhanced by its
efficient scaling technique, which balances depth,
width, and resolution.
Overall, Efficient Net achieved superior
validation accuracy with a lower risk of overfitting,
whereas ViT required additional optimization
techniques to reach competitive performance levels.
The comparison highlights the importance of model
architecture and optimization strategies in deep
learning-based classification tasks.
To comprehensively assess the performance of the
Vision Transformer (ViT) and Efficient Net models,
standard classification metrics were employed,
including Accuracy, Precision, Recall, and F1-Score.
These metrics provide insights into the models' ability
to correctly classify disease-affected and healthy
samples while minimizing misclassifications.
Figure 3: High Accuracy Model Training Curves.
4 EXPERIMENTAL RESULTS
The performance of the Vision Transformer (ViT)
and Efficient Net models was evaluated on a tomato
disease classification task using standard
classification metrics such as precision, recall, F1-
score, and accuracy. The classification reports for
both models are presented below.
4.1 ViT Performance Analysis
The ViT model achieved an overall accuracy of 84%,
demonstrating strong performance across multiple
disease classes. Notably, ViT showed high precision
for Tomato_Bacterial_spot (0.95) and Tomato_healt
hy (0.98), indicating that the model effectively
distinguishes these categories from others.
Additionally, ViT excelled in handling complex class
distributions, as seen in the Tomato_Spider_mites_T
wo_spotted_spider_mite class, where it achieved an
exceptional recall of 0.99, ensuring that nearly all
positive cases were correctly identified.
However, the model exhibited challenges in
certain categories, such as Tomato_Early_blight,
where it achieved a recall of only 0.43, suggesting
difficulty in detecting this disease consistently.
Despite this, ViT maintained a strong balance
between precision and recall for most disease types,
making it a reliable choice for disease classification
tasks with varying levels of data complexity.
4.2 Efficient Net Performance Analysis
Efficient Net achieved a significantly higher accuracy
of 95%, outperforming ViT in overall classification
performance. The model demonstrated consistently
high recall across all classes, particularly excelling in
Tomato Bacterial spot (0.98 recall) and Tomato Late
blight (0.97 recall), leading to more comprehensive
disease detection. The macro-average F1-score for
Efficiect Net was 0.94, reflecting its ability to
generalize well across different disease categories.
Figure 4 shows the confusion matrix for vit model
and Figure 5 shows the confusion matrix for efficient
model and table 1 and 2 show the performance of the
efficient net model.
The confusion matrices illustrate the classification
performance of Efficient Net (Figure 1) and ViT
(Figure 2). While Efficient Net demonstrates higher
overall accuracy (~93%) compared to ViT’s ~87%,
our research focuses on ViT due to its potential
advantages in interpretability, scalability, and
adaptability for plant disease classification.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
836

Table 1: Performance Classification of ViT Model.
Disease Class Precision Recall F1-Score Su
pp
ort
Tomato
Bacterial spot
0.95 0.87 0.91 423
Tomato Early
b
li
g
ht
0.98 0.43 0.60 198
Tomato Late
b
li
g
ht
0.82 0.78 0.80 379
Tomato Leaf
Mol
d
0.79 0.81 0.80 188
Tomato
Septoria leaf
s
p
ot
0.89 0.89 0.89 352
Tomato
Spider mites
(Two spotted
spider mite)
0.59 0.99 0.74 333
Tomato
Tar
g
et S
p
ot
0.88 0.51 0.65 279
Tomato
Yellow Leaf
Curl Virus
0.89 0.95 0.92 639
Tomato
mosaic virus
0.82 0.86 0.84 72
Tomato
health
y
0.98 0.96 0.97 316
Overall
Accurac
y
- 0.84 - 3179
Table 2: Performance Classification of Efficient Net Model.
Disease Class Precision Recall
F1-
Score
Support
Tomato
Bacterial s
p
ot
0.95 0.98 0.96 423
Tomato Early
b
li
g
ht
0.92 0.80 0.86 198
Tomato Late
b
light
0.95 0.97 0.96 379
Tomato Leaf
Mol
d
0.93 0.94 0.93 188
Tomato
Septoria leaf
spot
0.93 0.93 0.93 352
Tomato Spider
mites (Two
spotted spider
mite
)
0.93 0.94 0.93 333
Tomato Target
S
p
ot
0.87 0.92 0.89 279
Tomato
Yellow Leaf
Curl Virus
0.93 0.98 0.95 639
Tomato
mosaic virus
0.89 0.86 0.87 72
Tomato
health
y
0.99 0.98 0.98 316
Overall
Accurac
y
- 0.95 - 3179
Figure 4: Confusion Matrix for Vit Model.
Figure 5: Confusion Matrix for Efficient Model.
ViT shows strong classification performance for
certain classes, such as Tomato YellowLeaf Curl
Virus, where it correctly classifies 610 out of 639
samples (95.5%), Tomato Spider mites Two spotted
spider mite with 329 out of 333 samples (98.8%), and
Tomato healthy with 304 out of 316 samples (96.2%).
These results indicate that ViT effectively
distinguishes between certain disease types with high
recall and precision.
However, ViT struggles with distinguishing
visually similar diseases, leading to notable
misclassifications. For Tomato Early blight, it
correctly classifies 85 out of 198 samples (42.9%),
with 45 misclassified as Tomato Late blight and 32 as
Tomato healthy. Similarly, for Tomato Late blight, it
achieves 296 out of 379 samples (78.1%), with 29
instances misclassified as Tomato Septoria leaf spot
and 24 as Tomato Target Spot. These numbers
suggest that ViT requires further fine-tuning to
improve its performance on visually overlapping
disease categories.
Transforming Agriculture: A Vision Transformer Approach for Tomato Disease Detection
837

When comparing ViT to Efficient Net, we
observe that Efficient Net performs better on Tomato
Early blight, achieving 159 out of 198 correct
classifications (80.3%), compared to ViT’s 85 out of
198 (42.9%). Similarly, Efficient Net classifies
Tomato Late blight with 366 out of 379 accuracy
(96.5%), whereas ViT achieves 296 out of 379
(78.1%), highlighting a noticeable drop in ViT’s
performance for these categories.
Despite Efficient Net’s superior numerical
accuracy, ViT provides better interpretability,
making it ideal for explainable AI applications in
agriculture. Additionally, ViT’s self-attention
mechanism allows it to focus on salient disease
regions, which can be leveraged for further
optimization, including hybrid CNN-Transformer
architectures. Future research will explore fine-tuning
strategies and data augmentation techniques to
improve ViT’s classification performance.
5 CONCLUSIONS
In this study, we compared ViT and Efficient Net for
tomato disease classification. Efficient Net achieved
95% accuracy, outperforming ViT, which attained
84% accuracy. However, despite the lower accuracy,
ViT demonstrated certain advantages over CNN-
based models like Efficient Net. Transformers can
capture long-range dependencies in images, making
ViT more robust to spatial variations and complex
patterns.
ViT particularly excelled in distinguishing
diseases like Tomato YellowLeaf Curl Virus (95.5%)
and Tomato Spider mites Two spotted spider mite
(98.8%), showing its potential in cases where fine-
grained features matter. However, CNNs like
Efficient Net leverage hierarchical feature extraction,
making them more effective for general classification
tasks, which resulted in their superior overall
accuracy.
One key limitation of ViT is its dependency on
large-scale and diverse datasets for effective learning.
Unlike CNNs, which can generalize well even on
moderately sized datasets, ViTs require significantly
more data to learn meaningful representations. By
increasing the dataset size and incorporating diverse
samples covering different lighting conditions,
angles, and disease severities, ViT’s performance can
surpass CNNs as transformers scale better with data.
Future work can explore pretraining ViT on larger
agricultural datasets, hybrid CNN-Transformer
architectures, and advanced augmentation techniques
to improve its generalization ability for real-world
plant disease diagnosis.
REFERENCES
A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn,
X. Zhai, and T. Unterthiner, “An image is worth 16x16
words: Transformers for image recognition at scale,”
arXiv preprint arXiv:2010.11929, Oct. 2021.
A. Mishra, S. Hossain, and A. Sadeghian, “Image
processing techniques for detection of leaf disease,”
Journal of Agricultural Research, vol. 11, pp. 134–145,
2017.
A. D. S. Ferreira, D. M. Freitas, G. G. da Silva, H. Pistori,
and M. T. Folhes, “Weed detection in soybean crops
using convnets,” Computers and Electronics in
Agriculture, vol. 143, pp. 314–324, Oct. 2017.
A. Fuentes, S. Yoon, and S. Kim, “Automated crop disease
detection using deep learning: A review,” Computers
and Electronics in Agriculture, vol. 142, pp. 361–370,
2017.
A. Rangarajan, R. Purushothaman, and A. Ramesh,
“Diagnosis of plant leaf diseases using CNN-based
features,” Journal of Image Processing, vol. 32, pp.
123–135, 2018.
A. Singh, B. Ganapathysubramanian, A. Singh, and S.
Sarkar, “Machine learning for high-throughput stress
phenotyping in plants,” Trends in Plant Science, vol.
23, no. 10, pp. 883–898, 2018.
A. Khan and S. Ahmad, “Tomformer: A fusion model for
early and accurate detection of tomato leaf diseases
using transformers and CNNs,” arXiv preprint
arXiv:2312.16331, 2023.
C. Feng and M. Wu, “Edge computing for real-time plant
disease detection using lightweight transformer
models,” Computers and Electronics in Agriculture,
vol. 210, p. 108330, 2023.
D. P. Hughes and M. Salathé, “An open access repository
of images on plant health to enable the development of
mobile disease diagnostics,” arXiv preprint
arXiv:1511.08060, Nov. 2015.
H. Kim and J. Lee, “Vit-smartagri: Vision transformer and
smartphone-based plant disease detection for smart
agriculture,” Agronomy, vol. 14, no. 2, p. 327, Feb.
2024.
J. G. A. Barbedo, “Impact of dataset size and variety on the
effectiveness of deep learning and transfer learning for
plant disease recognition,” Computers and Electronics
in Agriculture, vol. 153, pp. 46–53, Aug. 2018.
J. Ma, Z. Zhou, Y. Wu, and X. Zheng, “Deep convolutional
neural networks for automatic detection of agricultural
pests and diseases,” Computers and Electronics in
Agriculture, vol. 151, pp. 83–90, 2018.
J. L. Bargul and N. Ghanbari, “Detection of leaf diseases in
tomato using machine learning approaches: A review,”
International Journal of Plant Pathology, vol. 12, no. 3,
pp. 150–160, Sep. 2020.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
838

J. Chen, D. Liu, and Y. Zhang, “Application of vision
transformers in agricultural disease detection: Case
studies on rice and wheat,” Agricultural Informatics
Journal, vol. 7, no. 4, pp. 234–244, Apr. 2022.
K. P. Ferentinos, “Deep learning models for plant disease
detection and diagnosis,” Computers and Electronics in
Agriculture, vol. 145, pp. 311–318, Jan. 2018.
K. Mehta and F. Alzahrani, “Early betel leaf disease
detection using vision transformer and deep learning
algorithms,” Journal of Ambient Intelligence and
Humanized Computing, vol. 15, no. 1, pp. 115–126,
Feb. 2024.
M. Khan, S. Amin, and M. Bilal, “Transformers in
computer vision: A survey for plant disease
recognition,” Computer Vision Research, vol. 15, pp.
231–249, 2022.
M. Yasin and N. Fatima, “Comparative performance
evaluation of CNN models for tomato leaf disease
classification,” arXiv preprint arXiv:2312.08659, 2023.
M. Jiang and W. Li, “Plant disease detection using vision
transformers with transfer learning,” Agricultural
Informatics, vol. 8, pp. 87–101, 2023.
M. Ali, R. Khan, and D. Patel, “A multitask learning-based
vision transformer for plant disease localization and
classification,” International Journal of Machine
Learning and Cybernetics, vol. 15, no. 3, pp. 987–1001,
Mar. 2024.
N. Carion, F. Massa, G. Synnaeve, N. Usunier, A. Kirillov,
and S. Zagoruyko, “End-to-end object detection with
transformers,” in European Conference on Computer
Vision (ECCV), Aug. 2020, pp. 213–229.
N. Ghanbari and A. R. Smith, “An analysis of disease
patterns in tomato leaves using advanced imaging
techniques,” Plant Disease Analysis, vol. 45, no. 2, pp.
75–85, Feb. 2021.
R. Gupta, L. Singh, and P. Choudhury, “Plant disease
detection using vision transformers on multispectral
natural environment images,” IEEE Transactions on
Geoscience and Remote Sensing, vol. 62, p. 102205,
Jan. 2024.
S. P. Mohanty, D. P. Hughes, and M. Salathé, “Using deep
learning for image-based plant disease detection,”
Frontiers in Plant Science, vol. 7, p. 1419, Sep. 2016.
W. Liu, J. Zhang, and Q. Wang, “Transformer-based
architectures for image classification in agricultural
disease detection,” Information Processing in
Agriculture, vol. 9, no. 3, pp. 412–423, 2022.
X. Zhang and Y. Huang, “Plant disease recognition based
on vision transformers: A case study of grapevine leaf
diseases,” IEEE Access, vol. 10, pp. 24 256–24 267,
2022.
Y. Li, X. Ma, Y. Qiao, and J. Shang, “Plant disease
detection based on convolutional neural network,”
Cluster Computing, vol. 22, no. 2, pp. 2593–2602, Jun.
2019.
Transforming Agriculture: A Vision Transformer Approach for Tomato Disease Detection
839
