
Melanoma Skin Cancer Detection: A Self‑Supervised Deep Learning
Approach
Syed Ismail A., Atif Alam Ansari, Amaan Hussain and Riyan Acharya
Data Science and Business Systems, SRMIST Kattankulathur, Chennai, Tamil Nadu, India
Keywords: Self‑Supervised Learning, Gen AI‑Powered Recipe Generation, Vegetable, BMI Detection, Computer Vision
in Nutrition, Personalized Dietary Recommendations, AI- Driven Food Recognition, Smart Cooking
Assistant.
Abstract: Early identification is essential for successful treatment outcomes because skin cancer remains one of the
most prevalent and potentially deadly cancers. Using deep learning methods and the ISIC dataset, this study
develops a model for automated skin lesion categorization with a focus on precisely identifying malignant
types. While pre-processing techniques like data augmentation are employed to address class imbalances in
the dataset, convolutional neural networks (CNNs) serve as the foundation for the model architecture. The
system’s performance was assessed using metrics such as F1-score, recall, accuracy, and precision; the results
demonstrated that the system was successful in comparison to alternative approaches. According to the
findings, deep learning may prove to be a helpful tool in dermatology, enhancing early intervention strategies
in clinical settings and increasing diagnostic accuracy.
1 INTRODUCTION
About 10 million deaths from cancer is predicted in
the year 2024, making it the second most common
cause of death worldwide. Skin, oral, and pancreatic
cancers are among the many cancer forms for which
early identification greatly improves prognosis;
survival rates for these tumors are above 90%. But
cancer is becoming more common, especially in
underprivileged areas of emerging countries. The
need for an easily accessible, precise, and reasonably
priced diagnostic tool is particularly pressing because
these communities frequently lack access to the
medical specialists required for prompt diagnosis.
Machine learning and artificial intelligence (AI
and ML) present intriguing answers that could meet
global healthcare demands and enable remote
diagnostics. These systems have the potential to
supplement or even improve clinical decision-
making, which would be extremely beneficial for
underserved areas, such rural locations with little
resources. Clinical photos can now be classified as”
malignant,”” benign,” or other pertinent categories
thanks to the recent surge in popularity of machine
learning applications for cancer diagnosis, including
lung and breast malignancies. However, in order to
perform at their best, deep learning models need big,
balanced datasets, which presents difficulties because
access to healthcare data is frequently restricted by
patient confidentiality and intricate information
sharing agreements. Furthermore, gathering data for
diagnostic imaging frequently necessitates
professional assessment, such biopsy, which raises
expenses and time. These barriers underscore the
need for accessible datasets to support ML research
for various cancer types, addressing a critical gap in
healthcare innovation.
2 LITERATURE SURVEY
Recent developments in deep learning have
profoundly influenced skin cancer diagnosis, as noted
by S. Inthiyaz et al. Convolutional neural networks
(CNNs) facilitate reliable diagnosis. Initial studies
employed various deep learning architectures, such as
AlexNet and ResNet, achieving significant
generalization outcomes for the classification of
dermoscopic images as benign or malignant. Recent
studies have developed hybrid approaches that
integrate machine learning classifiers with pre-
trained convolutional neural networks (CNNs) to
A., S. I., Ansari, A. A., Hussain, A. and Acharya, R.
Melanoma Skin Cancer Detection: A Self-Supervised Deep Learning Approach.
DOI: 10.5220/0013883900004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 2, pages
409-416
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
409

enhance diagnostic accuracy and minimize
computational costs.
The diagnosis of skin cancer primarily involves
the optimization of CNN architectures such as
Inception-V3 and InceptionResNet-V2, which
demonstrate significant depth and accuracy in image
classification tasks. The combination of
InceptionResNet-V2 and InceptionV3, along with
data augmentation, resulted in significant accuracy
improvements to address class imbalance in the
HAM10000 dataset. This method achieved diagnostic
accuracy comparable to that of dermatology
specialists through the fine-tuning of network layers.
Improving feature extraction to reduce reliance on
expert manual segmentation represents a significant
research focus. High-resolution image synthesis is
achievable through techniques such as Enhanced
Super-Resolution Generative Adversarial Networks
(ESRGANs), which have shown to improve CNN
performance for complex medical images, including
skin lesions. This method enhances the quality of the
input image, which is crucial for detecting subtle
morphological changes in lesions. Traditional
machine learning models for skin lesion data have
primarily relied on handcrafted feature extraction, yet
these approaches exhibit limitations in scalability and
adaptability. CNNs can automatically identify
complex patterns, which is particularly beneficial in
various datasets, including those related to melanoma
and basal cell carcinoma.
Numerous comparative studies indicate that deep
learning techniques generally outperform traditional
machine learning methods in the diagnosis of skin
cancer, particularly when substantial labeled datasets
are accessible. Transfer learning using pre-trained
weights from larger image datasets with models such
as DenseNet, Xception, and MobileNet is widely
adopted due to its ability to facilitate efficient
generalization with reduced data requirements. The
issues of class imbalance, data scarcity, and feature
complexity in dermoscopic datasets are being tackled
through the integration of robust CNN architectures
with GAN-based pre-processing techniques. Future
research should focus on enhancing high-accuracy
skin cancer diagnostic models for resource-limited
environments to improve global access.
(H. K. Gajera, et, al, 2022) notes that conventional
diagnostic techniques often rely on the subjective and
time-consuming evaluation of dermoscopy images by
experts. Convolutional neural networks (CNNs), a
category within deep learning (DL), have recently
gained prominence as an effective method for
automating the detection of skin cancer.
Convolutional Neural Networks (CNNs) have been
widely employed in the analysis of dermoscopy
images due to their ability to learn intricate patterns
that facilitate the differentiation between benign and
malignant tumors. CNNs face challenges stemming
from considerable intra-class variation and inter-class
similarity among skin lesion types, in addition to
insufficient and diverse training data. CNN-based
models typically necessitate a substantial number of
parameters, rendering them resource-intensive and
potentially inappropriate for practical clinical
applications.
Transfer learning employs feature representations
derived from large datasets to enhance performance
on smaller datasets, like those pertaining to
melanoma. Recent studies have utilized pretrained
CNN architectures to tackle some of these challenges.
To enhance accuracy, various CNN architectures,
including DenseNet, ResNet, and Inception, have
shown potential in classifying skin lesions. This is
particularly applicable when combined with other
classifiers, such as multi-layer perceptrons (MLPs).
The utilization of learned feature maps that capture
high-level visual cues relevant to melanoma detection
enables these pretrained models to mitigate issues
related to data scarcity. Research indicates that
employing image preprocessing techniques such as
normalization and boundary localization is crucial for
improving model performance. These methods
enhance the capacity of CNNs to identify and
distinguish subtle details in lesion images by reducing
noise and standardizing image quality.
Comprehensive comparisons of features from various
CNN architectures indicate that DenseNet-121 is
highly effective in melanoma detection. DenseNet-
121, in conjunction with MLP classifiers, attains
accuracy rates of 98.33%, 80.47%, 81.16%, and 81%
on benchmark datasets including PH, ISIC 2016, ISIC
2017, and HAM10000, demonstrating state-of-the-
art performance. The unique architecture of
DenseNet, which minimizes redundant feature
learning and enhances feature reuse among layers, is
responsible for this success.
The results underscore the importance of selecting
depend- able CNN architectures and effective
preprocessing methods for melanoma classification.
Anticipated advancements in the discipline will arise
from ongoing research into transfer learning, coupled
with comprehensive CNN feature analysis and
boundary-based preprocessing techniques.
Automated deep learning systems have the potential
to become an effective method for widespread
melanoma detection, provided that researchers
address existing challenges, thereby facilitating rapid
and straightforward diagnosis.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
410
(F. Alenezi, et, al 2022) proposed a wavelet
transform-based deep residual neural network (WT-
DRNNet) for the categorization of skin lesions. The
model employs wavelet transformation, pooling, and
normalization to emphasize critical features and
minimize irrelevant information in lesion images.
Global aver- age pooling is performed subsequent to
the extraction of deep features utilizing a residual
neural network through transfer learning. An Extreme
Learning Machine utilizing ReLU activation is
employed for training. The model surpassed earlier
models and facilitated automated skin cancer
classification in tests conducted on the ISIC2017 and
HAM10000 datasets, attaining an accuracy of
96.91% on ISIC2017 and 95.73% on HAM10000.
(R. K. Shinde et al. 2022) introduced Squeeze-
MNet, an efficient deep learning model designed
for the classification of skin cancer. Squeeze-MNet
integrates a MobileNet architecture with the Squeeze
algorithm and utilizes pretrained weights for digital
hair removal in the preprocessing phase. The Squeeze
algorithm captures essential image features, and a
black-hat filter is employed to eliminate noise.
MobileNet was optimized for classification
performance by utilizing the ISIC dataset alongside a
dense neural network. The model, designed for
lightweight deployment, underwent testing on an 8-
bit NeoPixel LED ring using a Raspberry Pi 4 IoT
device, following clinical validation by a medical
specialist. The model achieved an average accuracy
of 99.76% for benign instances and 98.02% for
malignant cases. The Squeeze technique enhanced
detection accuracy to 99.36%, reduced the dataset size
requirement by 66%, and achieved a 98.9% area
under the ROC curve.
(Q. Abbas, 2022) presented an automated method
for early melanoma detection based on deep transfer
learning. We utilize NASNet, a pre-trained neural
network model, to transfer learned features to a new
dataset for melanoma detection. The original design
incorporates global average pooling and tailored
classification layers. To address the challenge of
limited data, we enhance the photographs through
geometric transformations that maintain labels and
features. The model is trained utilizing dermoscopic
images from the International Skin Imaging
Collaboration (ISIC 2020) dataset. Our approach
demonstrates significantly greater efficiency than
prior methods, achieving state-of-the-art performance
with over 97.
3 PROPOSED METHOD
3.1 Data Description
The International Skin Imaging Collaboration (ISIC)
dataset is a recognized resource utilized mainly for
enhancing research in melanoma detection. The ISIC
Archive contains a dataset of dermoscopic images of
skin lesions, intended to support and evaluate
dermatology computer-aided diagnosis systems. This
study examines challenges in skin cancer diagnosis,
focusing on disparities in data representation across
lesion types, intra- class variability, and inter-class
similarities.
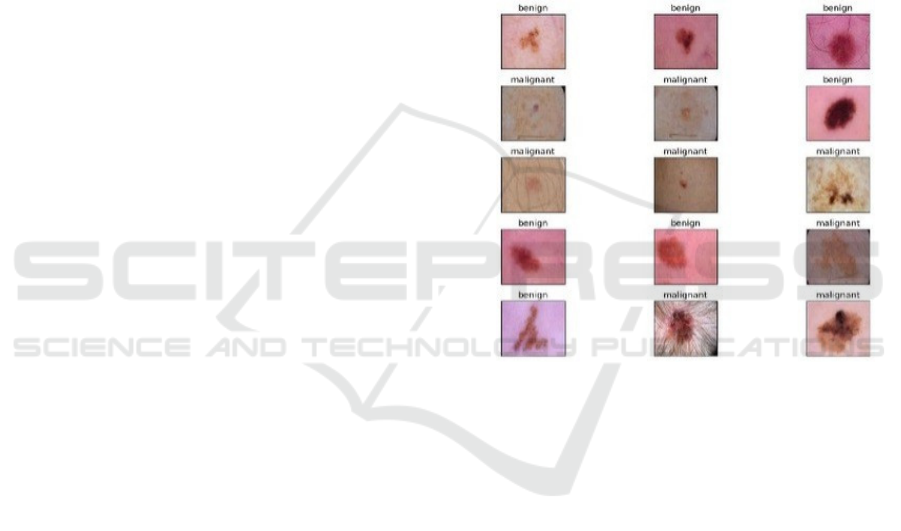
Figure 1: Data Sample.
Figure 1 demonstrates that ISIC images are
dermoscopic, utilizing either polarized or non-
polarized light to enhance visibility of details beneath
the skin’s surface that are not discernible to the naked
eye. This high-quality imagery enhances the models’
ability to detect subtle traits.
Fine lesion features are preserved in the dataset’s
generally high-quality images, which range in
resolution from 600x450 to 1024x1024 pixels.
We used 3297 photos from the ISIC dataset for
this study, of which 1977 were used to train the
model, 660 to test it, and 660 to validate it.
Consequently, the sample was separated into a 70%-
15%-15% segment.
Melanoma Skin Cancer Detection: A Self-Supervised Deep Learning Approach
411
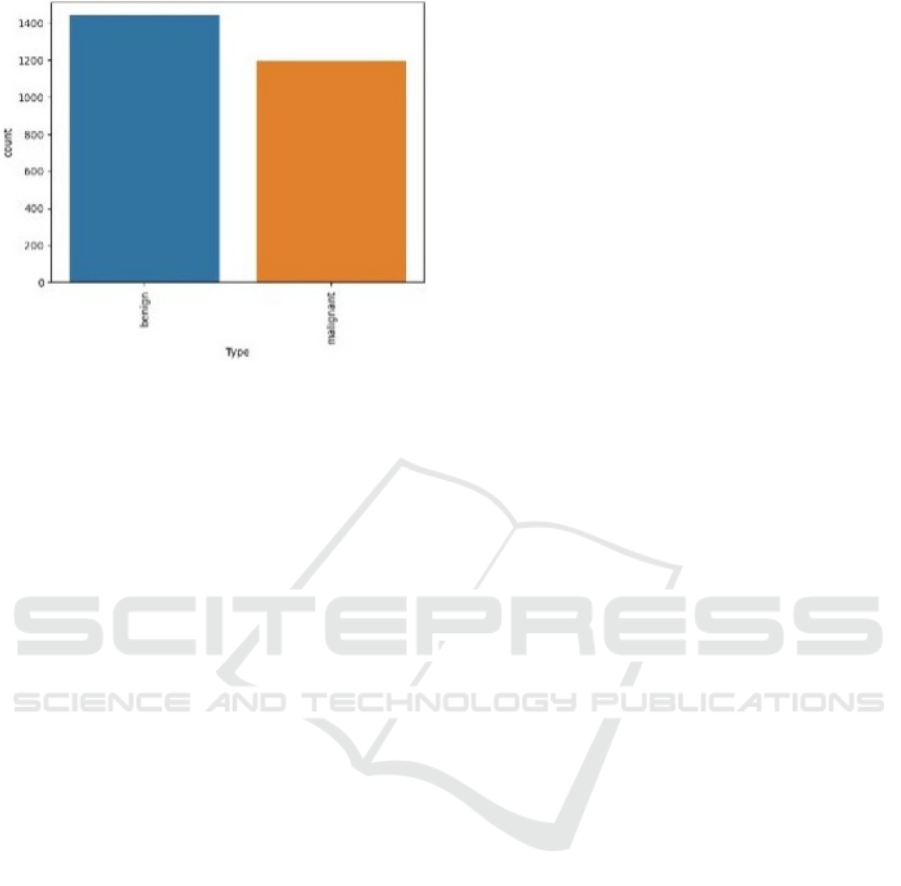
Figure 2: Data Distribution.
Figure 2 shows the dataset’s distribution of benign
and malignant data.
3.2 Data Splitting
The model utilizes a Data Frame containing
image file locations and corresponding labels. The
function flow_from_dataframe produces three data
generators: train generator, valid generator, and test
generator, which manage the data input for the model.
This approach employs predefined training and
testing datasets rather than executing an explicit train-
test split within the code.
The train Data Frame, which includes the file
directories and labels for training images, is where the
train generator gets its information. To increase
model generalization and introduce variability, this
generator incorporates data augmentations (specified
by train data generator). At the beginning of each
epoch, batches of 32 images are created and shuffled
for random ordering, with each image resized to (100,
100). The primary function of transgene is to provide
randomized, augmented data for model training.
A subset devoted to validation is the valid
generator, which also references the train Data Frame.
This generator provides consistent, non-augmented
data to assess model performance during training
because it does not use augmentation and is
configured to maintain images in a fixed sequence (no
shuffling).
The test generator supplies unaltered real-world
test data without augmentation, derived from the test
Data Frame. To maintain organization, shuffling is
disabled, similar to the validation generator. The
model’s performance on unseen data is assessed post-
training using test generator.
3.3 Data Preprocessing and
Augmentation
The model may be vulnerable to overfitting because
there aren’t many annotated medical images
available, particularly for skin cancer. In order to
solve this, we increased the dataset’s effective size by
using data augmentation tech- niques. The following
changes were made using Keras’s Image Data
Generator. In order to guarantee that every image
entered into the model has uniform dimensions that
correspond to the antic- ipated input shape, each
generator resizes images to a target size of (100, 100)
pixels.
• Flipping: Both vertical and horizontal flipping
were applied randomly to simulate various lesion
orientations.
• Rotation: To capture different viewpoints of skin
features, images were randomly rotated between
0 and 40 degrees.
• Zooming: Images were randomly zoomed in to
emphasize certain local characteristics within
lesions.
• Batching: For each of the three generators
(train_gen, valid_gen, and test_gen), images were
loaded in batches of 32. Batching allowed the
model to process input in manageable chunks,
improving computational efficiency and ensuring
memory constraints were met.
• Shuffling: To help the model learn more resilient
features rather than memorizing the order of the
data, the training generator (train_gen) ensured
that images were presented in a random order at
each epoch by setting shuffle=True. The
validation (valid_gen) and test (test_gen)
generators maintained a fixed or- der
(shuffle=False) to ensure consistent evaluation
metrics across epochs.
• Label Encoding: Labels were automatically one-
hot encoded using class_mode=’categorical’. This
ensured that every label was represented as a
binary vector, essential for multi-class
classification tasks.
• Preprocessing: The preprocess_input function of
ResNet50 was used to normalize pixel values,
scaling the images according to the input
expectations of the pretrained model. This step
ensured consistency with the ResNet50
architecture’s specifications.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
412
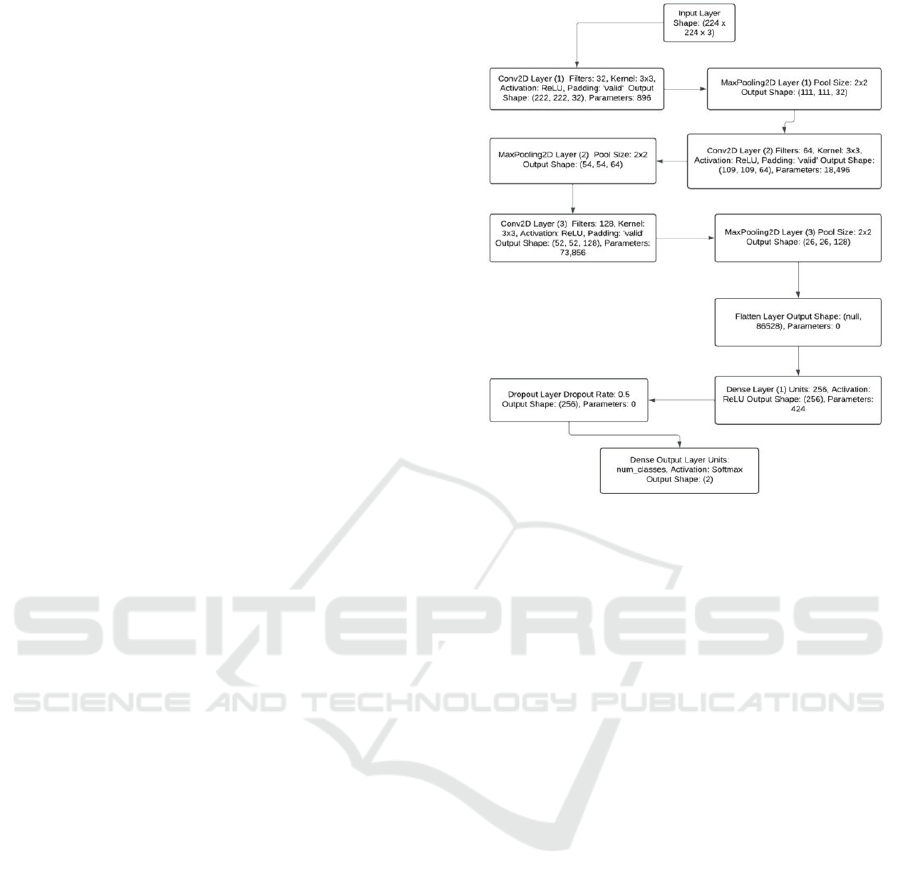
3.4 Model Architecture
A convolutional neural network (CNN) built on the
ResNet50 architecture was employed for this
investigation. A popular deep learning model,
ResNet50 has demonstrated excellent performance
across a range of picture categorization applications.
In order to overcome vanishing gradients in very deep
networks, as shown in Figure 3, it employs residual
learning to facilitate training deep networks by
permitting information to flow across shortcut links.
The basic model, which was pretrained on the
ImageNet dataset, is one of the main components of
the model architecture. We might profit from the
pretrained model’s capacity to identify intricate visual
patterns by utilizing transfer learning, which is
particularly helpful considering the tiny dataset.
A novel fully connected layer designed
specifically for binary classification has replaced the
top layer of ResNet50. A Global Average Pooling
(GAP) layer was employed as the classification head
to reduce the spatial dimensions of the feature maps,
followed by a Dense Layer that introduces non-
linearity through a ReLU activation function. A
Dropout layer that randomly omits neurons during
training with a probability of 0.5 to mitigate
overfitting.
The model comprises a sequential network
featuring an output layer for classification, a fully
connected layer for advanced learning, and
convolutional and pooling layers for feature
extraction.
3.5 Training Process
The model was trained with accuracy as the primary
metric, employing categorical cross-entropy as the
loss function. The following criteria are essential for
training:
Learning Rate: To start with a comparatively higher
learning rate and gradually reduce it as training
progressed, a learning rate scheduler was used. This
approach facilitated faster convergence in the early
stages while allowing fine-tuning in later epochs.
Early Stopping: Early stopping was employed to
prevent overfitting. Training was halted if the
validation accuracy did not improve after a
predetermined number of epochs, ensuring the model
remained optimal without excessive training.
Batch Size and Epochs: To balance memory usage
and training speed, the model was trained using a
moderate batch size (typically 16–32). Training could
continue for up to 50 epochs, depending on
convergence.
Figure 3: Architecture Diagram.
3.6 Evaluation Metrics
We utilized accuracy, precision, recall, and F1-score
to assess the model’s effectiveness. These measures
equilibrate both positive (malignant) and negative
(benign) categories, providing a thorough assessment
of the model’s performance.
3.7 Training Setup
The PC used for the studies has a Ryzen 5 5600H,
8GB RAM, a GTX 1650 graphics card, and Windows
11, which gave it the processing capability needed to
effectively train the deep learning model. Using the
Keras and TensorFlow frameworks, the ResNet50-
based model was trained, taking advantage of GPU
acceleration to achieve faster convergence. This is
how the dataset was split up, Seventy percent of the
entire dataset was in the Training Set.
Fifteen percent of the dataset comprised the
validation set, utilized for model adjustment and to
mitigate overfitting. To provide an objective
evaluation of the model’s performance, the test set,
comprising 15% of the dataset, was kept separate
form the training and validation data. A learning rate
scheduler was employed during the training process
to adjust the learning rate dynamically, facilitating the
identification of the optimal model with minimal
manual intervention, Early stopping was implemented
with a patience of 10 epochs, indicating that training
Melanoma Skin Cancer Detection: A Self-Supervised Deep Learning Approach
413

would cease if the validation accuracy did not improve
for ten consecutive epochs.
3.8 Evaluation Metrics
We utilized the following metrics to assess the
model’s efficacy: The Confusion Matrix, which
offers a comprehensive analysis of true positives, true
negatives, false positives, and false negatives, thereby
facilitating a complete understanding of classification
errors, as demonstrated in Figure 4. Accuracy,
defined as the percentage of correctly categorized
photos in the test set, functions as a general measure
of performance. Precision, indicative of the model’s
ability to minimize false positives, is defined as the
ratio of true positives (malignant lesions accurately
identified) to the total number of predicted positives.
The model’s capability to accurately identify
malignant cases is evidenced by recall (sensitivity),
defined as the ratio of true positives to the total
number of actual positives.
The F1-Score, defined as the harmonic mean of
precision and recall, offers a valid evaluation in
scenarios of class imbalance.
Figure 4: Confusion Matrix.
3.9 Result and Analysis
All assessment measures indicated that the model
exhibited strong performance on the test set, reflecting
its robustness and effective generalization
capabilities.
Accuracy: Provide an objective evaluation of the
model’s performance, the test set, comprising 15%
of the dataset, was kept separate.
𝐀𝐜𝐜𝐮𝐫𝐚𝐜𝐲
=
𝐓𝐏 + 𝐓𝐍
𝑻𝑷
+
𝑻𝑵
+
𝑭𝑷
+
𝑭𝑵
=
=
≈ 0.8485 or 84.85% (1)
The model achieved an accuracy of
approximately 84.85% on the test set, demonstrating
its overall ability to accurately categorize skin
lesions.
Precision: The model achieved an accuracy of
84.31%, effectively reducing false positives, which is
essential in medical applications to mitigate
unnecessary concern or intervention.
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 =
=
=
≈ 0.8431 𝑜𝑟 84.31% (2)
Recall: The model exhibits a recall rate of 83.23%,
indicating its proficiency in identifying malignant
lesions and minimizing the likelihood of overlooking
positive cases, both essential for early cancer detection.
𝑅𝑒𝑐𝑎𝑙𝑙 =
=
=
≈ 0.8323 or 83.23% (3)
F1-Score: The model demonstrates a balanced
performance between benign and malignant classes,
evidenced by an F1-score of 83.77%, which is
essential for addressing potential class imbalances.
𝐹1 =
2 𝑋 (𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 𝑋 𝑅𝑒𝑐𝑎𝑙𝑙)
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 + 𝑅𝑒𝑐𝑎𝑙𝑙
=
(. .)
..
≈ 0.8377 or 83.77% (4)
The analysis of the confusion matrix reveals a
relatively low false negative rate for the model.
Accurate cancer detection is crucial, as overlooked
malignant cases pose significant risks. Furthermore,
the reduction of false positives improved the model’s
practical applicability by decreasing the chances of
benign lesions being incorrectly identified as
malignant.
3.10 Visualizations
The model’s learning process was monitored by
plotting accuracy and loss, including training and
validation metrics, over epochs. The validation
measurements closely aligned with the training
metrics, and sustained convergence indicated the
effectiveness of regularization.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
414
3.11 Figures and Tables
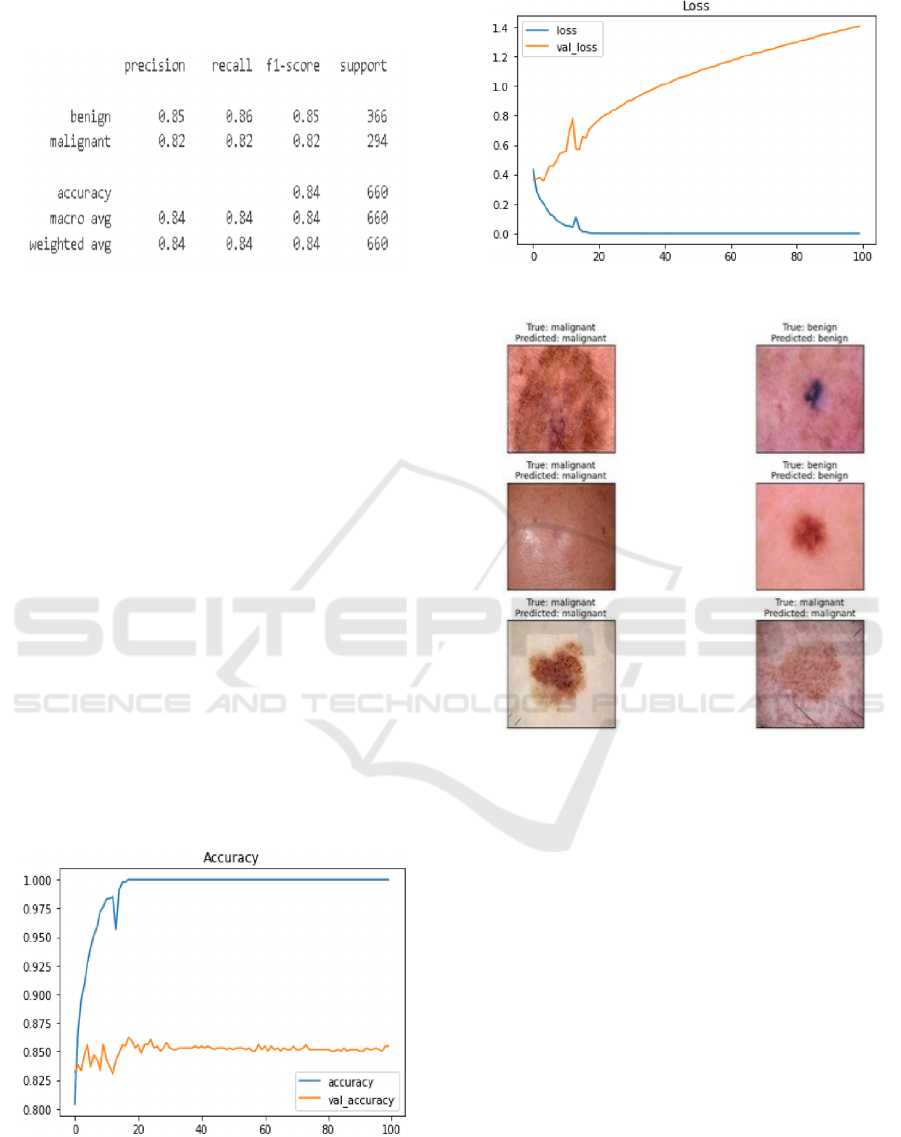
Figure 5: Classification Report.
The detailed results of the classification report are
presented in the figure above. This table aids in
understanding the model’s performance and final
outcomes. The table presents the model’s precision,
recall, F-1 score, and support for the two image
categories: malignant and benign. Furthermore, as
illustrated in Figure 5, it encompasses the weighted
average and macro average of the metrics.
The training and validation accuracy should
ideally increase over epochs and converge, as
illustrated in the accuracy and loss plot in Figure 6.
Overfitting is indicated by a continuous increase in
training accuracy while validation accuracy remains
constant. Training and validation loss should decrease
over time. Figure 7 indicates that overfitting may
occur if the validation loss increases while the
training loss decreases.
Figure 8 presents the labels predicted by the
model, along- side the test images that were
accurately identified, including their actual labels and
the corresponding predicted labels
.
Figure 6: Accuracy Plot.
Figure 7: Loss Plot.
Figure 8: Model Level Predictions.
4 CONCLUSIONS
This study demonstrates the effectiveness of
classifying skin lesion images through a
convolutional neural network (CNN) and its potential
role in facilitating early diagnosis of skin cancer. The
model was trained on a diverse set of thermoscopic
images using the ISIC dataset, achieving a high
degree of accuracy in distinguishing between
different types of skin lesions. The model’s
performance on unseen data illustrates its ability to
generalize effectively through data augmentation and
careful training, suggesting that CNNs are proficient
in identifying key features of skin lesions.
Although the model demonstrates promising
performance, it could be improved by fine-
tuning hyperparameters, exploring more
complex architectures such as transfer learning
Melanoma Skin Cancer Detection: A Self-Supervised Deep Learning Approach
415

with pre- trained models, or increasing the
dataset size. Additionally, validating the model
with real-world photographs in a clinical context
is crucial to demonstrate its efficacy. This
experiment demonstrates the potential
application of deep learning in dermatology,
facilitating the development of more effective
and accessible skin cancer screening tools.
REFERENCES
“A superior methodology for diagnosing melanoma skin
cancer through an explainable stacked ensemble of
deep learning models,” Multimedia Systems, vol. 28,
no. 4, pp. 1309–1323, Apr. 2021, doi: 10.1007/s00530-
021-00787-5.
A. C. Salian, S. Vaze, P. Singh, G. N. Shaikh, S.
Chapaneri and D. Jayaswal,” Deep learning architectur
es for skin lesion classification,” in IEEE CSCITA,
April 2020, doi: 10.1109/cscita47329.2020.9137810.
F. Alenezi, A. Armghan, and K. Polat, “Wavelet transform-
based deep residual neural network and ReLU-based
Extreme Learning Machine for skin
lesion classification,” Expert Systems with Applicatio
ns, vol. 213, p. 119064, Oct. 2022, doi: 10.1016/j.eswa
.2022.119064.
H. K. Gajera, D. R. Nayak, and M. A. Zaveri, “A
comprehensive analysis of dermoscopy images for
melanoma detection utilizing deep CNN features,”
Biomedical Signal Processing and Control, vol. 79, p.
104186, Sep. 2022, doi: 10.1016/j.bspc.2022.104186.
H. C. Reis, V. Turk, K. Khoshelham, and S. Kaya,
“InSiNet: A Deep Convolutional Approach to Skin
Cancer Detection and Segmentation,” Medical, 2022.
H.-C. Yi, Z.-H. You, D.-S. Huang, and C. K. Kwoh, “Graph
representation learning in bioinformatics: trends,
techniques, and applications,” Briefings in
Bioinformatics, vol. 23, no. 1, Aug. 2021, doi:
10.1093/bib/bbab340.
I. Kousis, I. Perikos, I. Hatzilygeroudis, and M. Virvou,”
Deep Learning Techniques for Accurate Skin Cancer
Detection and Mobile Application,” Electronics, vol.
11, no. 9, p. 1294, April 2022, doi: 10.3390/electronics
11091294.
IEEE,” ISIC Melanoma Dataset,” IEEE Data Portal, May
26, 2023. Online. Accessible at: https://ieeedataport.or
g.
M. Fraiwan and E. Faouri, “On the Automatic Detection
and Classification of Skin Cancer Utilizing Deep
Transfer Learning,” Sensors, vol. 22, no. 13, p. 4963,
June 2022, doi: 10.3390/s22134963.
Md. J. Alam et al.,” S2C-DeLeNet: A parameter transfer-
based integration of segmentation and classification
for the detection of skin cancer lesions in dermoscopic
images,” Computers in Biology and Medicine, vol. 150,
p. 106148, Sep. 2022.
P. Ghosh et al.,” SkinNet-16: A deep learning approach for
differentiating benign from malignant skin lesions,”
Frontiers in Oncology, vol. 12, Aug. 2022, doi:
10.3389/fonc.2022.931141.
Q. Abbas and A. Gul, “Detection and Classification of
Malignant Melanoma Using Deep Features of
NASNet,” SN Computer Science, vol. 4, no. 1, Oct.
2022, doi: 10.1007/s42979-022-01439-9.
R. K. Shinde et al., “Squeeze-MNet: A precise skin cancer
detection model for low-computing IoT devices
employing transfer learning,” Cancers, vol. 15, 2022.
S. Inthiyaz et al., “Deep learning for skin disease
detection,” Advances in Engineering Software, vol.
175, p. 103361, Nov. 2022, doi: 10.1016/j.advengsoft.
2022.103361.
W. Gouda, N. U. Sama, G. Al-Waakid, M. Humayun, and
N. Z. Jhanjhi,” Detection of Skin Cancer Based on
Skin Lesion Images Using Deep Learning,” Healthcar
e, vol. 10, no. 7, p. 1183, June 2022, doi: 10.3390/heal
thcare10071183.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
416
