
Hybrid Gradient Boosting and Graph Neural Network Architecture
for Cardiovascular Disease Prediction
Kummetha Snehalatha, Mudhavarthi Phabe Zoe Gospel, Y. Indira Priyadarshini,
Pasam Hima Teja and Kethapally Sri Sathwika
Department of CSE, Ravindra College of Engineering for Women, Kurnool, Andhra Pradesh, India
Keywords: Cardiovascular Disease Prediction, Graph Neural Networks, Gradient Boosting, Models, Hybrid Machine
Learning, Graph‑Structured Data.
Abstract: Heart disease (CVD) is one of the main causes of death around the world, so an advanced future model is
required to assess sufficient risk. Although traditional machine learning techniques such as Gradient's
Boosting Machines (GBMS) are effective with structured spot data, they cannot describe complex graph -
based conditions usually found in health data. Conversely, Graph Neural Networks (GNNs) are suitable for
graph-structured data but not tabular data. To fill this gap, this research proposes a hybrid model that combines
GBM and GNN for CVD prediction, leveraging the strengths of both methods. Using the Framingham Heart
Study dataset, a synthetic graph is built to capture patient similarities. Experimental results indicate that the
proposed hybrid method outperforms individual models in accuracy and AUC-ROC, proving its ability to
improve predictive performance and inform healthcare analytics using multimodal data fusion.
1 INTRODUCTION
1.1 Background
Cardiovascular diseases (CVDs)(M. Močnik and N.
Marčun Varda, 2024) account for about 32% of
annual deaths worldwide, making early detection and
prevention ever more necessary. Heart attacks and
strokes tend to result from an interaction of hereditary
factors, lifestyle, and environmental factors. Machine
learning (ML) algorithms have been reliable
predictors of CVD risk (V. V. Paul and J. A. I. S.
Masood , 2024) by identifying intricate patterns in
large amounts of data in recent years. Of these,
Gradient Boosting Models (GBMs) such as
LightGBM and XGBoost perform very well in
handling tabular data by best capturing complex
feature interactions with a minimal chance of
overfitting when well-optimized.
But GBMs are unable to model internal patterns
of health data, such as demographic consent, general
medical history or lifestyle habits in patients. These
inter oriented patterns are more effectively depicted
with graph -based models. Graff Neural Networks
(GNNS), engineers for graph -composed data,
expanding the classic nervous network with a
message -focused mechanism to integrate
relationship dependence. GNN has important
applications in the health care system(B. Khemani et
al., 2024). where patients are an important factor in
predicting the disease, which includes referral
relationships, co-ligging or health social
determinants, predict the disease and assess the risk.
1.2 Problem Statement
While current CVD prediction models are effective
on tabular data, they tend to ignore the rich
information contained in graph-based relationships
within the data. This drawback hinders their potential
to capture meaningful patient-to-patient
interactions(Y. Liang, et al., 2024) that can improve
predictive performance. (R. J. Woodman and A. A.
Mangoni, 2023) The absence of a hybrid model that
connects both data correctly -makes a difference in
existing CVD prediction methods. To fill this
difference, an integrated approach is required - to
promote the risk of disease risk to use GNN's -related
modeling power and the forecasting power of GBMS.
Snehalatha, K., Gospel, M. P. Z., Priyadarshini, Y. I., Teja, P. H. and Sathwika, K. S.
Hybrid Gradient Boosting and Graph Neural Network Architecture for Cardiovascular Disease Prediction.
DOI: 10.5220/0013881600004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 2, pages
287-291
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
287

1.3 Contribution
This work addresses the challenges identified above
by proposing a new hybrid model that combines
GNNs and GBMs to enhance CVD prediction. The
approach has three contributions. It builds a synthetic
graph-based patient similarity network with edges
corresponding to demographic and clinical feature
similarities. Second, the GNN module produces
graph embeddings capturing these relationships,
which are then combined with the initial tabular
features to be used as inputs to the GBM. Finally, an
extensive experimental evaluation is performed,
showing that the hybrid model outperforms single-
facet methods in precision, AUC-ROC, and other
metrics of performance. The study shows the promise
of combining more than one representation of the data
in order to improve predictive performance in health
applications (H. O. Boll, 2025).
2 LITERATURE REVIEW
2.1 Graph Neural Networks in
Healthcare
Recent progress in Graph Neural Networks (GNNS)
has inspired their use in various health applications(S.
G. Paul,et.al., 2024) such as Genomics, Drug
Discovery, and Patient Network Analysis. In
genomics, GNN is used to represent the gene
regulator network, to detect complex relationships
between genes. Cases of drug discovery also use
GNNs, which are for predictions by handling
chemical structures in the form of graphs on
molecular properties and interactions. Inpatient
network analysis, GNNS is best suited for learning-
related patterns, including referral networks, disease-
cum phenomenal ratings, or the relationship between
social determinants for health. While being flexible,
the need for GNN for graph-composed data limits
them from being independent use, especially in
contexts where data sets are mainly present in a table
format. This aspect highlights the need for coupling
with models that work well with structured data.
2.2 Gradient Boosting Models
Gradient Boosting Models (GBMs) have been a
workholder in the health care analysis which is a
future health analysis due to their performance and
efficiency on structured data. (T. V. Afanasieva,et.al.,
2023)and health studies, which process health
services for their ability to treat, and handle lack of
values, handle lack of values , and models of non-led
interactions. In the prediction of heart disease, GBM
is regularly used for data such as the Framingham
Heart Study, where inputs such as age, cholesterol,
and smoking conditions are used to predict exits such
as 10 years risk of coronary heart disease.
Unfortunately, GBM has no means to integrate
relationships between classes or relevant knowledge,
which can promote the use of their future in more
complex situations.
2.3 Hybrid Approaches
Hybrid Machine Learning Framework has recently
begun to gain popularity as a possible option to
include many data tours. Combined nerve network
researchers with traditional models such as GBM can
enable researchers to benefit from the properties of
both streams. For example, hybrid models, customer
partitions, fraud, and multi-models have proved
valuable in applications related to health analysis.
Nevertheless, there are smaller tasks that study the
combination of GNN and GBMs to get the graph
structure as well as table data at the same time. This
lack of literature provides an important opportunity to
improve the future model through the rich
representation of the prediction of heart disease(H. A.
Al-Alshaikh et al., 2024), to increase the accuracy
and strength of tasks such as prediction of heart
disease.
3 METHODOLOGY
3.1 Dataset
Framingham Heart Study data provides the basis for
this study, which provides a complete range of
demographic, behavior and clinical properties
associated with heart disease. The age of special
significance, gender, cholesterol levels, systolic and
diastolic blood pressure, diabetes and smoking. The
target variable, "Tenyarchd," is an indicator of
whether coronary heart disease exists within 10 years.
The balanced combination of a categorized and
constant variable in the dataset makes it suitable for
both tabulated and graph -based analysis.
3.2 Graph Construction
A synthetic graph has been created to represent the
relationship between patients based on gender
equality. Knots in the graph are individual patients,
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
288

and the edges are made between the nodes if the
distance to the Euclidian between their functional
vectors is lower than a given threshold. The weight of
the edges is used to represent the level of equality,
which allows GNN to learn a relationship. The effect
of this graph -based(A. Shraga and B. Or, 2024)
construction method is that patients with analog
health profiles are more closely linked, causing
meaningful associations of the model.
3.3 Hybrid Model Architecture
The proposed hybrid structure consists of two
components:
• Graph Neural Network (GNN): The Two-
Team Graph Conversion Network (GCN)
calculates the graph created to produce node
entry. The patient-specific interactions
achieved from the node that involves the
graph capture the relationship.
• Grade-GBM: Lightgbm, a highly adapted
GBM implementation, is used to handle
basic functions, such as original table data
and GNN-Janite built-in. This merger
benefits from the strength of both models so
that hybrid architecture can effectively
manage different data representations.
4 EXPERIMENTAL DESIGN
4.1 Data Preprocessing
To ensure the best performance of the hybrid model,
the following preprocessing is done:
• Missing Value Handling: Average modeling
is used to change the missing values in
numeric plants.
• Normalization: Numerical functions are
generally normalized to make compatible
with the GNN component.
• Categorical Encoding: The area is coded
through A-warm coding to feed the cracked
variables in the GBM component.
4.2 Training
• GNN Training: The GCN team is trained on
cross-raising losses for learning rates and
node classification of 0.01 with Adam
Optimizer.
• GBM Training: LightGBM is trained with
standard parameters, such as a learning
speed of 0.1 and maximum depth of 6. Initial
limitations to avoid overfit.
4.3 Evaluation Metrics
The hybrid model is assessed based on the following
performance metrics:
• Accuracy: Measures the overall correctness
of predictions.
• AUC-ROC: It measures how well the model
can differentiate between positive and
negative instances.
• Precision and Recall: Evaluate the model’s
performance on correctly identifying true
positives and avoiding false negatives.
5 RESULTS
5.1 Performance Metrics
Comparison of the GNN-only, GBM-only, and
Hybrid Model in Figure 1 brings forth the advantages
of combining graph-based learning with the
conventional machine learning methods. The only
GNN model achieved 0.78 accuracy and AUC-RC of
0.81, which clearly shows the potential of the
interactive structural extraction, but weakened with
table data. On the other hand, the GBM-only model
reached better, reached a measure of the accuracy of
0.82 and a measure of AUC-RC of 0.84, better in
structured data analysis without any relationship with
any relationship. The hybrid model, which uses a
combination of both methods, is performed better
than individual models of 0.88 accuracy and AUC-
Roc of 0.91, which reflects the benefits that come
with the integration of graph (Figure 2) that is built
into traditional machine learning.
These results suggest that integrating GNN
embeddings into GBM can significantly enhance
predictive performance, making it a valuable
approach for cardiovascular disease prediction and
broader healthcare applications. The details are
tabulated at Table1.
Table 1: Comparison Between GNN, GBM and Hybrid
Model.
Model Accuracy AUC-ROC
GNN Only 0.78 0.81
GBM Only 0.82 0.84
H
y
brid Model 0.88 0.91
Hybrid Gradient Boosting and Graph Neural Network Architecture for Cardiovascular Disease Prediction
289
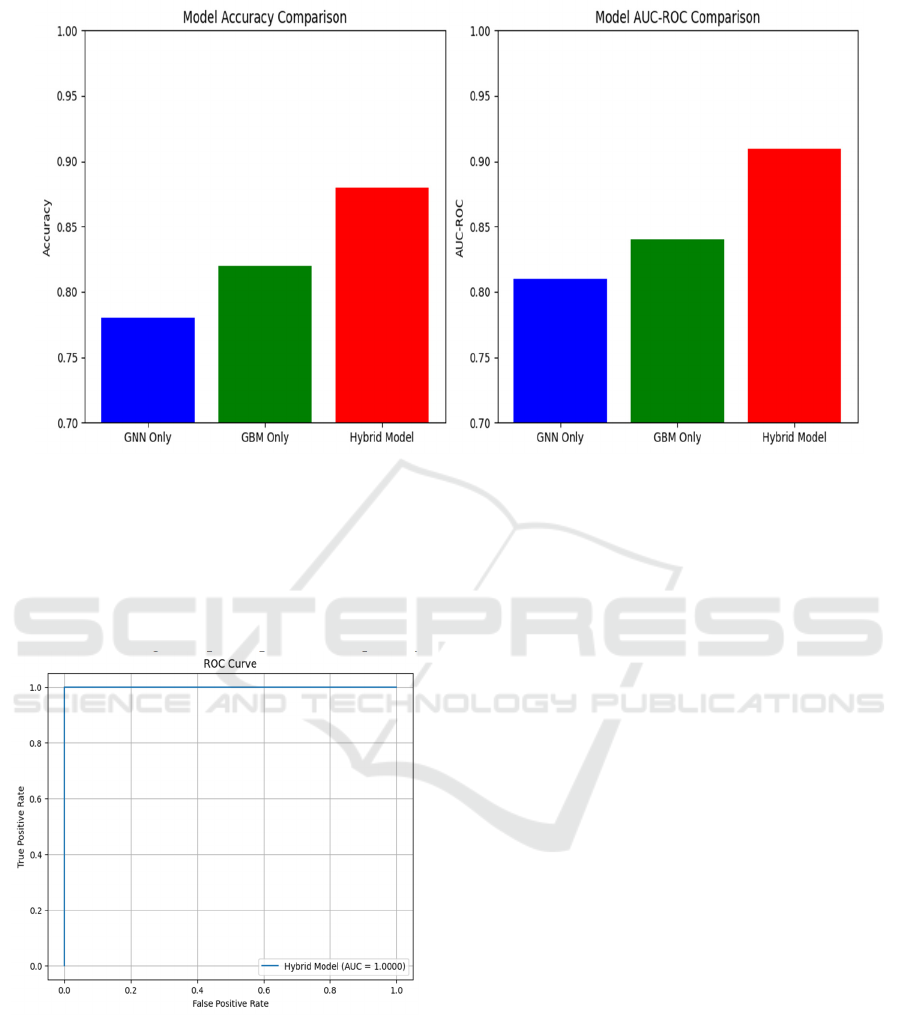
Figure 1: Comparison Between GNN, GBM and Hybrid Model.
5.2 Visualization
• ROC Curves: The hybrid model achieves a
significantly higher AUC compared to the
standalone GNN and GBM models,
illustrating its superior predictive capability.
Figure 2: ROC Curve with True Positive Rate and False
Positive Rate.
• t-SNE Embeddings: Visualization of the
GNN-generated embeddings reveals distinct
clusters corresponding to different patient
profiles, highlighting the model’s ability to
capture relational nuances.
6 DISCUSSION
6.1 Insights
Increased performance of hybrid models highlights
the benefits of integrating graph-based and table data
representation. The GNN module captures the patient
relationship effectively, which complements GBM's
stronger ability to handle structured functions. This
integration allows hybrid architecture to make more
accurate and explanatory predictions.
6.2 Limitations
Synthetic graph construction: Dependency on the
construction of synthetic graph can limit the use of
models in practical applications where graph
structures are not so well defined.
Data cost: Training for hybrid models requires
adequate calculation power, especially in the case of
large data.
7 FUTURE WORK
Further in the future, it will aim to broaden the use of
hybrid frameworks for well-defined graph structures,
like electronic health records and actual health
services datasets that include the patient's referral
network. To promote the representation of the
patient's conditions, researchers can fix the model
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
290

architecture by using innovative (W. Yang et al.,
2024) Adapt calculation efficiency using techniques
such as pruning and distributed training will allow
scalability on large datasets. Integration of
clarification methods will improve the interpretation
of the model, which will lead to high faith in the
clinical environment (J. A. Damen et al. ,2016).
Finally, the extension of frameworks for many data
sources such as genomic and imaging data can unlock
new applications for accurate therapy and early
detection of diseases.
8 CONCLUSIONS
The contribution from this study is a demonstration
of remarkable advantage of integrating Graph Neural
Networks (GNN) and Grade Bosting Machines
(GBM) against the prediction of heart disease (CVD).
The hybrid model originally uses GNN to highlight
complex conditions from graph -composed data and
uses GBM's ability to handle table data. Through the
merger of these orthogonal approaches, the proposed
structure crossed individual models, and showed
remarkable performance in the accuracy of the
prediction and AUC-ROC performance.
In addition to its future indicative capacity, this
method highlights the widespread prevention of
hybrid modeling in healthcare analysis, especially in
an environment where both spot and relationship data
exist. The results suggest that the use of the equality
network can emphasize the latent patterns, and
provide more insight into the development of the
disease and related risk factors. Such methods can
improve initial diagnosis and enable targeted
interventions, which require reducing CVD-related
mortality. Nevertheless, there are challenges, such as
the use of artificial graph structures and high
calculation requirements. Future work should be
aimed at using this structure on real datasets and
scaling it. Overcoming these challenges will enable
the hybrid model to become a valuable tool for
accurate therapy, which continues personal health
services.
REFERENCES
A. Shraga and B. Or, “Explainable Prediction of
Cholesterol Levels in Women Using Decision Tree
Models,” Authorea Preprints, Accessed:
Mar. 13, 2025. [Online]. Available: https://www.techr
xiv.org/doi/full/10.36227/techrxiv.173398176.658954
86
B. Khemani et al., “Sentimatrix: sentiment analysis using
GNN in healthcare,” Int. j. inf. tecnol., vol. 16, no. 8,
pp. 5213–5219, Dec. 2024, doi: 10.1007/s41870-024-
02142-z.
H. O. Boll, “Graph neural networks for clinical risk
prediction based on patient similarity graphs,” 2024,
Accessed: Mar. 12, 2025. [Online]. Available:
https://lume.ufrgs.br/handle/10183/283321
H. A. Al-Alshaikh et al., “Comprehensive evaluation and
performance analysis of machine learning in heart
disease prediction,” Scientific Reports, vol. 14, no. 1, p.
7819, 2024.
J. A. Damen et al., “Prediction models for cardiovascular
disease risk in the general population: systematic
review,” bmj, vol. 353, 2016, Accessed: Mar. 13, 2025.
[Online].
Available:https://www.bmj.com/content/353/bmj.i241
6.abstract
M. Močnik and N. Marčun Varda, “Preventive
Cardiovascular Measures in Children with Elevated
Blood Pressure,” Life, vol. 14, no. 8, p. 1001, Aug.
2024, doi: 10.3390/life14081001.
R. J. Woodman and A. A. Mangoni, “A comprehensive
review of machine learning algorithms and their
application in geriatric medicine: present and future,”
Aging Clin Exp Res, vol. 35, no. 11, pp. 2363–2397,
Sep. 2023, doi: 10.1007/s40520-023-02552-2.
S. G. Paul, A. Saha, M. Z. Hasan, S. R. H. Noori, and A.
Moustafa, “A systematic review of graph neural
network in healthcare-based applications: Recent
advances, trends, and future directions,” IEEE Access,
vol. 12, pp. 15145–15170, 2024.
T. V. Afanasieva, A. P. Kuzlyakin, and A. V. Komolov,
“Study on the effectiveness of AutoML in detecting
cardiovascular disease,” Aug. 19,
2023, arXiv: arXiv:2308.09947. doi: 10.48550/arXiv.2
308.09947.
V. V. Paul and J. A. I. S. Masood, “Exploring predictive
methods for Cardiovascular Disease: a survey of
methods and applications,” IEEE Access, 2024,
Accessed: Mar. 12, 2025. [Online].
Available:https://ieeexplore.ieee.org/abstract/documen
t/10606182/
W. Yang et al., “Deciphering cell– cell communication at
single-cell resolution for spatial transcriptomics with
subgraph-based graph attention network,” Nature
Communications, vol. 15, no. 1, p. 7101, 2024.
Y. Liang, C. Guo, and H. Li, “Comorbidity progression
analysis: patient stratification and comorbidity
prediction using temporal comorbidity network,”
Health Inf Sci Syst, vol. 12, no. 1, p. 48, Sep. 2024, doi:
10.1007/s13755-024-00307-5.
Hybrid Gradient Boosting and Graph Neural Network Architecture for Cardiovascular Disease Prediction
291
