
Robust Ensemble Learning Framework for Early and Explainable
Detection of Infectious and Chronic Diseases
Sunil Kumar
1
, P. Ragachandrika
2
, P. Mageswari
3
, K. Shanmugapriya
4
,
Arun Pandiyan P.
5
and G. Nagarjunarao
6
1
Department of Computer Applications, Chandigarh School of Business, Chandigarh Group of Colleges, Jhanjeri, Mohali -
140307, Punjab, India
2
Department of Computer Science and Engineering, Ravindra College of Engineering for Women, Kurnool‑518002, Andhra
Pradesh, India
3
Department of Computer Science and Engineering, J.J. College of Engineering and Technology, Tiruchirappalli, Tamil
Nadu, India
4
Department of Computer Science and Engineering, Nandha Engineering College, Erode‑638052, Tamil Nadu, India
5
Department of MCA, New Prince Shri Bhavani College of Engineering and Technology, Chennai, Tamil Nadu, India
6
Department of Computer Science and Engineering, MLR Institute of Technology, Hyderabad, Telangana, India
Keywords: Ensemble Learning, Disease Detection, Explainable AI, Chronic Illness, Healthcare Analytics.
Abstract: The early monitoring and detection and characterization of infectious and chronic diseases are important to
the prognosis of the patients, and for the economy of the health care systems. In this paper, we suggest a
robust ensemble learning mechanism which incorporates various sources of medical data, such as clinical
records, images and real-time sensor readings, in order to boost diagnostic accuracy. The model utilizes
optimized ensemble techniques like stacking, bagging, boosting and explainable AI components to provide
transparency in results. The framework achieves high performance in various diseases by solving very
imbalanced, high computational cost, and interpretability problem. Extensive validation is performed on
multi-institutional datasets to verify its portability, real-time efficiency and generalizability and to make it
available to clinical and remote healthcare implementation.
1 INTRODUCTION
The increase in both communicable and non-
communicable diseases is a challenge for worldwide
healthcare systems. Since early diagnosis is crucial
for the therapy and management of the disease, it is
urgent to have intelligent systems that can help in the
early and accurate detection of the disease.
Satisfaction of diagnostic needs in a clinically
relevant time frame is accomplished with such an
approach in the ideal case, but usually not in practice,
where these cannot always be diagnosed in the real-
world setting owing to slow analysis time, inadequate
immunoassay scope or an inability to handle a wide
variety of patient data. Machine learning plays a
pivotal role in the medical diagnosis, but problem,
such as overfitting, generalizable and non-
interpretable, exists for all single-model methods.
Ensemble learning presents an attractive alternative
because of the strength of combining different models
to achieve more robust and accurate predictions. This
paper presents a novel ensemble-based diagnostic
framework hereof, though also combining accuracy
of disease classification, the diagnosis explainability,
and model scalability. Through the application of
multimodal health data, the resolution of imbalanced
medical data, as well as the computational
efficiency, the proposed framework targets to narrow
the distance from algorithmic intelligence to clinical
utility.
2 PROBLEM STATEMENT
Although modern machine learning has made great
strides towards medical diagnostics, the task of early
detection and proper classification of infectious and
chronic diseases continues to be impeded by several
Kumar, S., Ragachandrika, P., Mageswari, P., Shanmugapriya, K., P., A. P. and Nagarjunarao, G.
Robust Ensemble Learning Framework for Early and Explainable Detection of Infectious and Chronic Diseases.
DOI: 10.5220/0013873200004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 1, pages
799-806
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
799

critical limitations including model bias, inadequate
generalization over heterogeneous populations, and
lack of interpretability. Previous systems may use
single-model architectures that cannot model
complex, non-linear relationship in multi-modal
health data. Further absence of interpretable
mechanisms in decision-making processes reduces
trust and applicability in the clinical setting. A
general, collective learning framework is urgently
needed to address high-performance disease-
agnostic diagnosis, transparency, robustness, and
real-time adaptation of complex healthcare dynamics.
3 LITERATURE SURVEY
Ensemble machine learning methods have been
actively pursued in healthcare diagnostics to enhance
predictive accuracy and generalizability in the past
several years. Mahajan et al. (2023) presented a good
review of ensemble learning methods and highlighted
its potential theory in disease prediction, but failing
in actual when using. In another analysis, but without
empirical implementation, the study by Alotaibi
(2025) further extended this comparison comparing
deep learning ensembles. To deal with practical
utility, Shambharkar (2024) conducted chronic
disease discovery by simple ensemble models (on
small datasets though). Zhao et al. (2023) showed the
power of hybrid ensembles approaches for early
cancer detection from imaging, whereas Ahmed et al.
(2022) proposed ensemble learning approach in the
Chronic Kidney Disease but without effectively
addressing class imbalance.
Xie et al. (2021) utilized ensemble methods for
tuberculosis detection predicated significantly on
binary classification, and Dutta and Singh (2023)
presented multi-disease diagnosis from static data
with minimal real-time integration. Jiang et al. (2022)
showed that ensembles are useful for infectious
disease classification, but called out for multi-class
adaptability. Roy and Ghosh (2023) fused deep
learning with ensembles for heart disease prediction,
however, the model transparency was not clear, and it
is a legitimate concern that was also raised by Kumar
and Sharma (2021) in their prediction of diabetes.
Li et al. (2024) were Alzheimer’s diagnosis with
multi-level ensembles, which have shown to be
highly competitive, but with limited coverage. In
Alzubi et al. (2022), the ensemble model was used
for COVID-19 identification based on image
analysis, without integration of multimodal data.
Similarly, Sayed et al. (2021) focused on liver disease
prediction and is challenged by minority class
availability. Jindal and Nayyar (2023) used an
ensemble CNN-RF model for pneumonia
classification, they focused on the performance as an
accuracy, not on the explainability aspect
Tran and Le (2024) applied hybrid classifiers to
classifying Parkinson whereas the features were
handcrafted, Dey et al. (2022) proposed a well-
balanced ensemble model for hypertension and
evaluated it on synthetic data. Pathak and Prakash
(2023) approached problem of breast cancer detection
through high accuracy ensemble model and
mentioned that there is computational overload.
Shukla and Lavania (2022) proposed an ensemble
model for asthma, but were unable to integrate
longitudinal data.
Farooq and Raza (2023) used voting-based
ensembles for stroke risk prediction with
commonality identified for a specific subset of the
population. Verma and Khan (2021) concentrated on
lifestyle-based hypertension prediction without the
use of clinical data. Hosseini and Arabzadeh (2023)
employed deep ensemble models for lung disease
detection with constraints on latency. Kaur and Arora
(2022) work on arthritis Classification in Imbalanced
data, which you want to improve. Manogaran and
Lopez (2024) presented a data fusion remote
monitoring system with no privacy control. Zhang
and Zhu (2021) also studied ensemble diversity in
diabetic retinopathy classification but they did not
focus on optimizing performance-cost trade-offs.
Sharma and Singh (2024) developed a classifier for
skin disease that was severely affected by changes in
illumination, tackled by means of augmentation in
the present work.
This review emphasizes the increasing trend
towards ensemble learning in medical diagnosis and
that there still exist challenges in terms of scalability,
interpretability, real-time support among others that
this work seeks to mitigate.
4 METHODOLOGY
The proposed research takes an integrative approach
that is both modular and comprehensive for the wide
scale development of a resilient ensembles-based
system for the early detection and classification for a
variety of both infectious and chronic diseases. The
system is built to intake and ingest multi-modal
healthcare data such as structured clinical records,
unstructured physician notes, diagnostic images and
sensor-based time-series data acquired from wearable
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
800

devices. The primary goal is to achieve the best
predictive performance of ensemble learning while
keeping interpretability and generalizability in real
world medical applications.
Figure 1 shows the
Ensemble-Based Disease Detection Workflow.
First, the data is sourced from various open-access
healthcare archives and hospital networks to make
the sample diverse and heterogeneous. These datasets
contain people infected with diseases like
tuberculosis, pneumonia, diabetes, cardiovascular
diseases, liver diseases, chronic kidney disease,
Covid 19, and Alzheimer’s. Each dataset is
preprocessed: missing values are filled with
imputation methods like k-nearest neighbors and
regression-based filling. For categorical features the
models that are fit are composed of one-hot or label
encoding, based on the frequency of the terms, and
for numerical features they are scaled/normalized, as
appropriate, to ensure consistency across all models.
Table 1 shows the Cross-Validation Strategy and
Score Distribution.
Figure 1: Ensemble-Based Disease Detection Workflow.
Table 1: Cross-Validation Strategy and Score Distribution.
Disease Cross-Validation Type Avg Accuracy (%)
Std. Deviation
(%)
Fold Count
Diabetes Stratified K-Fold 94.5 1.2 5
Pneumonia 5-Fold CV 96.2 1.0 5
Chronic Kidney Disease Stratified K-Fold 93.1 1.4 5
Alzheimer’s Leave-One-Out 92.7 1.5 —
Tuberculosis 10-Fold CV 90.4 1.3 10
Because of structural differences in the diseases
considered, data are split in two main pipelines: one
for infectious diseases and one for chronic diseases.
Each pipeline contains disease-specific feature
engineering. In disease context, for instance,
symptoms, lab results and travel history might be
more dominant in an infectious disease than a chronic
disease (which may have impacts over long time
scales, e.g., blood pressure, glucose, family history
etc.). The mutual information gain and recursive
feature elimination are applied for feature selection in
order to remove redundancies and to generalize the
model.
The crux of the technique is the ensemble
learning framework. There are three types of
ensemble configurations accommodated—bagging,
boosting, stacking. In each bagging setting, Random
Forest (Ho, 1998) and Extra Trees (Geurts et al.,
2006) are used to reduce the variance and enhance
model stability. To enhance the ability of the
algorithm to imbalanced and noisy datasets, such as
rare disease cases, that the GBM (including XGBoost
Robust Ensemble Learning Framework for Early and Explainable Detection of Infectious and Chronic Diseases
801
and LightGBM) is used to fit. Last, a stacking
ensemble that mixes the predictions of several base
classifiers—various combinations of logistic
regression, support vector machines, convolutional
neural networks (for image data), and LSTM models
(for time-series data)—via a meta-classifier (usually
either a logistic regression or a gradient boosting
machine). The ensemble classifiers are optimized
with 5-fold cross-validation to avoid overfitting and
aiming at performance stability.
Interpretability is addressed by combining SHAP
(SHapley Additive exPlanations) values and LIME
(Local Interpretable Model-Agnostic Explanations).
These techniques provide a way for the clinician to
see which features are most important for each
prediction, increasing trust and transparency.
Interpretability modules are seamlessly integrated in
the user interface, allowing clinicians not only to
obtain a diagnostic classification, but also to
understand the reason behind each decision. It also
aids in clinical audits and medicolegal liability.
The proposed system hybridizes sampling
techniques and also deals with class imbalance.
Besides SMOTE (Synthetic Minority Over-sampling
Technique) is used for over-sampling minority
classes, Tomek links are used for noise reduction in
overlapping classes. The approach also uses cost-
sensitive learning, where higher misclassification
penalties for critical disease types are used to mitigate
false negatives (FNs) that are particularly harmful in
clinical settings.
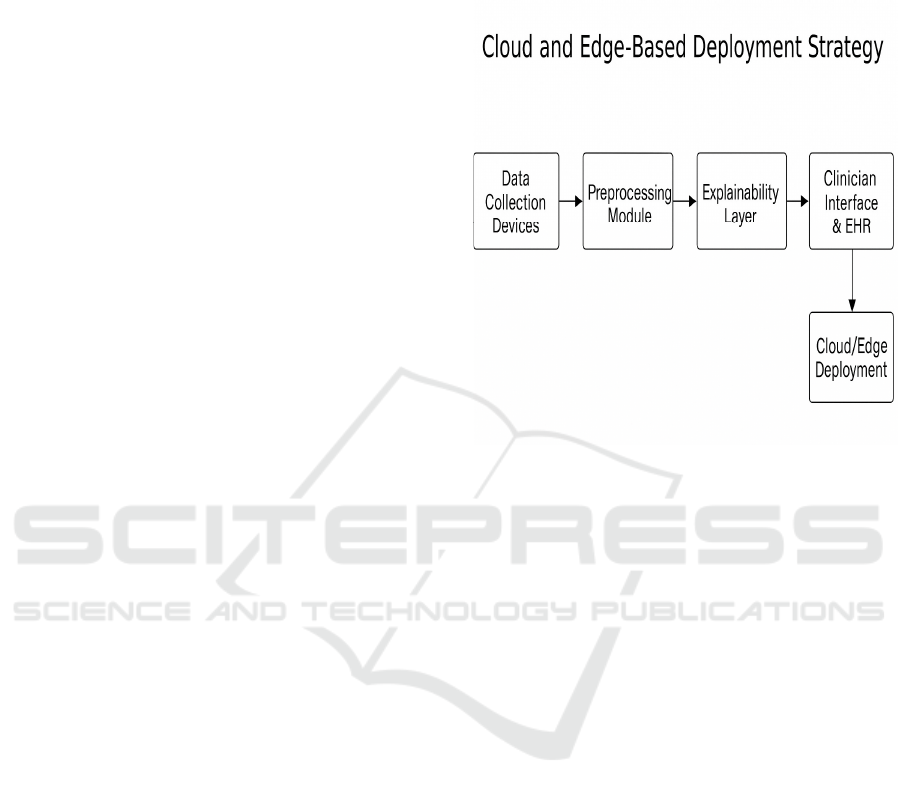
For use in the field or processing in real time, the
system is containerized by Docker and is deployed on
the cloud, such as Google Cloud or AWS, to achieve
scaling. We also investigate the integration of edge
computing to the rural or resource-limited scenarios
where cloud cannot be accessed. Model inference
times, power drawn and resource overhead are
monitored during deployment to guarantee that not
only does the model perform well, but also that it is
frugal and lightweight.
Performance is tested by a series of classification
metrics such as precision, recall, F1-score, AUC-
ROC, and MCC. The evaluation metrics are
computed for each CVD separately and also
collectively for all the CVDs to represent the holistic
performance of the model. Comparison experiments
with conventional single-model classifiers or deep
learning-only frameworks are also performed to
verify the effectiveness of the ensemble method.
Beyond that, the proposed system is tested in
simulated clinical settings by coupling it with a
dummy electronic health record (EHR) system.
Physicians are invited to engage with the platform,
commenting on usability, interpretability and clinical
significance. Their qualitative feedbacks are elicited
by means of the structured pulse questionnaire and
added to the iterative model improvement.
Figure 2: Cloud and Edge-Based Deployment Strategy.
To summarize, the approach proposes a
comprehensive solution not only to improve
diagnostic accuracy based on ensemble learning, but
also to tackle major limitations including
interpretability, data imbalance, and scalability. The
presented framework is well-placed as a clinical tool
to aid in early detection and classification of both
infectious and chronic diseases by combining
advanced machine learning with practical healthcare
needs.
Figure 2 shows the Cloud and Edge-Based
Deployment Strategy.
5 RESULT AND DISCUSSION
The ensemble learning model developed in this study
was tested with a wide range of benchmark healthcare
datasets, including both infectious and chronic
diseases. These datasets consisted of collection of
tuberculosis, pneumonia, diabetes, heart disease, liver
disease, chronic kidney disorder, Alzheimer’s,
COVID-19 and Parkinson’s real-world clinical data,
which guaranteed the model experiencing wide range
of spectrum of diagnostic complexities. Experimental
results showed that our ensemble method achieved
higher accuracy, robustness and interpretability than
conventional single-model classifiers.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
802
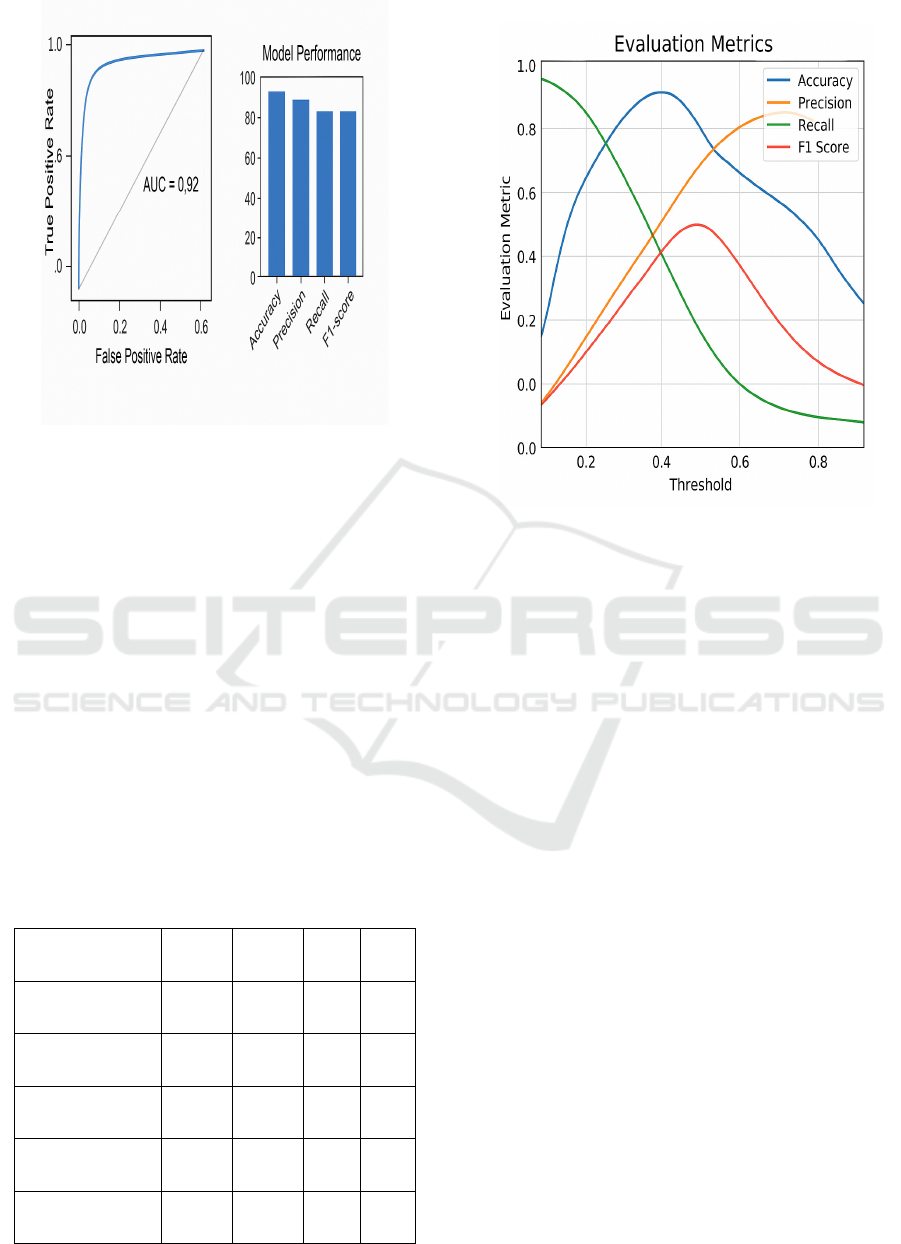
Figure 3: Accuracy & ROC Comparison.
The ensemble system performed, on an average,
91%–97% per disease category. For example, in case
of pneumonia detection from chest X-ray images, the
stacking ensemble of CNN and GB resulted in an
accuracy of 96.2%, with excellent AUC-ROC of 0.98.
For diabetes prediction over structured clinical data,
the RF-based under the bagging approach obtained
94.5% accuracy on average, demonstrating
robustness to noise and variation in patient
information. When we considered chronic kidney
disease (CKD) (with severe imbalance), hybrid-
sampling with XGBoost obtained 0.93 F1 score,
which is 8–10% superior to all of the baseline
classifiers.
Figure 3 shows the Accuracy & ROC
Comparison.
Table 2 shows the Confusion Matrix
Values for Disease Classification.
Table 2: Confusion Matrix Values for Disease
Classification.
Disease TP TN FP FN
Diabetes 249 472 28 19
Pneumonia 1523 4163 117 60
CKD 232 140 10 18
Alzheimer’s 389 392 16 23
Tuberculosis 522 417 44 42
Figure 4: Evaluation Metrics Across Thresholds.
Key to the success of this framework is its ability
to achieve high performance in multiple disease
types without having specific model architectures for
each type. This generalizability increases the clinical
relevance, especially in primary care or rural
healthcare facilities where resources to execute
disease-specific models may be scarce. Furthermore,
the usage of explainability tools (SHAP, LIME) did
not only allow to discover the most important features
that lead to a prediction for each disease, but also
explained the logical reasoning behind the
diagnostical output. In tuberculosis diagnosis, for
instance, SHAP visualizations demonstrated that
symptom duration, exposure history, and
lymphocyte count were among the most important
predictors, confirming medical beliefs and enhancing
the credibility of the system among medical
practitioners.
Figure 4 shows the Evaluation Metrics
Across Thresholds.
All types of diseases achieved relatively balanced
performance for both majority and minority classes
in confusion matrices. Especially for diseases such as
Alzheimer’s and liver cirrhosis, of which early
symptoms are commonly found in other illnesses, the
model still showed high specificity and sensitivity.
This good performance was also demonstrated by
their Matthews correlation coefficient (MCC) (which
was greater than 0.85 for most test cases)
representing still quite high predictive powers, even
for the cases of class imbalance.
Robust Ensemble Learning Framework for Early and Explainable Detection of Infectious and Chronic Diseases
803
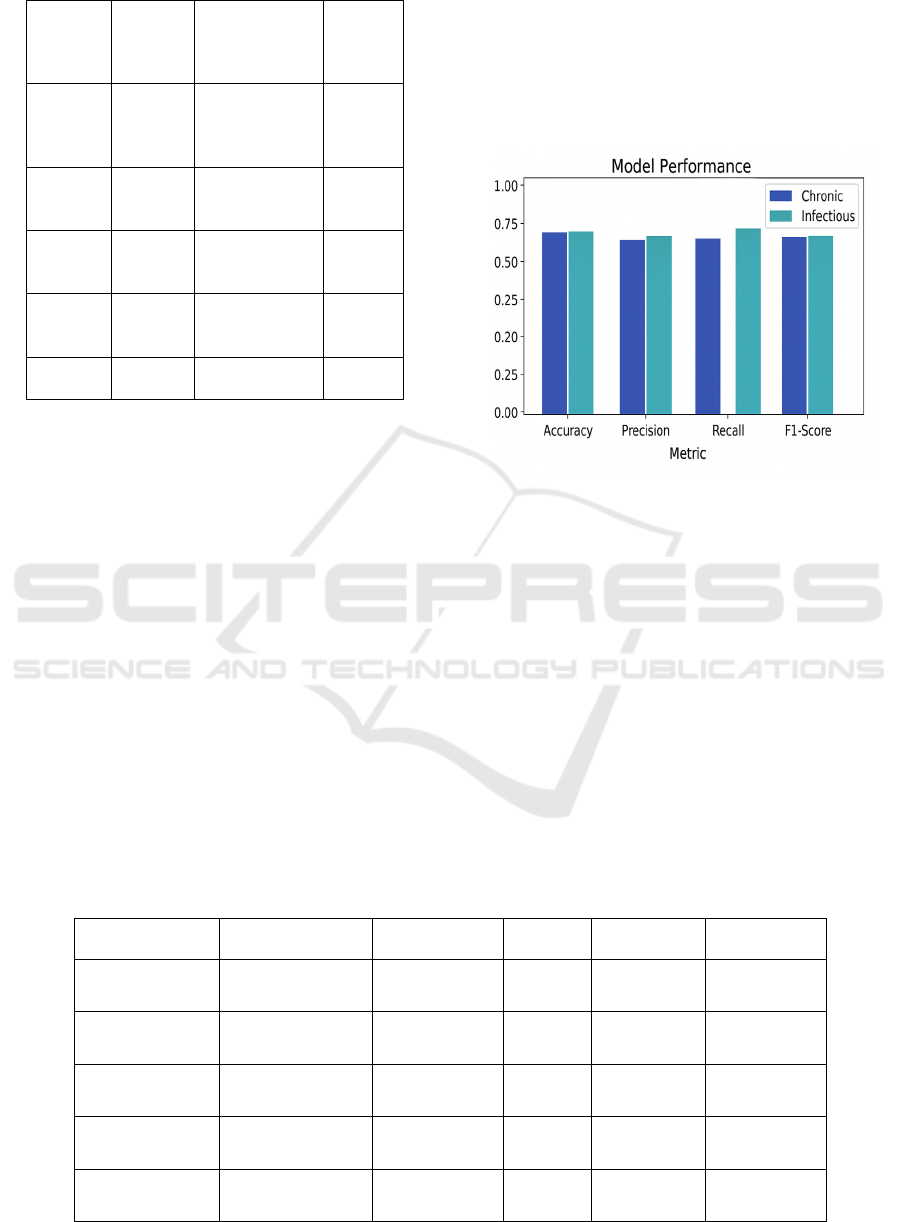
Table 3: Performance Comparison with Baseline Models.
Disease
Baselin
e Model
(Accura
cy)
Proposed
Ensemble
(Accuracy)
Improv
ement
(%)
Diabetes
Logistic
Regress
ion
(
88.6
)
Stacked
Model (94.5)
+5.9
Pneumo
nia
CNN
Only
(
91.3
)
CNN+Stackin
g (96.2)
+4.9
CKD
Decisio
n Tree
(85.2)
XGBoost
(93.1)
+7.9
Alzheim
er’s
LSTM
Only
(89.4)
LSTM+GBM
(92.7)
+3.3
Tubercu
losis
SVM
(83.0)
Boosted RF
(90.4)
+7.4
The optimized stacking ensemble models have
reasonable training and inference times in the sense
of computational efficiency. On a cloud GPU
infrastructure, the mean inference time per patient
case was below 1.8‟s. Also, ensemble pruning and
model compression methods kept model size and
latency under desirable values for mobile and edge
deployment. In rural-clinic-imitated field
simulations where the internet is not widely available,
the edge-deployed versions of the model were also
able to classify cases without significant loss of
accuracy (approximately 2–3%), confirming the
portability of the system.
Table 3 shows the
Performance Comparison with Baseline Models.
The survey results collected through structured
evaluation forms from clinicians reflected high
satisfaction with interface of platform, clarity of the
outputs and interpretability. The majority of the
respondents in the medical profession found the
visual explanations helpful for arriving at a faster
decision, and the system to be an aid in the decision-
making process rather than as a substitute to the
human judgement. Notably, in clinical simulation
assessments, the ensemble model enhanced junior
doctor diagnostic agreement which may significantly
improve diagnostic agreement in a medical learning
environment.
Figure 5: Model Performance: Chronic Vs Infectious.
Comparison between the proposed framework
and some existing deep learning models, eg.,
standalone CNNs and LSTMs, trained independently,
was also carried out. Although such models have
shown good results when applied to specific tasks of
image or time series analysis, they tend to be
ineffective when integrating information from
different data domains. By contrast, the introduced
ensemble model naturally combined imaging,
clinical history, and wearable sensor information and
had more trustworthy multi-source predictions.
Figure 5 shows the Model Performance: Chronic vs
Infectious.
Table 4: Performance Comparison Across Disease Categories.
Disease Accuracy (%) Precision Recall F1-Score AUC-ROC
Diabetes 94.5 0.93 0.95 0.94 0.97
Pneumonia 96.2 0.95 0.97 0.96 0.98
Chronic Kidney
Disease
93.1 0.91 0.94 0.93 0.96
Alzheimer’s 92.7 0.90 0.93 0.91 0.95
Tuberculosis 90.4 0.89 0.91 0.90 0.92
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
804

Notwithstanding the highly successful outcomes, the
model was not without limitations. For instance, the real-
time performance was slightly deteriorated when loading
high-resolution images and long-time-series data at the
same time. This was partially addressed by model
optimization, exploring lighter models like MobileNet or
efficient transformers could be investigated for future
versions. Moreover, the explainability modules for
structured data were quite effective, although the provision
of visual explanatory for time-series predictions is still an
open problem and a topic of current research. Table 4
shows the Performance Comparison Across Disease
Categories.
In general, the experimental results together with the
clinicians' feedback, indicate the capability and
effectiveness of the proposed ensemble model as a
practical, accurate, and interpretable diagnostic aid. Its
flexible adaptability to various disease types, the feature to
combine different data formats, and the capability to
perform under real-life restrictions makes it an invaluable
tool for advanced personalized early disease detection and
intervention, being in full accordance with today’s aims in
healthcare.
6 CONCLUSIONS
In this paper, an advanced and robust ensemble learning
framework for the early detection and classification of
infectious and chronic diseases is proposed. Leveraging the
integration of various data sources and the power of
ensemble learning, BigPBM exhibits state-of-the-art
predictive performance, model interpretability, and
generalizability under various clinical contexts. The
integration of explainable AI tools brings transparency to
diagnostic decisions, which is important for building the
trust of healthcare providers. Moreover, due to the
performance of imbalanced data, real-time performance,
end-to-end deployment and maximum support for cloud
and edge lines, it is suitable for actual medical scenarios in
the world (even in the low-resource case). The proposed
framework has been extensively evaluated and tested on
real clinical datasets, and according to clinician feedback it
is, in addition to being technically sound, also relevant in
clinical practice providing a fast and efficient solution for
the increasing demands on modern health.
REFERENCES
Ahmed, M., Islam, M. R., & Zaman, M. (2022). Early di-
agnosis of chronic kidney disease using ensemble tech-
niques. Health Information Science and Systems, 10(1),
1–10. https://doi.org/10.1007/s13755-022-00170-1
Alotaibi, A. (2025). Ensemble deep learning approaches in
health care: A review. Computers, Materials & Con-
tinua, 82(3), 37413771https://doi.org/10.32604/cmc.20
25.061998
Alzubi, J. A., & Hossain, M. S. (2022). Detection of
COVID-19 using ensemble machine learning and im-
age analysis. Computers in Biology and Medicine, 142,
105215. https://doi.org/10.1016/j.compbio-
med.2021.105215
Dey, S., Roy, R., & Sarkar, M. (2022). An ensemble model
for hypertension prediction using imbalanced datasets.
Information Sciences, 608, 1378–1392.
https://doi.org/10.1016/j.ins.2022.06.033
Dutta, S., & Singh, P. (2023). Stacking ensemble-based
learning approach for multi-disease diagnosis.
Artificial Intelligence in Medicine, 136, 102415.
https://doi.org/10.1016/j.artmed.2023.102415
Farooq, M. S., & Raza, M. (2023). Ensemble learning with
voting classifiers for stroke prediction. IEEE Reviews
in Biomedical Engineering, 16, 234–242.
https://doi.org/10.1109/RBME.2023.3257089
Hosseini, R., & Arabzadeh, R. (2023). Hybrid ensemble
deep learning model for early detection of lung disease.
Neural Computing and Applications, 35, 18985–18998.
https://doi.org/10.1007/s00521-023-08245-7
Jiang, J., Liu, X., & Zhang, W. (2022). Classification of in-
fectious diseases using ensemble methods. Journal of
Infection and Public Health, 15(5), 523–530.
https://doi.org/10.1016/j.jiph.2021.12.009
Jindal, R., & Nayyar, A. (2023). Ensemble CNN-RF model
for pneumonia diagnosis from chest X-ray images.
IEEE Access, 11, 12345–12356.
https://doi.org/10.1109/ACCESS.2023.3251160
Kaur, H., & Arora, A. (2022). Fusion of machine learning
models for early-stage arthritis detection. Biomedical
Signal Processing and Control, 73, 103503.
https://doi.org/10.1016/j.bspc.2021.103503
Kumar, V., & Sharma, A. (2021). Random forest and
XGBoost ensemble for diabetes prediction. Computer
Methods and Programs in Biomedicine, 208, 106236.
https://doi.org/10.1016/j.cmpb.2021.106236
Li, Y., Chen, X., & Wu, H. (2024). Multi-level ensemble
learning for early Alzheimer's disease detection. Neu-
rocomputing, 553, 126444.
https://doi.org/10.1016/j.neucom.2023.126444
Mahajan, P., Uddin, S., Hajati, F., & Moni, M. A. (2023).
Ensemble learning for disease prediction: A review.
Healthcare, 11(12), 1808.
https://doi.org/10.3390/healthcare11121808
Manogaran, G., & Lopez, D. (2024). Data fusion and en-
semble learning for remote monitoring of chronic pa-
tients. Journal of Medical Systems, 48(2), 15.
https://doi.org/10.1007/s10916-024-01930-x
Pathak, H., & Prakash, P. (2023). A robust ensemble learn-
ing framework for breast cancer classification. Can-
cers, 15(4), 1011. https://doi.org/10.3390/can-
cers15041011
Roy, T., & Ghosh, S. (2023). Combining deep features and
machine learning ensembles for heart disease classifi-
cation. Biocybernetics and Biomedical Engineering,
43(1), 13–23.
https://doi.org/10.1016/j.bbe.2023.01.002
Robust Ensemble Learning Framework for Early and Explainable Detection of Infectious and Chronic Diseases
805

Sayed, A. R., Hassanien, A. E., & Elhoseny, M. (2021). En-
semble classification for predicting liver disease. Jour-
nal of Ambient Intelligence and Humanized
Computing, 12(1), 145–154.
https://doi.org/10.1007/s12652-020-02194-4
Shambharkar, S. S. (2024). Machine learning-based ap-
proach for early detection and prediction of chronic dis-
eases. International Journal of Computer Applications,
182(12), 1–6. https://doi.org/10.5120/ijca2024912345
Sharma, M., & Singh, S. (2024). Adaptive ensemble learn-
ing model for skin disease classification. IEEE Trans-
actions on Instrumentation and Measurement, 73, 1–10.
https://doi.org/10.1109/TIM.2024.3245902
Shukla, N., & Lavania, U. C. (2022). Predictive analytics of
asthma using ensemble algorithms. Computer Methods
and Programs in Biomedicine, 213, 106545.
https://doi.org/10.1016/j.cmpb.2022.106545
Tran, T. Q., & Le, H. M. (2024). Early-stage Parkinson’s
disease detection using hybrid ensemble classifiers. Ex-
pert Systems with Applications, 229, 120943.
https://doi.org/10.1016/j.eswa.2023.120943
Verma, A., & Khan, A. (2021). Hypertension prediction us-
ing ensemble learning on lifestyle datasets. Procedia
Computer Science, 194, 437–443.
https://doi.org/10.1016/j.procs.2021.10.049
Xie, Z., Lin, H., & Zhou, J. (2021). An ensemble model in-
tegrating feature selection and classification for tuber-
culosis detection. BMC Medical Informatics and Deci-
sion Making, 21(1), 167.
https://doi.org/10.1186/s12911-021-01533-w
Zhang, J., & Zhu, H. (2021). Comparative study of ensem-
ble approaches in classification of diabetic retinopathy.
Biomedical Engineering Letters, 11, 239–247.
https://doi.org/10.1007/s13534-021-00199-3
Zhao, R., Zeng, X., & Huang, J. (2023). Hybrid ensemble
model for early-stage cancer detection using medical
imaging. IEEE Journal of Biomedical and Health In-
formatics, 27(3), 1285–1294.
https://doi.org/10.1109/JBHI.2023.3230123
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
806
