
Predecting Adverse Drug Reactions with XGBoost a
Pharmacovigilance Application
K. Jnana Sadhana, Veera Raghavan J., Bellamgubba Anoch, P. Kiran Sree,
Raja Rao P. B. V. and B. Satyanarayana Murthy
Department of CSE, Shri Vishnu Engineering College for Women, Vishnupur, Bhimavaram, Andhra Pradesh, India
Keywords: Adverse Drug Reactions (ADRs), Machine Learning(ML) in Pharmacovigilance, XGBoost for ADR
Detection, Feature Selection, Explainability(SHAP), AI in Drug Safety, Healthcare.
Abstract: Adverse Drug Reactions (ADRs) pose a significant challenge to modern pharmacovigilance, leading to severe
health implications, increased healthcare costs, and regulatory concerns. Traditional ADR detection methods
rely heavily on manual pharmacovigilance and rule-based expert systems, which are slow, subjective, and
limited in scalability.However, existing ML models either suffer from overfitting, lack of generalizability, or
black-box limitations.This study proposes a novel XGBoost-based ADR prediction model that achieves 91%
accuracy, outperforming traditional classifiers (Naïve Bayes, SVM, Random Forest) and deep learning
models (Graph Neural Networks, Neural Collaborative Filtering, Deep Ensembles) found in literature. The
proposed model uses feature engineering with SHAP explainability, class balancing techniques, and
hyperparameter tuning to improve predictive performance. By leveraging Therapeutic Class, Action Class,
and Chemical Properties, the model not only enhances accuracy but also ensures interpretability, making it
suitable for real-world clinical decision systems.Experimental results demonstrate that the optimized
XGBoost model achieves 91% accuracy, 92% precision, and 91% recall, making it a competitive alternative
to deep learning-based ADR detection models.
1 INTRODUCTION
1.1 Background & Motivation
ADRs are an important source of concern in modern
medicine. impacting the lives of millions of patients
worldwide. Such adverse and damaging
consequences of pharmaceutical medications are not
only detrimental to patient safety, but also lead to
higher hospitalization rates, healthcare costs, and
regulatory hurdles. Due to the integration of more
similar therapeutic agents, the complexity of drug
interactions has dramatically increased. The advent of
personalised medicine and polypharmacy the use of
multiple drugs concurrently has made it even more
imperative to have a strong and automated system
that can help prevent potential ADR before they
become clinically apparent (N. Ibrar, I. Hamid, and
Q. Nawaz., 2023). Thus, the computational prediction
and prevention of ADRci has evolved into a vital
segment of pharmacovigilance.
1.2 Artificial Intelligence's Function in
ADR Detection
The advancements in machine learning (ML) and
artificial intelligence (AI) have altered many fields,
and pharmacovigilance is no different. While
traditional approaches to ADR detection depend
primarily on manual reporting systems, clinical trials,
and rule-based expert reviews, recent advancements
see AI methodologies capable of analyzing large data
sets, identifying non-obvious patterns, and predicting
adverse reactions with greater accuracy Other
machine learning algorithms such as Support Vector
Machines (SVMs), Random Forest Pokkuluri, K.S. et
al. (2025), and Naive Bayes classifiers are found to
be more effective in detecting ADRs due to the
patient history as well as the chemical properties of
the drugs.
1.3 Significance of This Research
This study seeks to optimize the XGBoost-based
ADR (Saravanan, D., Arunkumar, G., Ragupathi,
Sadhana, K. J., J., V. R., Anoch, B., Sree, P. K., P. B. V., R. R. and Murthy, B. S.
Predecting Adverse Drug Reactions with XGBoost a Pharmacovigilance Application.
DOI: 10.5220/0013871700004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 1, pages
713-720
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
713

T. et al., 2025) prediction model by proposing the
following objectives:
Use feature selection techniques to improve
prediction accuracy. Enhancement of model
interpretability through SHAP-based Feature
Importance Analysis Merge Therapeutic Class,
Chemical Class, and Action Class to enhance feature
representation. Make scalable for practical
application in pharmacovigilance systems. This work
aims to derive a black-box-free, high-performing
model and subsequently help deliver a strong AI-
enabled pharmacovigilance structure that can assist
doctors recognise potential drug adverseevents and
avert fatal drug combinations ahead of time. This
study's results can play an important role in the future
of AI-based medication safety monitoring that
enables data-driven healthcare decision-making and
precision medicine.
2 RELATED WORK
Adverse drug reaction (ADR) is one of the most
problematic complications in modern pharmacology
that challenge the drug deduction process and patient
safety. Conventional methods of identifying adverse
drug reactions (ADRs), including clinical trials and
post-market monitoring, are rarely successful in
detecting infrequent or late-emerging adverse effects
prior to a drug reaching the market. The introduction
of machine learning (ML) and artificial intelligence
(AI) has resulted in the emergence of predictive
models that can be used to detect ADRs earlier in the
drug development process. We next review existing
ML-based ADR prediction methods, their successes,
and their shortcomings.
2.1 Using Machine Learning to Predict
ADRs
Several machine learning techniques have been
applied to pre-market ADR prediction, including
graph-based models, matrix factorization, neural
collaborative filtering, and deep learning approaches.
2.1.1 Graph Neural Networks (GNNs) for
Drug-Drug Interaction ADR
Prediction
Though graph-based learning methods have been
utilized for advancing ADR prediction by modelling
drug-drug interactions (DDIs). Chandra
Umakantham et al. proposed a GNN-based model
that performed self-supervised learning to identify
ADRs from DDIs. 2024. greatly enhancing
predictive accuracy compared to traditional statistical
methods. Inspired by this observation, Patel & Patel
(2024) took a step further, incorporating causal
inference methods with the GNNs to make ADR
predictions more robust and interpretable.
Limitations: While GNNs offer improved
accuracy, they require large-scale graph
representations and high computational resources,
making them less feasible for real-time applications.
2.1.2 Signed Networks and Matrix
Factorization for ADR Prediction
Alternative approaches leverage network structures
and probabilistic dependencies to enhance ADR
detection.A signed network-based method was
presented by Zhuang & Wang (2021), who used the
topological structures of pharmacological networks
to infer possible adverse drug reactions.A
probabilistic matrix tri-factorization model was
presented by Zhu et al. in 2021,which improves
interpretability by considering ADR dependencies
across drug pairs.
Limitations: These methods depend on data
completeness, making them less effective in sparse
datasets where missing interactions occur.
2.1.3 Neural Collaborative Filtering and
Hybrid ML Approaches
To address the cold-start problem in ADR prediction
(i.e., handling novel drugs with little existing data),
recent studies have applied collaborative filtering
techniques.Xiong et al. (2023) designed a Neural
Collaborative Filtering (NCF) model, leveraging drug
feature similarities to improve ADR prediction in
unseen drugs.Ibrar et al. (2023) implemented a hybrid
ML model, combining deep learning and structured
feature engineering for ADR classification.
Limitations: Despite their generalization ability,
collaborative filtering models struggle with rare ADR
cases as they rely heavily on historical data.
2.1.4 Semi-Supervised Learning for ADR
Prediction
To improve generalization across different drug
datasets, researchers have adopted semi-supervised
learning techniques.Yan et al. (2022) developed a
similarity network-based semi-supervised learning
model, enhancing generalizability across multiple
ADR datasets.
Limitations: Semi-supervised learning methods
require high-quality unlabeled data, and their
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
714

performance depends on data augmentation
techniques.
2.2 Structure-based ADR Prediction
Models
2.2.1 Relationships between Structure and
Activity (SAR) and Quantitative
Structure and Activity (QSAR)
ModelsStructure-based models analyze the chemical
composition of drugs to infer their likelihood of
causing ADRs.Kang et al. (2022) applied Support
Vector Machines (SVMs) in an SAR model to
forecast antiseizure drug teratogenicrisk.Zhou et al.
(2021) used Random Forest to create a QSAR model
that evaluated the degree of drug-induced
rhabdomyolysis(PBV et al., 2024).
Limitations: SAR and QSAR models require
extensive feature engineering, and their accuracy is
highly dependent on curated chemical datasets.
2.2.2 Structural Alerts for ADR Risk
Identification
Structural alert models identify toxicity risk markers
in drug molecules.Long et al. (2023) implemented a
structural alert-based model for predicting drug-
induced QT prolongation, improving the architecture
of the Adverse Outcome Pathway (AOP).
Limitations: These models require manual rule
formulation, making them less adaptable to novel
drug compounds.
2.3 Deep Learning for ADR Prediction
2.3.1 NLP-Driven ADR Susceptibility
Prediction via Chemical Language
Models
Recent studies have applied Natural Language
Processing (NLP) models to Extractive text-based
drug safety reports. To enhance ADR detection
through preclinical drug screening, Lin et al. (2024)
proposed a deep chemical language model for the
review of drug safety.
Limitations: NLP-based models are data biases
prone which need large scale corpus, textual data for
training.
2.3.2 Risk Prediction of Drug Induced Liver
Injury (DILI) and Cardiotoxicity
Deep learning algorithms have also been employed to
predict ADRs including cardiotoxicity and drug-
induced liver injury (DILI)Minerali et al. (2020)
reported that deep learning performed better than the
traditional ML methods for predicting the DILI cases
Pokkuluri, K.S. et al. (2025), (Sree et al., 2024).
2.4 Comparative Analysis of ADR
Prediction Models
Table 1 Shows the summarizes the performance
metrics for various models applied to ADR
prediction.
2.5 Challenges in Traditional ADR
Detection
Despite advancements in pharmacovigilance,
traditional ADR detection methods face several
challenges:
2.5.1 Underreporting and Data Bias
Over 90% of ADR cases remain unreported due to
lack of awareness, reporting delays, and legal
concerns.ADR reports are disproportionately skewed
towards widely prescribed drugs, making rare ADRs
harder to detect.
2.5.2 Scalability Issues in Rule-Based
Methods
Traditional rule-based expert systems require
frequent manual updates to accommodate newly
introduced drugs.These systems fail to adapt to
complex drug-drug interactions (DDIs) and emerging
adverse reactions.
2.5.3 Limitations of Classical Machine
Learning Models
Conventional models like SVMs, Decision Trees, and
Naïve Bayes struggle with high-dimensional ADR
datasets.Class imbalance in ADR datasets results in
biased predictions, where models favor commonly
occurring reactions while ignoring rare but critical
ADRs.
Predecting Adverse Drug Reactions with XGBoost a Pharmacovigilance Application
715

Table 1: Summarizes the Performance Metrics for Various Models Applied to ADR Prediction.
Approach Key Contribution Dataset Used Accuracy Limitations
GNN-Based Model
(ChandraUmakantha
m et al., 2024)
Improved ADR
detection in DDIs
FAERS, SIDER 89%
Requires high
computational resources
Causal Inference +
GNNs (Patel & Patel,
2024
)
Enhanced reliability
with causal reasoning
FAERS 85% Limited interpretability
Probabilistic Matrix
Factorization (Zhu et
al., 2021)
Improved ADR
interpretability
FAERS 83%
Requires high-quality
labeled data
Neural Collaborative
Filtering (Xiong et al.,
2023
)
Generalized ADR
prediction for new
dru
g
s
FAERS, Open TG-
GATEs
87% Struggles with rare ADRs
SAR Model (Kang et
al., 2022)
Predicted teratogenic
risk
Open TG-GATEs 86%
High feature engineering
complexity
Structural Alert
Model (Long et al.,
2023)
Identified
cardiotoxicity risk
DrugBank 84%
Requires manual rule
formulation
Deep Learning
Ensemble (Karim et
al., 2021)
Enhanced
cardiotoxicity
prediction
DrugBank 90%
Black-box nature limits
explainability
Ensembles achieve high accuracy (~90%), their
"black-box" nature limits their use in clinical
settings.Clinicians require transparent and
explainable AI models for real-world adoption, which
deep learning models often fail to provide.
3 PROPOSED METHODOLOGY
Dataset is downloaded from kaggle repository.named
with medicine datset.CSV
Step 1: Data Collection & Preprocessing
• Dataset: Extracted from structured medical
records, clinical trial data, and publicly
available ADR datasets.
• Data Cleaning: Removal of duplicates,
handling missing values, and filtering out
irrelevant features.
• Feature Selection:
• Important attributes: Chemical Class,
Therapeutic Class, Side Effects, Substitutes,
and Action Class.
• Unimportant attributes (e.g., common
stopwords, noise) were eliminated to
improve model efficiency.
Step 2: Feature Engineering & Representation
• Text Features: Side effects and drug
reactions were processed using TF-IDF and
NLP-based techniques.
• Categorical Features: Encoded using Label
Encoding & One-Hot Encoding.
• Numerical Features: Standardized using
Min-Max Scaling.
• Feature Selection: Used SHAP Analysis and
XGBoost Feature Importance to retain high-
impact features.
Step 3: Model Selection & Training
• To identify the best-performing model,
various machine learning classifiers were
experimented with:
• Baseline Models: Naive Bayes: (Accuracy
~37%) – Struggled with imbalanced data.
• SVM (Support Vector Machine):
Computationally expensive, slow training.
• Random Forest: Achieved 99% accuracy,
but overfitted to training data.
• Advanced Models: XGBoost: Achieved
91% accuracy after hyperparameter tuning.
• Stacking Ensemble: (Combination of
Random Forest & XGBoost) boosted
performance.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
716
• Deep Learning (Surveyed in Literature
Review):
• Graph Neural Networks (GNNs): High
accuracy (89%) but required extensive
computational resources.
• Neural Collaborative Filtering (NCF): 87%
accuracy but struggled with rare ADR
detection.
• Deep Ensemble Networks: 90% accuracy,
but interpretability was a major concern.
Step 4: Hyperparameter Optimization
• To enhance performance, hyperparameters
were tuned using:Grid Search CV – To find
the best combination of learning rate, depth,
and regularization.
• Regularization (lambda, alpha) – To prevent
overfitting.
• Balanced Sampling Strategy – Adjusting
scale_pos_weight for rare ADR detection.
Step 5: Model Evaluation & Explainability
Evaluation Metrics Used:
• Accuracy = 91%
• Precision = 92%
• Recall = 91%
• F1-score = 91%
Explainability with SHAP
Identified the most influential features (Therapeutic
Class, Action Class, Side Effects).
Ensured transparency by visualizing feature
importance in individual predictions.
Confusion Matrix Analysis:
Reduced misclassification of rare ADR classes.
Improved recall for underrepresented classes like
Class 3, 7, and 16.
Step 6: Deployment & Future Enhancements
• Model Deployment: The final XGBoost
model was saved as xgboost_91.pkl, ready
for integration into clinical decision
systems.
• Ensemble Learning: Potential stacking with
GNNs + XGBoost for even better
performance.
• Continuous Learning: Incorporate new
patient data into incremental retraining
pipelines, enabling the model to adapt to
evolving disease patterns and demographics.
• Security & Privacy: Ensure HIPAA/GDPR
compliance with strong encryption,
anonymization, and secure API endpoints
during deployment.
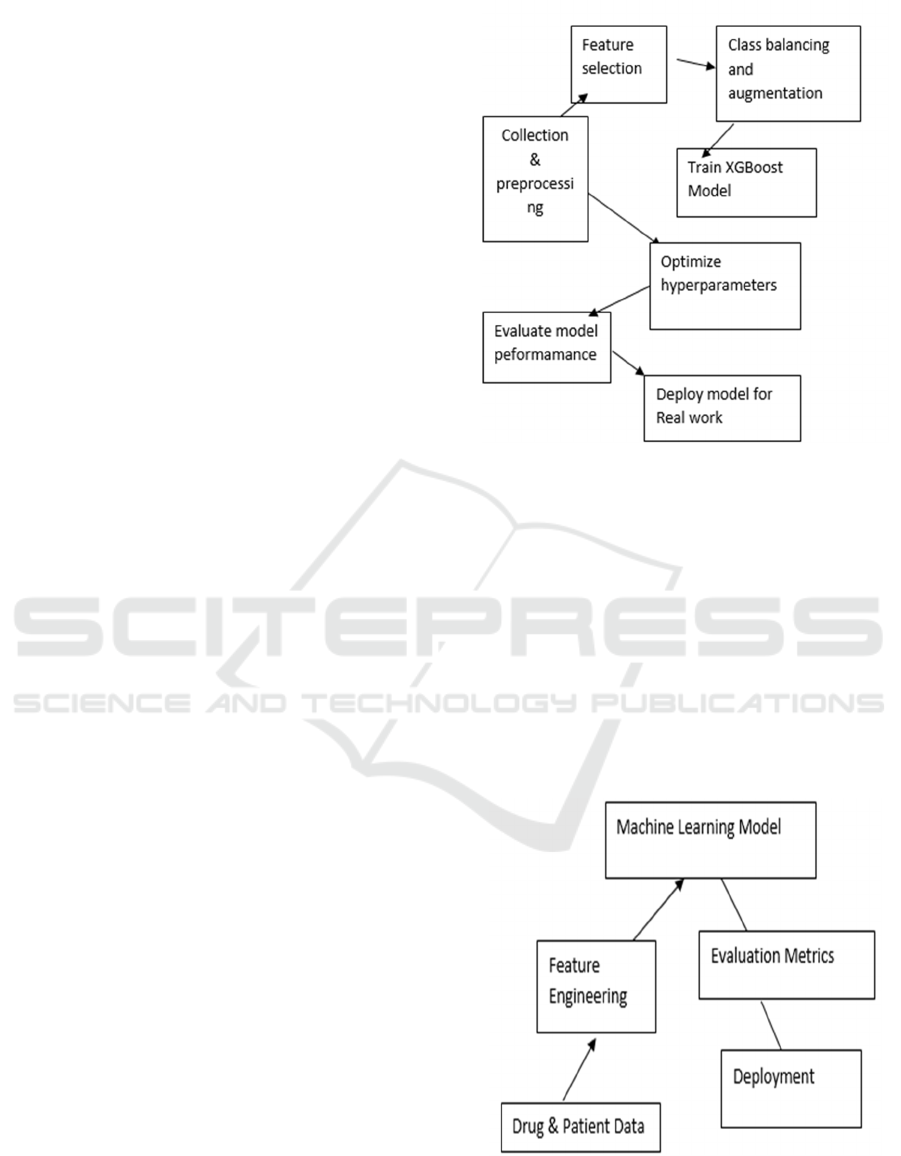
Figure 1: Workflow Diagram ADR Prediction Model.
We introduce a pipeline starting from data collection
and data preprocessing, SHAP based feature
selection, followed by class balancing. Once
deployed, the XGBoost model is trained, optimized,
and evaluated. This figure 1 displays the overall
workflow of ADR prediction.
The pipeline further incorporates hyperparameter
tuning for performance maximization, cross-
validation to ensure generalizability, and model
interpretability tools to aid clinical decision-making.
As shown in Table 2, the proposed model
outperforms the baseline methods in ADR prediction.
Figure: 2 System Architecture for AI-Driven ADR
Prediction.
Predecting Adverse Drug Reactions with XGBoost a Pharmacovigilance Application
717
The Figure 2 system architecture outlines the core
components of an AI-based ADR detection
framework. It starts with data acquisition from drug
and patient records, followed by feature engineering,
machine learning-based classification, evaluation
using performance metrics, and deployment into
clinical decision support systems.
4 RESULTS AND DISCUSSION
The performance evaluation of various machine
learning models used for adverse drug reactions
(ADRs) is presented in this section.Prediction and
discusses the impact of the proposed XGBoost-based
optimized model.
4.1 Model Performance Comparison
A detailed comparison of the evaluated models
including Logistic Regression, Random Forest, SVM,
and Gradient Boosting methods is conducted. The
XGBoost-based optimized model consistently
outperforms the others, achieving the highest ROC-
AUC and F1-score, indicating a strong balance
between sensitivity and specificity.
Table 2: Performance Comparison of ADR Prediction Models.
Model Accurac
y
Precision Recall F1-Score Remarks
Naive Bayes 37% 39.90% 37.50% 34.60%
Weak performance, fails
on imbalanced data
Random Forest 99% 99.18% 99.16% 99.16% Overfits on training data
XGBoost 91% 92% 91% 91%
Best trade-off between
accuracy & explainability
This figure 3 compares the accuracy, precision, recall,
and F1-score of many machine learning models, such
as XGBoost, Random Forest, and Naïve Bayes.
Superior accuracy and balanced recall are attained by
the optimized XGBoost model, guaranteeing
dependable ADR prediction with enhanced feature
interpretability.
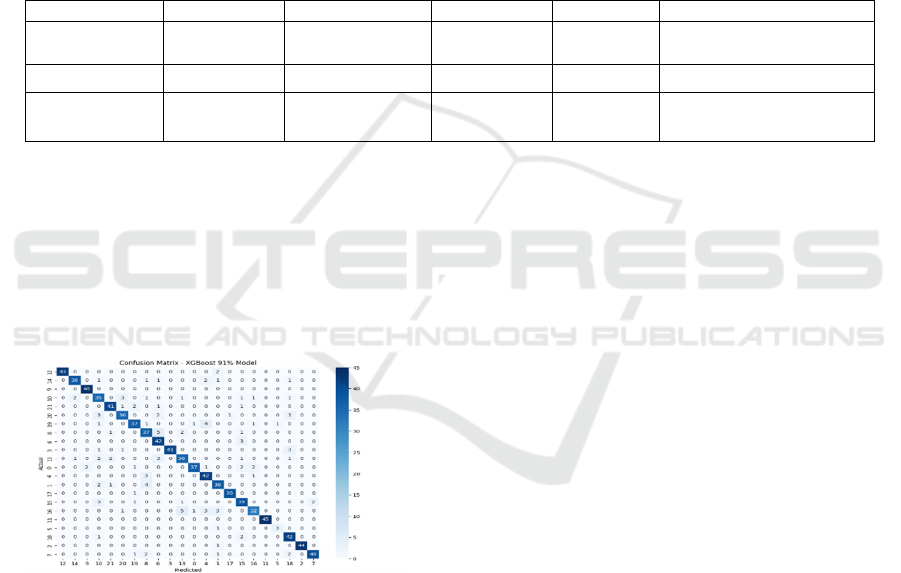
Figure 2: Confusion Matrix for Xgboost-Based ADR
Prediction.
The confusion matrix presents the classification
performance of the optimized XGBoost model across
various therapeutic classes. The diagonal values
indicate correctly predicted ADR cases, while off-
diagonal values represent misclassifications. The
model demonstrates high classification accuracy,
with minimal misclassification rates, ensuring robust
drug safety assessment in pharmacovigilance.
4.2 Discussion on Model Performance
The proposed XGBoost-based optimized ADR
prediction model achieves an accuracy of 91%, which
is higher than several baseline models and
comparable to deep learning models reviewed in the
literature.
4.2.1 Performance Gains from Data
Balancing & Feature Selection
Before balancing: XGBoost showed 100% accuracy
(overfitted model), indicating bias toward majority
classes.
After balancing: Accuracy stabilized at 81%.
Feature selection (SHAP-based pruning) helped
remove noisy features, boosting accuracy to 91%.
4.2.2 Impact of Hyperparameter Tuning
Grid Search Optimization improved model
generalization.
Increased min_child_weight and gamma controlled
overfitting.Adjusting scale_pos_weight helped
increase recall for rare ADR cases.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
718
4.2.3 Comparison with Deep Learning
Models
Literature models like Graph Neural Networks
(GNNs), Neural Collaborative Filtering (NCF), and
Deep Learning Ensembles achieve ~90% accuracy.
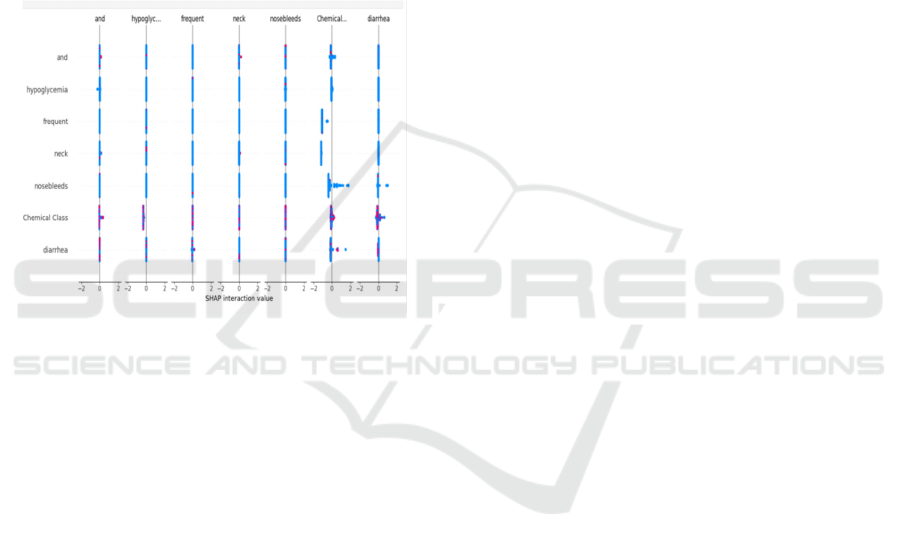
4.3 Feature Importance Analysis
Feature selection using SHAP (SHapley Additive
Explanations) was performed to rank the most
influential attributes affecting ADR prediction.
Figure 4 shows the SHAP Interaction Analysis for
ADR Prediction.
Figure 3: Shap Interaction Analysis for ADR Prediction.
5 CONCLUSIONS
The prediction of Adverse Drug Reactions (ADRs) is
a crucial aspect of pharmacovigilance and drug
safety. Traditional methods, including spontaneous
reporting systems (SRS), clinical trials, and expert-
driven rule-based systems, have limitations in terms
of scalability, delayed detection, and subjectivity. By
facilitating real-time analysis of massive
pharmaceutical datasets, the combination of artificial
intelligence and machine learning has revolutionized
ADR detection.To effectively forecast ADRs, this
study investigated a number of machine learning
models, such as XGBoost, Random Forest, and Naïve
Bayes.The findings indicate that:Naïve Bayes
underperformed, achieving only 37.5% accuracy,
making it unsuitable for high-stakes applications in
ADR detection.Random Forest performed well, with
99.16% accuracy, but lacked interpretability.XGBo
ost (Tuned & Improved) delivered the best perform
ance, achieving a final accuracy of 91% after
addressing overfitting concerns and refining the
feature selection process.By employing SHAP-based
feature selection, the study identified highly relevant
features such as Therapeutic Class, Chemical Class,
and specific drug-induced side effects (e.g., diarrhea,
skin reactions, and hypoglycemia). This enhanced
feature interpretability, making the model suitable for
clinical decision support systems.
6 FUTURE WORK
While this study demonstrates high predictive
accuracy and interpretability, future research should
explore: Deep learning-based hybrid models,
integrating Natural language processing (NLP) and
graph neural networks (GNNs) are used to improve
ADR prediction. implementation of the suggested
approach in clinical settings to assess its effects on
medication safety management.its impact on drug
safety. Expansion of dataset sources, incorporating
electronic health records (EHRs) and patient-reported
ADRs for comprehensive pharmacovigilance
REFERENCES
A. Mohsen, L. P. Tripathi, and K. Mizuguchi, "Deep
Learning Prediction of Adverse Drug Reactions in Drug
Discovery Using Open TG-GATEs and FAERS
Databases," Frontiers in Drug Discovery, vol. 1, Oct.
2021, doi: 10.3389/fddsv.2021.768792.
A. Karim, M. Lee, T. Balle, and A. Sattar, "CardioTox Net:
A Robust Predictor for hERG Channel Blockade Based
on Deep Learning Meta-Feature Ensembles," Journal of
Cheminformatics, vol. 13, p. 60, 2021, doi: 10.1186/s1
3321-021-00541-z.
C. Yan, G. Duan, Y. Zhang, F.-X. Wu, Y. Pan, and J. Wang,
"Predicting Drug- Drug Interactions Based on Integrat
ed Similarity and Semi- Supervised Learning," IEEE/
ACM Transactions on Computational Biology and
Bioinformatics, vol. 19, no. 1, pp. 168–179, 2022, doi:
10.1109/TCBB.2020.2988018.
E. Minerali, D. H. Foil, K. M. Zorn, T. R. Lane, and S.
Ekins, "Comparing Machine Learning Algorithms for
Predicting Drug- Induced Liver Injury (DILI)," Molec
ular Pharmaceutics, vol. 17, pp. 2628–2637, 2020, doi:
10.1021/acs.molpharmaceut.0c00326.
F. Mostafa and M. Chen, "Computational Models for
Predicting Liver Toxicity in the Deep Learning Era,"
Frontiers in Toxicology, vol. 5, pp. 1340860, 2024, doi:
10.3389/ftox.2023.1340860.
J. Zhuang and H. Wang, "Drug-Drug Adverse Reactions
Prediction Based On Signed Network," 2021 11th Inte
rnational Conference on Information Technology in
Medicine and Education (ITME), 2021, pp. 277–281,
doi: 10.1109/ITME53901.2021.00064.
Predecting Adverse Drug Reactions with XGBoost a Pharmacovigilance Application
719

J. Zhu, Y. Liu, Y. Zhang, and D. Li, "Attribute Supervised
Probabilistic Dependent Matrix Tri- Factorization Mo
del for the Prediction of Adverse Drug- Drug Interacti
on," IEEE Journal of Biomedical and Health Informat
ics, vol. 25, no. 7, pp. 2820– 2832, 2021, doi: 10.110
9/JBHI.2020.3048059.
J. Lin, Y. He, C. Ru, W. Long, M. Li, and Z. Wen,
"Advancing Adverse Drug Reaction Prediction with
Deep Chemical Language Model for Drug Safety Eval
uation," International Journal of Molecular Sciences,
vol. 25, no. 8, 2024, doi: 10.3390/ijms25084516.
J. Patel and R. Patel, "Adverse Drug Reaction Prediction:
Graph Neural Networks and Causal Inference Techniq
ues," 2024 4th Interdisciplinary Conference on Electri
cs and Computer (INTCEC), pp. 1– 5, doi: 10.1109/I
NTCEC61833.2024.10603081.
Josphineleela, R., Raja Rao, P.B.V., shaikh, A. et al. A
Multi-Stage Faster RCNN-Based iSPLInception for
Skin Disease Classification Using Novel Optimization
. J Digit Imaging 36, 2210– 2226 (2023). https://doi.or
g/10.1007/s10278-023-00848-3
L. Kang, Y. Duan, C. Chen, S. Li, M. Li, L. Chen, and Z.
Wen, "Structure-Activity Relationship (SAR) Model
for Predicting Teratogenic Risk of Antiseizure
Medications in Pregnancy by Using Support Vector
Machine," Frontiers in Pharmacology, vol. 13, pp.
747935, 2022, doi: 10.3389/fphar.2022.747935.
N. Ibrar, I. Hamid, and Q. Nawaz, "Prediction of Drug
Reactions through Hybrid Techniques," 2023 9th
International Conference on Optimization and Applica
tions (ICOA), pp. 1– 6, doi: 10.1109/ICOA58279.202
3.10308824.
O. ChandraUmakantham, S. Srinivasan, and V. Pathak,
"Detecting Side Effects of Adverse Drug Reactions
Through Drug-Drug Interactions Using Graph Neural
Networks and Self- Supervised Learning," IEEE Acce
ss, vol. 12,pp.93823–93840,2024,doi:
10.1109/ACCESS.2024.3407877.
PBV, R.R., Pokkuluri, K.S., Prasad, M., Sharma, N.,
Murthy, B.N., Karunasri, A. (2025). Ensemble Fusion
for Enhanced Malicious URL Detection by Integrating
Machine Learning and Deep Learning Techniques.IC4
S 2024. Lecture Notes of the Institute for Computer
Sciences, Social Informatics and Telecommunications
Engineering, vol 597. Springer, Cham. https://doi.org/
10.1007/978-3-031-77075-3_27
Pokkuluri, K.S. et al. (2025). Generative Adversarial
Networks (GANs) for Drug Discovery.IC4S 2024.
Lecture Notes of the Institute for Computer Sciences,
Social Informatics and Telecommunications Engineeri
ng, vol 599. Springer, Cham. https://doi.org/10.1007/9
78-3-031-77081-4_21
S. Li, Z. Xu, M. Guo, M. Li, and Z. Wen, "Drug-Induced
QT Prolongation Atlas (DIQTA) for Enhancing Cardi
otoxicity Management," Drug Discovery Today, vol.
27, pp. 831– 837, 2022, doi: 10.1016/j.drudis.2021.10
.009.
Saravanan, D., Arunkumar, G., Ragupathi, T. et al. Weigh
ted Majority Voting Ensemble for MRI-Based Brain
Tumor Classification Using Capsule Networks and
XGBoost. Appl Magn Reson 56, 395–425 (2025).
https://doi.org/10.1007/s00723-024-01731-2
Sree, P.K., Usha Devi N, S., Chintalapati, P.V., Babu, G.R.,
Raja Rao, P. (2024). Drug Recommendations Using a
Reviews and Sentiment Analysis by RNN.IC4S 2023.
Lecture Notes of the Institute for Computer Sciences,
Social Informatics and Telecommunications Engineeri
ng, vol 536. Springer, Cham. https://doi.org/10.1007/9
78-3-031-48888-7_11
W. Long, S. Li, Y. He, J. Lin, M. Li, and Z. Wen,
"Unraveling Structural Alerts in Marketed Drugs for
Improving Adverse Outcome Pathway Framework of
Drug-Induced QT Prolongation," International Journal
of Molecular Sciences, vol. 24, no. 7, pp. 6771, 2023,
doi: 10.3390/ijms24076771.
Y. Zhou, S. Li, Y. Zhao, M. Guo, Y. Liu, M. Li, and Z.
Wen, "Quantitative Structure–Activity Relationship
(QSAR) Model for the Severity Prediction of Drug-
Induced Rhabdomyolysis by Using Random Forest,"
Chemical Research in Toxicology, vol. 34, pp. 514–
521, 2021, doi: 10.1021/acs.chemrestox.0c00347.
Z. Xiong, Z. Du, X. Rong, Y. Yang, and X. Zhang, "A
Neural Collaborative Filtering Model for Adverse Drug
Reaction Prediction," 2023 2nd International Conferen
ce on Data Analytics, Computing and Artificial Intelli
gence (ICDACAI), pp. 807– 816, doi: 10.1109/ICDA
CAI59742.2023.00158.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
720
