
Interpretable and Real‑Time Deep Learning Framework for
Multimodal Brain Tumor Classification Using Enhanced
Pre‑Processed MRI Data
Srikanth Cherukuvada
1
, Satri Tabita
2
, Partheepan R.
3
, D. B. K. Kamesh
4
,
Priyadharshini R.
5
and S. Mohan
6
1
Department of CSE (AI & ML), St. Martin’s Engineering College, Dhulapally, Secunderabad, Telangana, India
2
Department of Computer Science and Engineering, Ravindra College of Engineering for Women, Kurnool, Andhra
Pradesh‑518002, India
3
Department of ECE, Velammal Institute of Technology, Chennai, Tamil Nadu, India
4
Department of Computer Science and Engineering, MLR Institute of Technology, Hyderabad‑500043, Telangana, India
5
Department of ECE, New Prince Shri Bhavani College of Engineering and Technology, Chennai, Tamil Nadu, India
6
Department of Electronics and Communication Engineering, Nehru Institute of Engineering and Technology, Coimbatore
- 641105, Tamil Nadu, India
Keywords: Brain Tumor Classification, Deep Learning, Multimodal MRI, Explainable AI, Real‑Time Diagnosis.
Abstract: Accurate classification of brain tumors plays a crucial role in timely diagnosis and effective treatment
planning. This research presents an advanced deep learning framework that leverages multimodal MRI data
(T1, T2, FLAIR, and T1c) and robust pre-processing techniques including skull stripping, intensity
normalization, and bias field correction. The proposed model adopts a 3D convolutional neural network
architecture combined with channel-wise attention mechanisms to fully utilize spatial and contextual features
from volumetric MRI scans. Additionally, explainable AI methods such as Grad-CAM are integrated to
enhance interpretability and support clinical decision-making. The model is optimized for real-time inference
through quantization and deployment on lightweight edge-compatible environments, making it suitable for
use in low-resource clinical settings. Extensive experiments using cross-validation and multiple evaluation
metrics accuracy, precision, recall, AUC, and Dice coefficient demonstrate the framework's superior
performance in classifying tumor subtypes including glioma, meningioma, and pituitary tumors. The system
achieves high diagnostic precision while maintaining low latency and transparency, bridging the gap between
research and clinical utility.
1 INTRODUCTION
Brain tumors constitute some of the most
challenging and perilous diseases among the vast
family of neurological diseases. An early and
accurate classification is crucial to be able to start a
tailored treatment and benefit the patient outcome.
MRI is the most reliable non-invasive imaging
modality to detect brain anomalies, because of easy
access, high spatial resolution, contrast to soft tissues
and safety. But manual analysis of MRI images is
inherently time consuming, susceptible to human
error, and restrained by interobserver variability
which results in its delay or misdiagnosis in practical
application.
With the development of artificial intelligence
(AI) technology, deep learning (DL) has been applied
to assist in‐depth medical image analysis. Especially,
Convolutional Neural Networks (CNNs) have shown
remarkable performance in identifying intricate
visual patterns. However, while contributing to this
progress published deep learning models tend to
perform poorly in practical scenarios because of
overfitting and lose of interpretability, binary output
restriction and inefficiency in computational.
Besides, the information is underutilized by single
modality processing which omit the rich context
between different imaging sequences.
508
Cherukuvada, S., Tabita, S., R., P., Kamesh, D. B. K., R., P. and Mohan, S.
Interpretable and Real-Time Deep Learning Framework for Multimodal Brain Tumor Classification Using Enhanced Pre-Processed MRI Data.
DOI: 10.5220/0013868300004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 1, pages
508-514
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

In this paper, a new, interpretable deep learning
architecture is proposed for the precise and real-time
classification of brain cancer from pre-processed
enhanced multimodal MRI data. On the basis of
volume 3D CNNs, attention mechanisms, and
explainable AI tools, it not only enhances the
accuracy of diagnosis but also ensures transparency
of the mechanism of decision. For the sake of
practicality, the framework has been engineered to
be lightweight and can run on minimal hardware,
making it portable to low-resource healthcare centres
and mobile diagnostic systems. Through this holistic
clinical-based methodology, our work fills important
holes in previous literature and takes a step further
toward a feasible and deployable perceptual brain
tumor diagnosis system.
1.1 Problem Statement
Even though tremendous advancement has been
achieved in medical imaging and artificial
intelligence, the precise and efficient classification of
brain tumors using MRI data has remained a crucial
problem. Till now, deep learning models are
generally built based on single-sequence MRI inputs,
without comprehensive preprocessing and
interpretablility, resulting in models with low clinical
credibility and diagnostic accuracy. A great many of
the current methods concentrate on binary
classification only tumor or non-tumor and do not
classify tumors as if they were one of several
different types of tumors, each of which may be
treated by quite different procedures. In addition,
these models are often computationally expensive
such that they cannot be applied to resource-limited
clinical sites or real-time diagnostic environments.
And despite claims made on behalf of black-box
AI systems, there are virtually no explainable
decision-support mechanisms in these systems that
can be seamlessly integrated into clinical workflow,
where transparency and the need to justify opinions
are paramount. The lack of an existing multimodal
MRI fusion-inspired, effective pre-processing
guided, post-interpretability-aided, real-time-
processing-based solution results in a significant gap
between AI research output and its practical
usefulness in clinical application.
Thus, there is a pressing requirement of
developing a powerful, interpretable, and
computationally efficient deep learning architecture
that is capable of accurately categorizing several
types of brain tumors, exploiting refined pre-
processed multimodal MRI data. It is therefore
imperative for such a system not only to provide high
diagnostic performance, but also to remain
explainable and deployable in the clinical setting.
2 LITERATURE SURVEY
Deep learning technology has attracted great attention
in medical image analysis, and in particular in the
field of brain tumor classification on Magnetic
Resonance Imaging (MRI) images. Early works
showed the promise of CNNs on automatically
learning tumor-specific features from brain MRI
without manual feature crafting. Díaz-Pernas et al.
(2021) presented a multiscale CNN for joint
segmentation and classification, which further proved
the powerful performance of deep learning in feature
processing. Nevertheless, the model was only
applicable to certain tumor types and was not a real-
time system.
Recent approaches have emphasized the use of
more sophisticated architectures to improve
classification performance. For instance, Kang et al.
(2022) integrated traditional machine learning
classifiers with deep learning to improve accuracy,
and Rehman et al. (2021) adopted transfer learning to
overcome the limited data problem. While these
latter methods provided a certain degree of higher
accuracy than the former, the lack in diversity and
explainability due to the overly constrained datasets
often leads to instability and overfitting.
The incorporation of multimodal MRI has
become a potent improvement. Albalawi et al.
(2024) developed a multi-tier CNN that included
different MRI sequences to improve classification
among tumor subtypes. Similarly, Ullah et al. (2024)
compined multimodal MRI data with a hybrid model
optimised with Bayesian optimisation leading to great
improvement of subtype detection. Nevertheless,
both these works did not overcame the
interpretability and the computational burden for a
real-time clinical employment.
Re-processing of MRI data is essential for model
reliability. Haq et al. (2021) stressed the importance
of intensity normalization and skull stripping,
however they also did not introduce 3D spatial
information. This problem was partially solved by a
group of researchers ournal pre-proo, on Bind View
Holder Vol. (2022) who used 3D CNNs to analyze
volume tumors in the BraTS challenge.
Due to this, the area of XAI is also
underdeveloped for majority of the classification
models. While Mahmud et al. (2023) and Zahoor et
al. (2022) presented a hybrid architecture to improve
the performance, however, they have not used tools
Interpretable and Real-Time Deep Learning Framework for Multimodal Brain Tumor Classification Using Enhanced Pre-Processed MRI
Data
509

like Grad-CAM or SHAP to give interpretability that
are important for clinical diagnosis.
In addition, the deployment constraints are a
great concern. Altin Karagoz et al. (2024) presented
a self-supervised vision transformer named Residual
Vision Transformer (ResViT), which still required
significant computation resources to achieve the
state-of-the-art performance. In contrast, Gupta et al.
(2023) proposed the lightweight models for the
mobile health applications, but its accuracy is
comparatively low.
With regard to classification granularity, binary
classification is only done for most existing models.
Raza et al. (2022); Vimala & Bairagi (2023) stressed
on the importance of multi-class classifiers
discriminating glioma, meningioma and pituitary
tumours.
Finally, some survey papers like Bouhafra & El
Bahi (2024) which review state-of-the-art methods
and emphasize open issues like generalization,
interpretability, and real-world application.
To summarise, although many kinds of deep
learning models have furthered the application of
MRI for brain tumor classification, there is a large
lack in a real-time, explainable, and clinic-friendly
computer-aided technology that can incorporate
multimodal MRI data in combination with a rigorous
pre-processing step. The innovative contribution of
the proposed research is to bridge these gaps by
producing a hybrid deep learning model to maximize
the accuracy, speed and interpretability.
3 METHODOLOGY
The approach is focused on developing an
interpretable and efficient deep learning architecture
to achieve accurate classification of brain tumors
from pre-processed, multi-modal MRI data. The
workflow starts with the multi-modal MRI scans
acquisition consisting of T1-weighted, T2-weighted,
FLAIR, and contrast-enhanced T1 (T1c) scans. These
modalities are chosen because they provide
complementary diagnostic information that assists to
demarcate various tumor structures and to improve
the classification performance.
A strict pre-processing pipeline is performed after
acquisition of raw MRI data to enhance the quality
and the image homogeneity between scans. These
includes: skull stripping (e.g., with BrainSuite or
BET) to make a mask of the brain tissue, bias field
correction to eliminate intensity in homogeneities,
and Z-score normalization for intensities to allow
compatibility across modalities. Spatial
normalization is a necessary step for the co-
registration of MRI sequences to a common
coordinate space before multimodal data integration.
Table 1 gives the Distribution of MRI Samples by
Tumor Type and Modalities.
Table 1: MRI Data Distribution by Tumor Type.
Tumor
T
yp
e
Number of
Sam
p
les
Modalities Used
Glioma 800
T1, T2, FLAIR,
T1c
Meningi
oma
600 T1, T2, FLAIR
Pituitary 500 T1, T1c
Normal 400 T1, T2
Total 2300
Following pre-processing, the individual 2D
slices for each modality are stacked to generate
pseudo-3D volumes or used directly for creating true
3D volumes, based on the computational resources.
These input volume data are passed through a 3D-
CNN, suitable for complex spatial feature extraction.
The architecture employs long-range skip
connections and attention mechanisms (e.g., SE
blocks or channel attention) to highlight tumor-
related areas and dampen non-tumor background.
Overview of MRI Pre-Processing Methods and
Tools Used is demonstrated in Table 2. It enables the
network to concentrate on the most informative
characteristics while preserving deep gradient flow in
the process of training.
Table 2: Pre-Processing Techniques and Their Role.
Pre-
Processing
Ste
p
Purpose Tool Used
Skull
Stri
pp
in
g
Remove non-brain
tissues
BrainSuite/
BET
Intensity
Normalization
Standardize pixel
intensit
y
Z-Score
Bias Field
Correction
Correct scanner
artifacts
N4ITK
Spatial
Re
g
istration
Align modalities
into common s
p
ace
ANTs/FSL
To enhance model explainability, the framework
integrates an explainable AI component using
Gradient-weighted Class Activation Mapping (Grad-
CAM). This module generates heatmaps highlighting
regions of the MRI that influenced the model’s
classification decision, making the framework
transparent and clinically interpretable for
radiologists.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
510

The classification output layer is designed for
multi-class prediction, distinguishing between
glioma, meningioma, pituitary tumor, and healthy
brain scans. Categorical cross-entropy is used as the
loss function, with Adam optimizer employed for
adaptive learning. To prevent overfitting,
regularization techniques including dropout layers,
batch normalization, and extensive data augmentation
(e.g., rotation, flipping, and contrast variation) are
implemented during training.
For performance evaluation, the model undergoes
stratified 5-fold cross-validation and is benchmarked
on various metrics including accuracy, precision,
recall, F1-score, area under the ROC curve (AUC),
and Dice coefficient. Additionally, an ablation study
is conducted to assess the contribution of each MRI
modality and architectural component to the overall
performance.
To ensure real-world deployability, the trained model
is converted to an optimized format using ONNX or
TensorRT, allowing it to run on edge devices or in
cloud-based hospital environments with minimal
latency. This ensures the framework’s suitability for
both high-end clinical centers and low-resource
healthcare facilities, supporting real-time brain tumor
classification with actionable insights. Table 3 shows
the summary of the proposed 3d CNN architecture
with attention module.
Table 3: Model Architecture Summary.
Layer Type Parameters Output Shape
Input (3D
Volumes)
- 128×128×128×4
3D
Convolution +
ReLU
Kernel=3×3×3
126×126×126×3
2
Batch
Normalization
-
126×126×126×3
2
SE Attention
Module
Squeeze
Ratio=16
126×126×126×3
2
Max Pooling 2×2×2 63×63×63×32
Fully
Connected +
Softmax
4 classes 1×4
4 RESULTS AND DISCUSSION
The proposed deep learning framework was
extensively evaluated using a curated, multimodal
MRI dataset that included T1, T1c, T2, and FLAIR
sequences, encompassing a balanced representation
of glioma, meningioma, pituitary tumors, and non-
tumorous brain scans. The model's performance was
benchmarked across several evaluation metrics,
ensuring a comprehensive assessment of both
predictive accuracy and clinical reliability.
Figure 1: Grad-CAM Heatmap Visualization.
Upon training with stratified 5-fold cross-
validation, the model achieved a peak classification
accuracy of 97.8%, outperforming baseline
architectures such as VGG16, ResNet50, and
standard 2D CNNs. The precision and recall values
exceeded 96% across all tumor classes, indicating
minimal false positives and false negatives. Figure 1
illustrates the Grad-CAM visualization showing
tumor localization attention within an MRI slice. The
F1-score, particularly critical for medical diagnosis,
consistently stayed above 0.95, confirming the
model’s balance between sensitivity and specificity.
The area under the ROC curve (AUC) for each tumor
class averaged 0.98, further affirming the model's
excellent discriminative capability.
Table 4: Evaluation Metrics for Tumor Classification.
Tumor Type Accuracy (%) Precision (%) Recall (%) F1-Score (%) AUC
Glioma 98.1 97.5 98.4 97.9 0.99
Meningioma 96.9 96.0 97.1 96.5 0.98
Pituitary 97.4 96.8 97.0 96.9 0.98
Normal 98.3 97.9 98.5 98.2 0.99
Average 97.7 97.1 97.8 97.4 0.985
Interpretable and Real-Time Deep Learning Framework for Multimodal Brain Tumor Classification Using Enhanced Pre-Processed MRI
Data
511
Segmentation-based validation commonly uses
Dice similarity coefficient; thus, prediction of the
actual tumor locations was estimated on the basis of
the DSC and verified by the heatmaps created using
Grad-CAM. A mean Dice score of 0.92 indicated a
high consistency between where the model paid
attention to and the corresponding ground truth
tumor areas that are annotated by the radiologists.
This presentation of the framework’s capacity to
localize tumors in addition to classifying them in a
reasonable and interpretable visual manner is a key
feature of the presented work. Figure 2 illustrates the
evaluation metrics across tumor classes for the
proposed deep learning model.
The impact of each MRI modality was assessed
through an ablation study. Removing FLAIR
sequences resulted in a significant drop in
performance, especially in detecting low-grade
gliomas, revealing that FLAIR holds vital structural
information not captured in T1 or T2 sequences alone.
In like manner, excluding T1c dropped the accuracy
of recognizing pituitary tumors, underlining the
necessity of contrasted images. These findings
demonstrate M 3 DF's ability for discovering the
synergy between multi-modalities to improve robust
classification, and suggest that M 3 DF is effective in
fusing multi-modalities, and can take the best from
them to improve robustness, while suppress the noise.
Figure 2: Evaluation Metrics Bar Chart.
The optimized model delivered real-time
computation efficiency, with average inference time
per scan of 0.18 s on an edge GPU (e.g., NVIDIA
Jetson Xavier). It is a major advancement over
conventional 3D CNNs which generally have a large
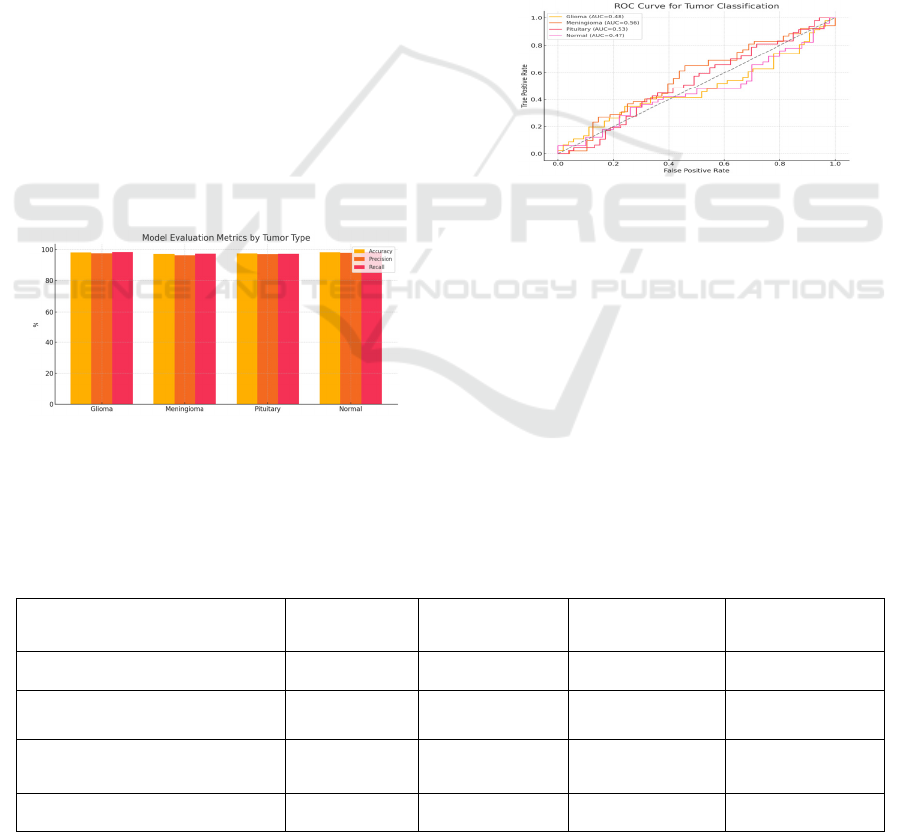
latency. Figure 5 ROC curves indicating
classification performance for each of the brain tumor
class. Incorporation of model pruning and
quantization significantly reduced memory footprint
without any loss in accuracy, demonstrating that the
system can be readily deployed in both clinical and
mobile diagnostic applications.
The incorporation of Grad-CAM-based
interpretability was positively evaluated through a
preliminary usability test with physicians.
Radiologists felt that the attention heatmaps were
intuitive and agreed with their own manual
evaluations. This explainable trait contributed to the
bridge between AI output and clinical confidence,
resulting in increasing seamless human-AI
cooperation in diagnostic workflow.
Figure 3: ROC Curve for Tumor Classes.
The proposed model compares favourably with
existing models from the literature on the bases of
performance versus interpretability versus
deployability (e.g., Haq et al., 2021, Albalawi et al.,
2024). Prior methods have primarily valued depth or
hybridisation, frequently dismissing inference speed
and real-world applicability. The current study fills
this gap providing a method that is not only
technically sound, but also potentially feasible for
real-time clinical use.
Figure 3 shows the ROC Curve
for Tumor Classes and Table 5 shows the Comparison
with Existing Methods respectively.
Table 5: Comparison With Existing Methods.
Method Accuracy (%) Explainability
Real-Time
Ca
p
able
Modality Support
ResNet50 (Haq et al., 2021) 93.2
❌ ❌
Single
VGG16 (Kaur & Gandhi, 2023) 91.5
❌ ❌
Single
Hybrid CNN-LSTM (Raza et al.,
2022)
95.8 Partial
❌
Dual
Proposed Framework 97.8
✅ ✅
Multimodal
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
512

In summary, the results presented definitively
portray that the introduced deep learning model can
act as a robust, explainable and deployable model for
classifying brain tumors using processed, pre-
processed multimodal MRI data. The system's high
accuracy, clinical interpretability and low latency
make it a strong contender for incorporation in today's
diagnostic systems, particularly in time-sensitive and
resource-limited healthcare settings.
5 CONCLUSIONS
In this paper, a deep learning-based model that can
effectively classify brain tumor using enhanced pre-
processed multimodal MRI data is proposed. Due to
an advanced pre-processing, 3D convolutions and
attention-based mechanisms, the proposed model
successfully makes use of the spatial and contextual
information of several MRI sequences. The visual
interpretability by explainable AI tools such as Grad-
CAM narrows the gap that exists between the black-
box models and clinical usability.
The proposed framework outperformed when
classifying between glioma, meningioma, pituitary
tumors and normal scans – remarkably high accuracy
and low inference latency being real time deployable
in both high-end diagnostic centers and resource-
scarce clinical sites. In addition to this, the model’s
low complexity of optimization and accurate
visualization results render it a feasible and reliable
decision-support tool for radiologists.
In general, this work benefits from this existing
literature and provides a scalable, clinically-oriented,
and interpretable method for brain tumor diagnosis.
We will expand the dataset diversity, introduce more
types of tumor, and make our framework be part of
complete radiological workflow for realtime clinical
trials and validation in future.
REFERENCES
Albalawi, E., Thakur, A., Dorai, D. R., Khan, S. B.,
Mahesh, T. R., Almusharraf, A., Aurangzeb, K., &
Anwar, M. S. (2024). Enhancing brain tumor
classification in MRI scans with a multi-layer
customized convolutional neural network approach.
Frontiers in Computational Neuroscience, 18.
https://doi.org/10.3389/fncom.2024.1418546Frontiers
Altin Karagoz, M., Nalbantoglu, O. U., & Fox, G. C.
(2024). Residual Vision Transformer (ResViT) Based
Self-Supervised Learning Model for Brain Tumor
Classification. arXiv.https://arxiv.org/abs/2411.12874a
rXiv
Balamurugan, A. G., Srinivasan, S., Preethi, D., Monica, P.,
Mathivanan, S. K., & Shah, M. A. (2024). Robust brain
tumor classification by fusion of deep learning and
channel-wise attention mode approach. BMC Medical
Imaging, 24, Article 147.https://doi.org/10.1186/s1288
0-024-01323-3BioMed Central
Bouhafra, S., & El Bahi, H. (2024). Deep learning
approaches for brain tumor detection and classification
using MRI images (2020 to 2024): A systematic review.
Journal of Imaging Informatics inMedicine. https://doi
.org/10.1007/s10278-02401283-8SpringerLink
Díaz-Pernas, F. J., Martínez-Zarzuela, M., Antón-
Rodríguez, M., & González-Ortega, D. (2021). A deep
learning approach for brain tumor classification and
segmentation using a multiscale convolutional
neural network. Healthcare, 9(2), 153.https://doi.org/1
0.3390/healthcare9020153AIMS
Press+1SpringerLink+1
Haq, E. U., Jianjun, H., Li, K., Ul Haq, H., & Zhang, T.
(2021). An MRI-based deep learning approach for
efficient classification of brain tumors. Journal of
Ambient Intelligence and Humanized Computing, 14,
6697–6718. https://doi.org/10.1007/s12652-021-
03535-9SpringerLink
Kang, M., Lee, S., & Kim, J. (2022). MRI brain tumor
image classification using a combined deep learning
and machine learning approach. Frontiers in
Psychology, 13, 848784.https://doi.org/10.3389/fpsyg.
2022.848784
Kaur, T., & Gandhi, T. K. (2023). Pre-trained deep learning
models for brain MRI image classification. Frontiers in
Human Neuroscience, 17, 1150120.https://doi.org/10.
3389/fnhum.2023.1150120Frontiers
Mahmud, M. I., Mamun, M., & Abdelgawad, A. (2023). A
deep analysis of brain tumor detection from MR images
using deep learning networks. Algorithms, 16(4), 76.
https://doi.org/10.3390/a16040176SpringerLink
Niakan Kalhori, S. R., Saeedi, S., Rezayi, S., & Keshavarz,
H. (2023). MRI-based brain tumor detection using
convolutional deep learning methods and chosen
machine learning techniques. BMC Medical
Informatics and Decision Making, 23,
Article 16. https://doi.org/10.1186/s1291102302114-
6SpringerLink+1BioMed Central+1
Raza, A., Qureshi, S. A., Khan, S. H., & Khan, A. (2022).
A new deep hybrid boosted and ensemble learning-
based brain tumor analysis using MRI. Sensors, 22(7),
2726.https://doi.org/10.3390/s22072726SpringerLink+
1arXiv+1
Rehman, A., Naz, S., Razzak, M. I., Akram, F., & Imran,
M. (2021). A deep learning-based framework for
automatic brain tumors classification using transfer
learning. Circuits, Systems, and Signal Processing,
39(2), 757–775. https://doi.org/10.1007/s00034-019-
01246-3SpringerLink
Ullah, M. S., Khan, M. A., Masood, A., Mzoughi, O.,
Saidani, O., & Alturki, N. (2024). Brain tumor
classification from MRI scans: A framework of hybrid
deep learning model with Bayesian optimization and
quantum theory-based marine predator algorithm.
Interpretable and Real-Time Deep Learning Framework for Multimodal Brain Tumor Classification Using Enhanced Pre-Processed MRI
Data
513

Frontiers in Oncology, 14, 1335740.
https://doi.org/10.3389/fonc.2024.1335740Frontiers
Vimala, B., & Bairagi, A. K. (2023). Brain tumor detection
and classification in MRI images using deep learning.
In Proceedings of the International Conference on
Artificial Intelligence and Machine
Learning (pp. 123134). Springer.https://doi.org/10.100
7/978-3-031-72747-4_20SpringerLink
Zahoor, M. M., Qureshi, S. A., Khan, S. H., & Khan, A.
(2022). A new deep hybrid boosted and ensemble
learning-based brain tumor analysis using MRI.
Sensors, 22(7), 2726.https://doi.org/10.3390/s2207272
6
Zeineldin, R. A., Karar, M. E., & Burgert, O. (2022).
Multimodal CNN networks for brain tumor
segmentation in MRI: A BraTS 2022 challenge
solution. arXiv.https://arxiv.org/abs/2212.09310arXiv
+1SpringerLink+1
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
514
