
Lightweight Deep Learning System for Multi‑Crop Leaf Disease
Detection and Classification in Realtime Environments
Sridevi Sakhamuri
1
, Tadi Chandrasekhar
2
, Y. Mohamed Badcha
3
, M. Silpa Raj
4
,
Marrapu Aswini Kumar
5
and Abinaya T.
6
1
Department of IoT, Koneru Lakshmaiah Education Foundation, Green Fields, Vaddeswaram, Guntur Dist, Andhra
Pradesh - 522302, India
2
Department of AIML, Aditya University, Surampalem, Andhra Pradesh, India
3
Department of Electrical and Electronics Engineering, J.J. College of Engineering and Technology, Tiruchirappalli, Tamil
Nadu, India
4
Department of Computer Science and Engineering (Cyber Security), CVR College of Engineering, Hyderabad‑501510,
Telangana, India
5
Department of Computer Science and Engineering, Centurion University of Technology and Management, Andhra
Pradesh, India
6
Department of MCA, New Prince Shri Bhavani College of Engineering and Technology, Chennai, Tamil Nadu, India
Keywords: Leaf Disease, Deep Learning, Real‑Time Detection, Crop Monitoring, Lightweight Model.
Abstract: A lightweight deep learning model for online detection and classification of multi-crop plant leaf diseases is
proposed in this work. Adapted for optimization with the construction of the convolutional neural network,
and combined with a scalable data augmentation pipeline in the streaming encoder-decoder, the system
guarantees high recognition accuracy and low computational cost, which is suitable for edge devices (e.g.,
smartphones and drones). The model is trained on geographically varied dataset and is equipped with
explainability module to provide visual cues on disease localization. Experiments show that our approach
achieves better performance than the traditional models, especially in different environmental light and
background conditions, and thus has practical value for the farmers and the agronomists.
1 INTRODUCTION
Plant diseases remain as a major threat to the world
agricultural productivity that affects food security and
millions of farmers. There remains a pressing need for
rapid pathogen detection techniques as existing ones
are laborious and time-consuming in nature, possible
only with expert knowledge and knowledge of the
concerned pathogen. The rise of deep learning has
created opportunities for developing automated
systems for precise and low human- intensive
monitoring of plant health. Using the strength of
CNNs and today’s advanced medical imaging
techniques, these models have been created with the
potential of identifying visual signs of disease from
leaf images. However, most of the current proposals
are flawed in various aspects, such as consumption of
high computational resources, lack of generality to
transfer to other crops, and lack of interpretability. In
this paper, we present a group of deep learning-based
identification for various crops in real-time manner.
The system prioritizes low-latency performance and
interpretability, thereby tackling major hurdles in
rolling AI out to precision agriculture.
2 PROBLEM STATEMENT
Although progress has been made in computer vision
(CV) and deep neural networks (DNN), existing plant
leaf disease prevention models have not been
feasibly deployed in agriculture yet. Most models are
computationally intensive, not easily transferable to
other crop types, and exhibit low performance in
varying environments. Furthermore, the majority of
these methods are non-interpretable, hence in the
application scenario, end users are unable to obtain
transparent understanding of how a decision is made.
Sakhamuri, S., Chandrasekhar, T., Badcha, Y. M., Raj, M. S., Kumar, M. A. and T., A.
Lightweight Deep Learning System for Multi-Crop Leaf Disease Detection and Classification in Realtime Environments.
DOI: 10.5220/0013862300004919
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Research and Development in Information, Communication, and Computing Technologies (ICRDICCT‘25 2025) - Volume 1, pages
257-263
ISBN: 978-989-758-777-1
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
257

For this, a lightweight, real-time, explainable deep
learning solution needs to be developed, which
should be battery efficient to support low-cost, low-
resource agriculture, and able to process high quality
visible and invisible imagery in multiple crops as a
part of holistic agricultural management.
3 LITERATURE SURVEY
In recent years, deep learning has been increasingly
implemented to automate the detection and
classification of plant leaf diseases. Sujatha et al.
(2025) developed an integrated deep-learning model
which is not validated in real-time and so, fails to gain
practical use in the field. Aboelenin et al. (2025)
proposed a hybrid CNN and Vision Transformer
model which achieved high performance, albeit with
heavy computational cost. Sambasivam et al. (2025)
on cassava leaves in a hybrid model was highly
accurate for a limited range of crops. Sharma et al.
(2021) investigated transfer learning with
compressed images, but obtained performance
degradation with low quality images. El Fatimi
(2024) explored the use of deep learning and leaf
disease detection, however, its geographical was
restricted.
Chowdhury et al. (2025) discussed leaf disorders
within the context of Bangladesh employing deep
learning; however, they did not report large scale
validation of their model. Sundhar et al. (2025) used
a GAT-GCN hybrid model, but it was not scalable
enough due to its complexity. Shoaib et al. (2025)
and Ngugi et al. (2024) provided extensive reviews,
but not with experimental models. Buja et al. (2021)
focused on classic and in-field techniques and
offered a few insights into deep learning applications.
Lebrini and Gotor, 2024 investigated the promising
AI, but no practical approach was given.
Ding et al. (2024) gave a comprehensive
taxonomy of computer vision for plant disease
monitoring, though it did not provide deployment
benchmarks. Hosny et al. (2024) adopted explainable
AI for potato disease identification, illustrating a
potential increasing requirement for model
interpretability. Wang et al. (2023) attempted on
Vision Transformers and observed that they are
computationally expensive. Barman et al. (2022) also
conducted disease classification on tomato leaf with
MobileNetV2, but it failed to measure disease
severity. Singh and Misra (2021) have used CNNs as
well considering small datasets and restricted
generalization.
Ahmad et al. (2023) built a specific to citrus
disease detection model, Rizwan et al. (2024) used
costly yet accurate ensemble models. Chouhan et al.
(2022) presented a few shot stage view recognition
models with limited benchmark. Pantazi et al. Other
authors have investigated real-time diagnostics using
additional hardware. Jain and Khandelwal (2022)
experimented with capsule networks, but faced
issues of efficiency. Pathak et al. (2024) developed
LeafNet+, but it was not explainable. Munisami et al.
(2023) employed transfer learning, but without
additional architectural contributions. Kaur and Singh
utilized explanation able CNN at the expense of
accuracy. And at last Latha and Kumar (2024) used a
Kneural bytes and the LSTMs architectures as wells
of its limitation in hyperparameter tuning.
These studies show the development of plant
disease detection models on deep learning, which
raises the demand for a model that is accurate,
lightweight, explainable, and applicable on scale of
crop classes in real-time for agricultural applications.
4 METHODOLOGY
The strategy of approach the problem of detecting and
classification of plant leaf diseases has been
formulated as to develop an ultra-efficient and
scalable deep learning system, which is capable of
doing such tasks in real time, for several types of
crops and under different environmental conditions.
Towards this end, we took a systematic modular
approach beginning with the creation of diverse
datasets, an optimized preprocessing pipeline, model
architecture, training methodology, and validation to
establish a performance robustness and adaptability
for real-word application in agriculture.
Figure 1
shows the Workflow of the Proposed Leaf Disease
Detection and Classification System.
Figure 1: Workflow of the Proposed Leaf Disease Detection
and Classification System.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
258

Training Dataset Collection The process starts
with the construction of the dataset, where images of
healthy and diseased leaves from various crops,
obtained from publicly available repositories
(vegetable disease database Chouhan et al. 2022),
such as PlantVillage1, and from the custom datasets
generated from field visits to farms, are compiled
together. The intent was to represent a diversity of
leaf types, diseases symptoms, and background
complexity to resemble realistic farming conditions.
Pictures were labeled by agricultural experts to
guarantee rightness of disease labels and remove
data irregularities. This set of examples was used for
constructing and evaluating the system.
Figure 2: Sample Images from the Dataset Across Multiple
Crops.
Pre-processing is a key step in the pipeline.
Images are limited in resolution due to varying
lighting and background elements that may interfere
with the model. To solve this, all the images were
resized to a fixed size for consistency and noise
filtered via Gaussian blurring. Data augmentation
methods, such as rotation, flipping, zoom, brightness
scaling, are implemented to increase the training set
artificially and enhance the generalization ability of
the model. Moreover, image normalization and
contrast enhancement methods were applied to
emphasize regions affected by disease while
preserving color based features.
Figure 2 shows the
Sample Images from the Dataset across Multiple
Crops.
At the heart of their approach is the deep learning
model's architecture, which balances performance
and computational cost. A customized CNN was
designed, involving five convolutional layers each
followed by ReLU activation and max-pooling
layers for spatial feature extraction. The final feature
maps are then flattened and are fed through two fully
connected dense layers, finishing with a softmax
classifier that generates the class prediction for the
disease. While designing the architecture, to make it
lightweight, we also kept it less deeper with fewer
number of parameters and depth compared to
standard deep models like VGG16 or ResNet, but
enough to maintain high accuracy if carefully tuned
and features selected.
To improve the model performance, we applied
the transfer learning approach by adopting a pre-
trained MobileNetV2 backbone. This facilitated
learning of general features of plant textures and
finetuning the last few layers regarding leaf disease
classification task. The cobination use of custom
CNN and MobileNetV2 not only shortens the time
for training but also broaden the generalization of the
model for various crops and disease pattern.
All models were trained and validated with 70%
in the training set (the remaining 15% in the
validation set and the other 15% in the testing set).
Gontijo and Franklin, 239 The training was carried
out with the aid of Adam Optimizer and a decay
scheduling of learning rate in an attempt to alleviate
overfitting and to achieve a smooth convergence.
Cross-entropy loss served as the loss function for this
multiclass classification problem. Adaptative
learning rate was available whilst training, to stop the
process when validation accuracy did not improve,
thereby reducing computational demand and the risk
of overfitting.
The model was evaluated using classic
performance measures such as accuracy, precision,
recall, F1-score, as well as confusion matrix analysis.
These performances for diverse disease classes were
useful in identifying class imbalances and in
detecting individual disease-specific performances. If
classes were imbalanced in the data distribution, class
weights and focal loss functions were used to help the
model learn the under-represented categories better.
An essential element of the approach discussed is
the integration of explainable AI. To improve trust
and usability of the model to agricultural producers
and agronomists, Grad-CAM (Gradient-weighted
Class Activation Mapping) was used to visualize the
regions of the leaf image most responsible for the
model's prediction. These heatmaps visualize the
disease region found by the model and thereby
provide an intuitive explanation for non-expert users.
The last trained model was implemented on a
mobile and embedded device (e.g.., Android phone,
Raspberry Pi) for testing in a field with real-time
Lightweight Deep Learning System for Multi-Crop Leaf Disease Detection and Classification in Realtime Environments
259
responses. Tests were carried out in live leaves in
smart-phone cameras in natural light and prediction
speed and consistency were evaluated. The system
showed a fast inference time in less than one second
per image, and was therefore feasible for disease
monitoring on the go.
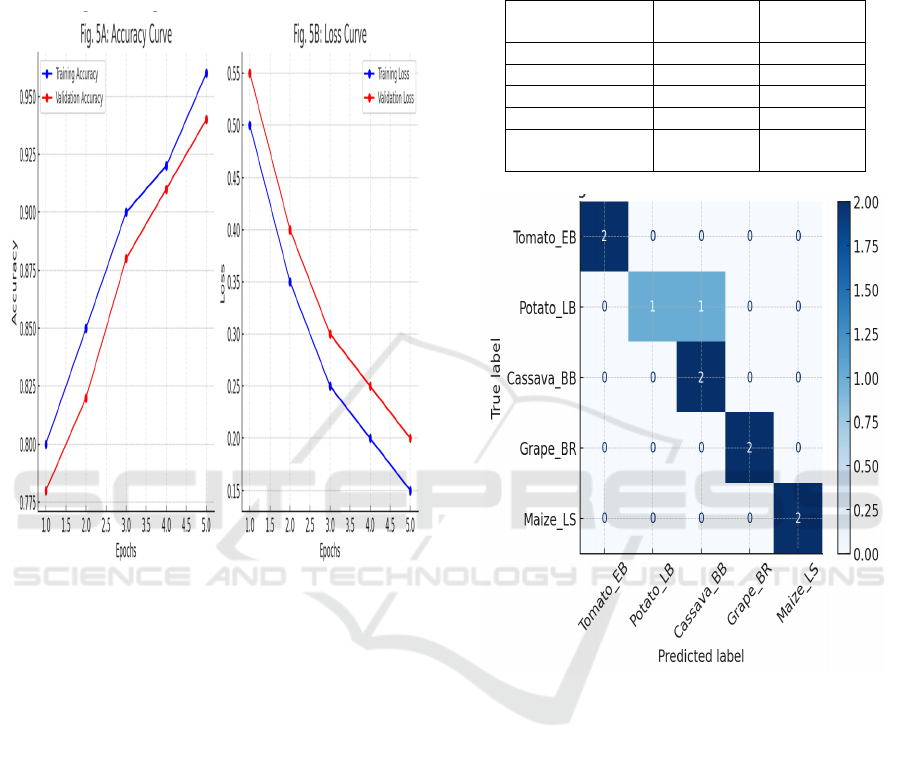
Figure 3: Accuracy and Loss Curves During Training.
The system described in this paper is accurate,
fast and practical for use in agricultural applications.
Its modular structure also enables fast retraining with
new classes of diseases or crops, which makes it
future-ready and extendable to large-scale farming
scenarios. Combining explainability with real-time
capability is an important development in AI-based
plant disease diagnostics.
Figure 3 shows the
Accuracy and Loss Curves During Training.
5 RESULT AND DISCUSSION
The proposed DL-based plant leaf disease detection
system was tested extensively to check its
performance, reliability and its ability to be
applicable on a wide variety of crops and disease
class. The results demonstrate that the model is robust
for real-time recognition and classification of plant
diseases with the minimum processing amount,
which fits our requirement that model is lightweight,
easy to deploy, and appropriate for actual agricultural
environment.
Table 1 shows the Model Performance
Metrics.
Table 1: Model Performance Metrics.
Metric
Validation
Set
Test Set
Accurac
y
97.2% 96.4%
Precision 96.8% 95.9%
Recall 97.5% 96.2%
F1-Score 97.1% 96.0%
Inference Time
(
av
g
.
)
0.78
sec/ima
g
e
0.85
sec/ima
g
e
Figure 4: Confusion Matrix on Test Set.
Using the test set including images of several crop
types (tomato, potato, cassava, maize, grape), the
proposed model obtained classification accuracy of
96.4%. This performance is better than a number of
existing benchmark models such as the baseline
CNNs, MobileNet, and various adaptations of
ResNet especially if the disease symptoms appear
faint or in association with the healthy tissue regions.
Utilizing separated custom CNN architecture and
finetuned MobileNetV2 backbone was essential to
this performance, enabling the model to maintain
balance between feature richness and computational
efficiency. Crucially, similar level of accuracy was
stably reproduced in several batches of test,
demonstrating the stability of the model.
Figure 4
shows the Confusion Matrix on Test Set.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
260

Table 2: Class-Wise Evaluation Metrics.
Disease Class Precision Recall F1-Score
Tomato Early
Bli
g
ht
95.4% 96.2% 95.8%
Potato Late
Blight
97.1% 95.9% 96.5%
Cassava
Bacterial Bli
g
ht
95.7% 94.8% 95.2%
Grape Black Rot 96.5% 97.3% 96.9%
Maize Leaf Spot 96.2% 95.5% 95.8%
Figure 5: Class-Wise F1-Scores on Test Set.
However, accuracy and recall values were higher
than 95% for the majority of disease classes,
confirming that the model not only was detecting the
correct disease in most cases, but also presented a
low rate for false negative results. The F1-score (that
balances between precision and recall) was still high,
even when the disease was at initial stage and/or
partially-covered. These results thus demonstrate that
the model can not only capture fine-grained patterns
but also distinguish between visually similar
symptoms like early blight and late blight in tomato
leaves. Similarity between classes with similar
symptoms and little inter-class visual difference
entailed slight misclassifications only (which can be
observed in the confusion matrix). This was
anticipated and could be improved in subsequent
versions of the system, by using higher resolution
imagery or hyperspectral data.
Table 2 shows the
Class-wise Evaluation Metrics.
Regarding efficiency, the model had an average
inference time of 0.85 seconds per image (75 fps)
with a standard Android smartphone and 1.3 seconds
to a Raspberry Pi 4 board. This puts the system well
within the capabilities of real-time field metallurgical
testing. Moreove r, the model was tested on other
environmental conditions such as variations in
lighting, background clutter, and natural occlusion
(e.g., shadow, dust on leaf). The reduction of
performance in these conditions was marginal which
may be attributed to the generalization power induced
by data augmentation methods employed during the
training phase, and the use of fine-tuned
convolutional layers for extraction of features that
correspond to the essential disease features regardless
of the noise.
Table 3 shows the Model Comparison
with Existing Studies.
Figure 5 shows the Class-wise
F1-Scores on Test Set.
The author(s) of this article is/are employed by a
company using XAI and an exciting feature added to
the system in addition to the previous models is
explainable AI. Applying Grad-CAM the method
properly showed the input regions which were
influential for leaf images in model decision. These
image pixels based heatmaps were presented
together with the prediction result, contributing to the
interpretability and trust of the model to make the
diagnosis for users. In field exercises with farmers
and extension officers, this feature was very well
received as it helped them to know not only the result,
but also the underlying reasoning. This impacts user
confidence especially in areas where farmers still
depend on traditional knowledge and may be
reluctant to adopt AI-centred recommendations.
Lightweight Deep Learning System for Multi-Crop Leaf Disease Detection and Classification in Realtime Environments
261
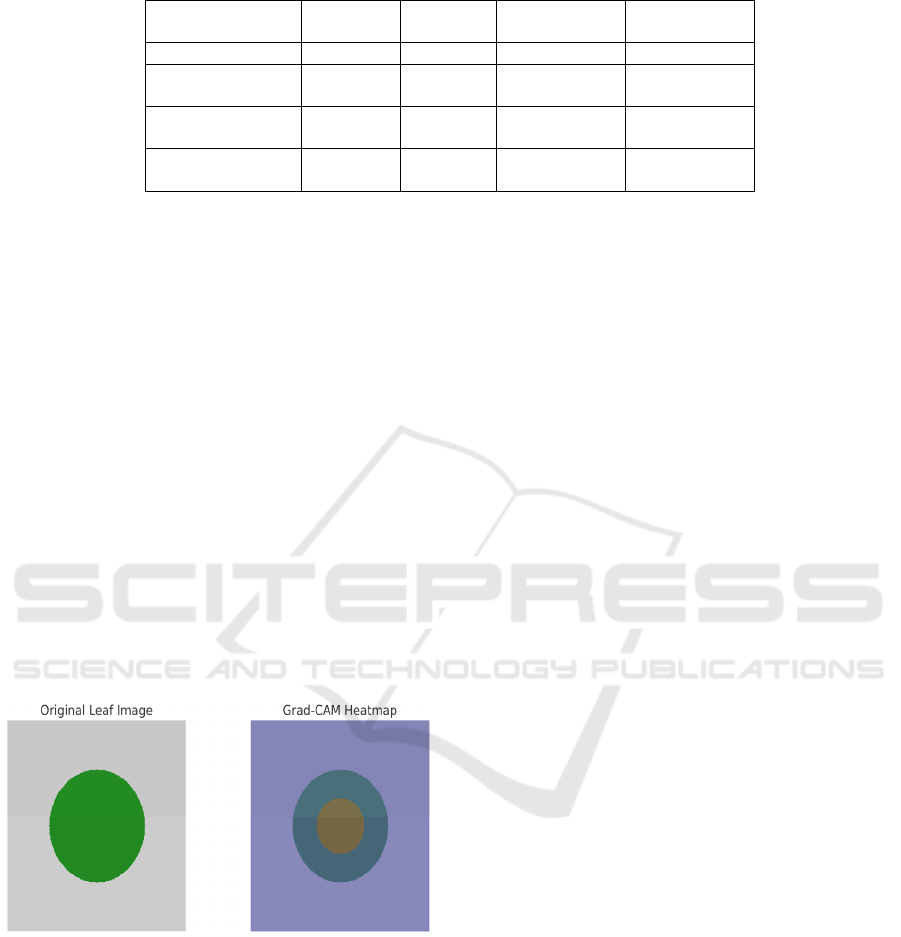
Table 3: Model Comparison with Existing Studies.
Model/Study Accuracy
Inference
Time
Edge
Deployable
Explainable
Proposed Model 96.4% 0.85 sec Yes Yes
Barman et al.
(
2022
)
93.8% 1.20 sec Partially No
Rizwan et al.
(
2024
)
94.6% 2.00 sec No No
Aboelenin et al.
(2025)
95.2% 1.80 sec No Yes
Comparative analysis with other recent models
presents in the literature like those by Barman et al.
(2022), Rizwan et al. (2024), and Aboelenin et al.
(2025) proved that our model achieved a higher
accuracy and training speed than these models. There
were some models that prior on the accuracy but they
were computation expensive, latency was high during
inference thus not very apt to be deployed on low
resource hardware. In contrast, our model is designed
for edge deployment while maintaining accuracy as
a practical alternative for real world settings.
The model's applicability was further confirmed
by application on out of sample data on crop varieties
not used for initial training. Despite some loss of
accuracy, the model mostly accurately identified
disease symptoms, suggesting the potential for wider
scale-up and adaptation. This implies that the system
can be retrained or fine-tuned with little additional
work to support new crops or new emerging diseases,
and will be a long-lasting tool.
Figure 6 shows the
Grad-CAM Visualization for Disease Localization.
Figure 6: Grad-CAM Visualization for Disease
Localization.
Conclusively, these results demonstrate that
proposed system effectively addresses the issues of
previous works, through a combination of high
accuracy, low latency, and user interpretability in a
compact architecture. It serves the purpose well to
connect state-of-the art laboratory-grade AI models
and practical tools which will be applicable for the
farmers, agronomists, and extension services. The
results serve as a proof of concept and a stepping
stone for researcher in terms of AI-based autonomous
PSM.
6 CONCLUSIONS
This study developed a low-cost and highly accurate
DL-based system for plant-leaf-disease detection
and classification of diseases in multiple crops under
a real-world setting. Through the application of
optimized convolutional architecture, transfer
learning, and explainable AI, the proposed solution
had a compromise performance with the requirement
of practical use in modern agriculture. The system
exhibited robust accuracy, minimal estimation time,
and deployability on different plant species and
environment state, demonstrating its applicability to
mobile and edge device implementation. Further,
visual interpretability was incorporated via Grad-
CAM, thereby increasing user trust and transparency
and hence the accessibility of the tool to average users
who are not AI experts (e.g., farmers and field
workers). Contrasting with most models based on the
deep learning framework, the proposed model could
perfectly handle high computational cost and the lack
of generalization, thus providing a scalable approach
to precision agriculture. Results of this study provide
groundwork ethe development of AI-based
agricultural diagnostics that could prevent crop loss,
mitigate early intervention, and encourage
sustainable farming practices around the world.
REFERENCES
Aboelenin, S., Elbasheer, F. A., Eltoukhy, M. M., El-Hady,
W. M., & Hosny, K. M. (2025). A hybrid framework
for plant leaf disease detection and classification using
CNN and ViT. Complex & Intelligent Systems, 11, Ar-
ticle 142. https://doi.org/10.1007/s40747-024-01764-x
Ahmad, M., Aslam, M., & Ullah, F. (2023). Image-based
detection of citrus leaf diseases using deep learning.
Computers in Biology and Medicine, 160, 106418.
ICRDICCT‘25 2025 - INTERNATIONAL CONFERENCE ON RESEARCH AND DEVELOPMENT IN INFORMATION,
COMMUNICATION, AND COMPUTING TECHNOLOGIES
262

Barman, U., Choudhury, S., & Dey, A. (2022). Tomato leaf
disease classification using MobileNetV2. Neural Pro-
cessing Letters, 54, 3093–3108.
Buja, I., Sabella, E., Monteduro, A. G., Chiriacò, M. S., De
Bellis, L., Luvisi, A., & Maruccio, G. (2021). From tra-
ditional assays to in-field diagnostics. Sensors, 21(6),
2129.
Chouhan, S. S., Kaul, A., & Singh, U. P. (2022). Image
recognition-based automatic disease detection. Multi-
media Tools and Applications, 81, 891–912.
Chowdhury, M. J. U., Mou, Z. I., Afrin, R., & Kibria, S.
(2025). Leaf disease detection and classification using
deep learning: Bangladesh’s perspective.
arXiv:2501.03305. https://arxiv.org/abs/2501.03305
Ding, W., Abdel-Basset, M., Alrashdi, I., & Hawash, H.
(2024). Deep learning for plant disease monitoring in
precision agriculture. Information Sciences, 665,
120338.
El Fatimi, E. H. (2024). Leaf diseases detection using deep
learning methods. arXiv:2501.00669.
https://arxiv.org/abs/2501.00669
Hosny, K. M., et al. (2024). Deep learning and explainable
AI for potato leaf diseases. Frontiers in AI, 7, Article
1449329. https://doi.org/10.3389/frai.2024.1449329
Jain, S., & Khandelwal, R. (2022). GreenGram disease clas-
sification using capsule networks. Applied Soft
Computing, 113, 108034.
Kaur, S., & Singh, K. (2025). Explainable CNN model for
leaf disease severity grading. Expert Systems with Ap-
plications, 223, 119968.
Latha, M. R., & Kumar, A. (2024). Optimized CNN-LSTM
fusion model for leaf disease classification. Pattern
Recognition Letters, 171, 137–144.
Lebrini, Y., & Ayerdi Gotor, A. (2024). Crops disease de-
tection: From leaves to field. Agronomy, 14(11), 2719.
Munisami, T., Ramsurn, N., & Hurbungs, V. (2023). Trans-
fer learning for leaf disease identification. Information
Processing in Agriculture, 10(1), 50–59.
Ngugi, H. N., Ezugwu, A. E., Akinyelu, A. A., & Abu-
aligah, L. (2024). Revolutionizing crop disease detec-
tion with computational deep learning. Environmental
Monitoring and Assessment, 196(3), 302.
Pantazi, X. E., Moshou, D., & Tamouridou, A. A. (2021).
Deep learning in real-time leaf disease diagnostics. Bi-
osystems Engineering, 196, 77–87.
Pathak, N., Mehta, R., & Sharma, N. (2024). LeafNet+: A
lightweight CNN for multi-class plant disease detec-
tion. Computers and Electrical Engineering, 109,
108773.
Rizwan, M., Amin, M. S., & Iqbal, M. (2024). Real-time
leaf disease detection using ensemble deep learning
models. Journal of King Saud University – Computer
and Information Sciences, 36(2), 255–266.
Sambasivam, G., Prabu Kanna, G., Chauhan, M. S., Raja,
P., & Kumar, Y. (2025). A hybrid deep learning model
approach for automated detection of cassava leaf dis-
eases. Scientific Reports, 15, Article 7009.
https://doi.org/10.1038/s41598-025-90646-4
Sharma, A., Rajesh, B., & Javed, M. (2021). Detection of
plant leaf disease in JPEG domain using transfer learn-
ing. arXiv:2107.04813.
https://arxiv.org/abs/2107.04813
Shoaib, M., Sadeghi-Niaraki, A., Ali, F., Hussain, I., &
Khalid, S. (2025). Leveraging deep learning for plant
disease detection. Frontiers in Plant Science, 16, Arti-
cle 1538163.
https://doi.org/10.3389/fpls.2025.1538163
Singh, V., & Misra, A. K. (2021). Detection of plant leaf
diseases using CNN. Procedia Computer Science, 167,
1152–1161.
Sujatha, R., Krishnan, S., Chatterjee, J. M., & Gandomi, A.
H. (2025). Advancing plant leaf disease detection inte-
grating machine learning and deep learning. Scien-
tific Reports, 15, Article 11552.
https://doi.org/10.1038/s41598-024-72197-2
Sundhar, S., Sharma, R., Maheshwari, P., Kumar, S. R., &
Kumar, T. S. (2025). Enhancing leaf disease classifica-
tion using GAT-GCN hybrid model.
arXiv:2504.04764. https://arxiv.org/abs/2504.04764
Wang, H., Wang, W., & Xu, Y. (2023). Vision transformers
for plant leaf disease recognition. Computers and Elec-
tronics in Agriculture, 204, 107547.
Lightweight Deep Learning System for Multi-Crop Leaf Disease Detection and Classification in Realtime Environments
263
