
From Free Text to Upper Gastrointestinal Cancer Diagnosis:
Fine-Tuning Language Models on Endoscopy and Histology Narratives
Kazhan Misri
1 a
, Leo Alexandre
2 b
and Beatriz De La Iglesia
1 c
1
University of East Anglia, School of Computing Science, U.K.
2
University of East Anglia, Norwich Medical School, U.K.
Keywords:
Transformer Models, Upper GI Cancer, Clinical Text Classification.
Abstract:
Clinical free text reports from endoscopy and histology are a valuable yet underexploited source of information
for supporting upper gastrointestinal (GI) cancer diagnosis. Our initial learning task was to classify procedures
as cancer-positive or cancer-negative based on downstream registry-confirmed diagnoses. For this, we devel-
oped a patient-level dataset of 63,040 endoscopy reports linked with histology data and cancer registry out-
comes, allowing supervised learning on real-world clinical data. We fine-tuned two transformer-based models:
general-purpose BERT and domain-specific BioClinicalBERT and evaluated methods to address severe class
imbalance, including random minority upsampling and class weighting. BioClinicalBERT combined with up-
sampling achieved the best recall (sensitivity) of 85% and reduced false negatives compared to BERT’s recall
of 78%. Calibration analysis indicated that predicted probabilities were broadly reliable. We also applied
SHapley Additive exPlanations (SHAP) to interpret model decisions by highlighting influential clinical terms,
fostering transparency and trust. Our findings demonstrate the potential of scalable, interpretable natural lan-
guage processing models to extract clinically meaningful insights from unstructured narratives, providing a
foundation for future retrospective review of cancer diagnosis and clinical decision support tools.
1 INTRODUCTION
Upper gastrointestinal (GI) cancers (affecting the oe-
sophagus, stomach, and duodenum) remain a leading
cause of cancer-related mortality worldwide (World
Health Organization (WHO) 2025; Cancer Research
UK 2025). Upper GI endoscopy is the gold standard
diagnostic test; however, findings are often reported
in semi and unstructured free text reports, which are
rich in detail but difficult to exploit systematically for
research or clinical tools.
Rule-based methods often struggle with inconsis-
tencies in terminology, structure, and ambiguity, such
as multiple biopsy sites or subtle benign–malignant
distinctions. With proper preprocessing and anno-
tation, however, these narratives can support super-
vised machine learning to detect clinically relevant
outcomes.
This work focuses on classifying upper GI pro-
cedures as cancer-positive or cancer-negative using
a
https://orcid.org/0009-0002-7548-9755
b
https://orcid.org/0000-0003-2618-7128
c
https://orcid.org/0000-0003-2675-5826
free text reports, with the goal of identifying poten-
tial missed diagnostic opportunities. Such classifica-
tion is especially relevant for post-endoscopy upper
GI cancers (PEUGIC), defined as cancers diagnosed
within three years of a negative endoscopy, which ac-
count for roughly 10% of cases (Wani et al. 2022;
Beg et al. 2017; Alexandre et al. 2022) and provide
important context for quality monitoring and future
research.
The study has two main objectives: (i) to con-
struct a temporally aligned, patient-level dataset link-
ing endoscopy and histology reports with registry-
confirmed cancer outcomes, and (ii) to evaluate
transformer-based NLP models for predicting con-
firmed upper GI cancer from historical records. The
dataset comprises routine, unlabeled endoscopy and
histology reports from 44,152 patients at Norfolk and
Norwich University Hospital (NNUH) collected be-
tween January 2015 and December 2021, created
through clinical linkage and temporal alignment.
Two transformer models, BERT (Devlin et al.
2019) and BioClinicalBERT (Lee et al. 2020), were
fine-tuned for this task. To address the extreme class
imbalance, four strategies were assessed: baseline
Misri, K., Alexandre, L. and De La Iglesia, B.
From Free Text to Upper Gastrointestinal Cancer Diagnosis: Fine-Tuning Language Models on Endoscopy and Histology Narratives.
DOI: 10.5220/0013836200004000
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2025) - Volume 1: KDIR, pages 501-508
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
501

training, class weighting, minority upsampling, and
combined weighting with upsampling, using patient-
level stratified splits (60% training, 20% validation,
20% test) with early stopping.
Interpretability is critical in clinical AI. We use
SHapley Additive exPlanations (SHAP) (Lundberg
and Lee 2017) to highlight influential words in predic-
tions, supporting retrospective analysis and potential
integration into decision support workflows.
The contributions of this work are:
• Development of a high-quality, linked dataset with
confirmed cancer outcomes.
• Evaluation of general and clinical-domain trans-
former models on endoscopy and histology narra-
tives.
• Comparison of class imbalance mitigation strate-
gies for cancer classification.
To address these objectives, Section 5 outlines the
experimental design, and Section 6 reports and dis-
cusses the findings, focusing on the role of model
choice and strategies for class imbalance mitigation.
2 RELATED WORK
Applying NLP to clinical free text has grown rapidly
for tasks such as disease classification, risk pre-
diction, and decision support. Transformer-based
models, such as BERT (Devlin et al. 2019), have
advanced contextual understanding of unstructured
medical narratives. Domain-adapted variants, includ-
ing BioBERT (Lee et al. 2020) and BioClinicalBERT,
pretrained on biomedical literature and clinical notes,
improve performance across healthcare tasks by inte-
grating domain knowledge and reducing reliance on
manual feature engineering.
In gastrointestinal cancer, traditional and deep
learning NLP approaches have identified relevant
concepts from pathology or endoscopy records.
Oliwa et al. (2019) used named entity recognition and
support vector machines on pathology reports, but
relied on extensive hand-crafted features and small
datasets. More recent transformer-based studies show
improved performance: Wang et al. (2024) used a
multi-branch BERT to classify gastroscopy findings,
and Iyer et al. (2023) applied BERT on structured
and unstructured EHRs to predict oesophageal cancer
risk. Syed et al. (2022) incorporated clinical embed-
dings into hybrid networks, though such approaches
often require complex designs and multimodal data
not always available in routine care.
For endoscopy, Pan et al. (2020) trained a neural
network classifier to detect gastric cancer, but without
contextual language representations and on a limited
dataset. In histology, Cheng (2022) applied CNNs to
classify malignancy from pathology reports, but gen-
eralisability across sites may be limited due to text
variability.
Class imbalance is a persistent challenge in these
studies, as cancer-positive cases are rare. Strategies
such as oversampling, class weighting, and focal loss
(Lin et al. 2017) have been proposed, yet their real-
world impact remains underexplored (Johnson and
Khoshgoftaar 2019).
Our study builds on this prior work by systemati-
cally evaluating imbalance mitigation on upper GI en-
doscopy and histology narratives, comparing general
(BERT) and clinically pre-trained (BioClinicalBERT)
transformers in a single-centre, secondary care setting
with confirmed registry-based cancer outcomes. To
our knowledge, few studies have modelled combined
endoscopy and histology reports or benchmarked im-
balance handling in this context.
3 DATA AND PREPARATION
We used pseudonymised Electronic Health Records
(EHR) from Norfolk and Norwich University Hospi-
tal (NNUH), part of the UK National Health Service
(NHS), spanning January 2015 to December 2021.
Our dataset combined three main data sources, linked
at the patient-level via pseudonymised NHS and hos-
pital numbers:
• Endoscopy reports: 65,084 procedure records
from 44,152 patients, with structured metadata and
free text clinical descriptions.
• Histology reports: 13,306 biopsy records from
10,479 patients, linked temporally to endoscopy
procedures.
• Cancer registry data: Structured records of can-
cer diagnoses (dates, morphology, staging) from the
Somerset Cancer Registry.
Statistics refer to the population undergoing en-
doscopy. Records were approximately balanced by
gender (52% female, 48% male). Patient ages had a
mean of 64 ± 17 years, reflecting the endoscopy co-
hort rather than cancer cases specifically. Over 80%
of procedures were performed in patients aged 50
years or older, reflecting national trends in endoscopy
(Beaton et al. 2024) and consistent with increased up-
per GI pathology incidence in older adults. Proce-
dure volumes declined markedly in 2020 due to the
COVID-19 pandemic, mirrored in biopsy submission
counts between March and August 2020. By October
2022, 74% of patients were alive and 26% deceased,
with a mean age at death of 80 ± 12 years among de-
ceased patients, aligning with expected clinical trajec-
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
502

tories.
3.1 Data Linking and Labelling
Histology records were matched to preceding en-
doscopy procedures per patient using pseudonymised
identifiers within a 0-9 day window, reflecting typi-
cal biopsy turnaround times. Cancer registry records
were retrospectively linked to endoscopy and histol-
ogy records per patient, with endoscopy defining the
baseline population. For labelling purposes, only can-
cer registry diagnoses were used as the gold stan-
dard. For patients with a cancer diagnosis, the pro-
cedure closest to and within three months prior to the
registry date was labelled cancer-positive, while all
other procedures before this date, as well as all proce-
dures from patients without a cancer diagnosis, were
labelled cancer-negative. Procedures occurring after
a cancer-positive event were treated as post-cancer
follow-up and excluded (1,922 records) to avoid bias
from post-diagnostic notes. This temporal alignment
reflects the diagnostic workflow, allowing the model
to learn from initial detection rather than subsequent
treatment or surveillance.
3.2 Text Cleaning and Standardisation
The clinical text fields in both endoscopy and his-
tology reports exhibited significant variability in for-
matting, typographical errors, and frequent use of
domain-specific abbreviations and shorthand. To en-
sure data quality and improve model training, we ap-
plied a comprehensive set of preprocessing steps:
• Removal of extraneous whitespace, repeated
spaces, and special characters, including common
encoding artefacts such as “
ˆ
A” and “
ˆ
aC™”, which
often arise from text extraction processes.
• Expansion of abbreviations and acronyms using a
manually curated dictionary tailored to each report
type, converting terms like “OGD” to “oesopha-
gogastroduodenoscopy” and “Ca” to “cancer” stan-
dardising clinical shorthand.
• Standardisation and unification of synonymous di-
agnostic terms and anatomical locations to reduce
vocabulary fragmentation, for example, grouping
various descriptions of gastric biopsies under a sin-
gle “stomach” category.
• Unicode normalisation and application of regular
expressions to correct encoding errors and remove
or replace non-ASCII (American Standard Code for
Information Interchange) characters, ensuring con-
sistent character encoding throughout the dataset.
These preprocessing steps enhanced the consis-
tency and clarity of the clinical narratives, reduced
noise caused by misspellings and shorthand, and sim-
plified the vocabulary, ultimately facilitating more ef-
fective and robust model training.
3.3 Balancing Classes by Sampling
The prepared dataset consisted of 63,040 procedure
records from 44,258 patients, including 994 cancer-
positive and 43,264 cancer-negative patients. The
dataset was highly imbalanced. To manage compu-
tational load and improve class balance, we retained
all cancer-positive patients and applied weighted ran-
dom sampling to the cancer-negative group.
Sampling was performed at the patient-level to
preserve longitudinal intra-patient variation. Higher
sampling weights were assigned to procedures from
more recent years (2020–2021) to reflect improved
data quality and clinical relevance. This resulted
in a more balanced dataset comprising 994 cancer-
positive and 3,429 cancer-negative procedures, pre-
serving realistic prevalence while maintaining suffi-
cient negative examples for robust model training.
Overall, the dataset included 4,423 procedures
from 3,123 unique patients, with cancer-positive
records representing approximately 22.5% of the
data. Yearly distributions and label proportions were
reviewed to confirm temporal and class balance, min-
imising systematic bias across the study period. The
cohort remained approximately balanced, with 2,356
(53%) male and 2,067 (47%) female patients. The
mean patient age at procedure was 67 ± 16 years,
with 85% of procedures performed in patients aged
50 years or older, reflecting typical demographics for
upper gastrointestinal pathology.
Regarding survival status, 2,615 patients (59%)
were alive and 1,808 patients (41%) were deceased at
the end of follow-up in October 2022. Among cancer-
positive patients, 5-year survival was approximately
22%, aligning with national averages reported by
Cancer Research UK (Cancer Research UK 2024a,b,
2025). Among deceased patients, the mean age at
death was 77±12 years, consistent with expected sur-
vival profiles in this clinical context. These descrip-
tive statistics confirm the dataset’s representativeness
and provide a foundation for subsequent modelling
and analysis.
4 MODEL TRAINING
4.1 Model Overview
To classify cancer from clinical text narratives, we
fine-tuned two transformer-based language models
From Free Text to Upper Gastrointestinal Cancer Diagnosis: Fine-Tuning Language Models on Endoscopy and Histology Narratives
503

under controlled conditions. The first, BERT-base
(uncased) (Devlin et al. 2019), is a general-purpose
model pretrained on Wikipedia and BookCorpus,
providing a strong baseline for downstream NLP
tasks. The second, BioClinicalBERT (Alsentzer et al.
2019), was further pretrained on biomedical litera-
ture (PubMed) and clinical notes (MIMIC-III), en-
abling deeper understanding of domain-specific lan-
guage and documentation style. Prior work suggests
BioClinicalBERT often outperforms general models
on clinical NLP tasks (Alsentzer et al. 2019; Si et al.
2022).
Both models share the same architecture: 12 trans-
former layers, 12 attention heads, a hidden size of
768, and a maximum sequence length of 512 tokens.
Input texts were tokenised with the corresponding
model tokenizer and truncated to fit the token limit.
For model input, histology text was prioritised
when available, as biopsies provide definitive diag-
noses. For each endoscopic procedure, matched his-
tology and endoscopy records were linked. If histol-
ogy was present, the input consisted exclusively of
histology text, with the endoscopy report used only
for linkage and contextual information. When no
biopsy was available, the endoscopy report was used
as input. This hierarchical approach mirrors routine
clinical workflows and ensures the model is trained
on the most clinically relevant information.
4.2 Handling Class Imbalance
Class imbalance is common in clinical datasets,
where positive cases are much rarer than negatives.
This can bias models towards the majority class, re-
ducing sensitivity to the clinically important minority
class.
We evaluated several approaches: (i) baseline
training without special handling of class imbalance;
(ii) class-weighted loss functions, assigning a higher
penalty to misclassified cancer-positive examples to
encourage focus on the minority class; (iii) ran-
dom oversampling of the minority class, duplicat-
ing cancer-positive samples to balance class repre-
sentation; (iv) a combined approach applying both
class weighting and oversampling; and (v) focal loss,
which reduces the contribution of well-classified ex-
amples and emphasises harder-to-classify minority
samples, helping the model focus on challenging
cases.
Even after downsampling non-cancer patients,
cancer-positive cases remained a small minority.
Comparing these methods allowed exploration of the
individual and combined effects of resampling and
loss function modifications, guiding identification of
the most effective strategy for this clinical task.
4.3 Hyperparameter Settings
All models were fine-tuned using the HuggingFace
Transformers library (Wolf et al. 2020) with the
AdamW optimiser (Loshchilov and Hutter 2017), a
learning rate of 2 × 10
−5
, and a batch size of 16 for
training (32 for validation and testing). Training ran
for up to 10 epochs with early stopping based on val-
idation F1-score (patience: 2 epochs), retaining the
checkpoint with the highest validation F1. Mixed pre-
cision training on GPUs was used to improve com-
putational efficiency without affecting model perfor-
mance.
To address class imbalance, a stratified batch sam-
pler ensured that each batch contained approximately
equal numbers of cancer-positive and cancer-negative
samples, improving learning from the minority class
and enabling a fair comparison between BERT and
BioClinicalBERT. For focal loss models, the γ param-
eter was set to 1.0 based on preliminary experiments.
4.4 Post-Training Probability
Calibration and SHAP
Reliable probability estimates are essential for clin-
ical decision support (Niculescu-Mizil and Caruana
2005). We assessed calibration by plotting curves on
the test set to evaluate alignment between predicted
and observed event rates. Analyses indicated reason-
able calibration, so no post-training recalibration (e.g.
logistic regression) was applied (Section 6).
To interpret model predictions, we applied SHAP
(SHapley Additive exPlanations), which assigns
token-level attribution scores indicating each word’s
contribution to classification. This supports valida-
tion of model reasoning, highlights clinically relevant
language, and helps identify potential causes of mis-
classification.
5 EXPERIMENTAL SETUP AND
EVALUATION
This study had three core objectives: (i) to construct
a high-quality, temporally aligned dataset linking en-
doscopy, histology, and cancer registry records (Sec-
tion 3.1); (ii) to fine-tune transformer-based models
for cancer classification using free text clinical re-
ports; and (iii) to systematically assess how different
class imbalance strategies affect model performance
(Section 4.2).
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
504

Table 1: Test set classification performance across models and imbalance-handling strategies.
Model Imbalance Strategy Precision (%) Recall (%) F1-score (%) F2-score (%) Accuracy (%)
BERT No Handling 0.0 0.0 0.0 0.0 77.3
BERT Class Weight 88.7 74.5 81.0 76.9 92.1
BERT Upsampling 87.6 78.0 82.5 79.7 92.5
BERT Upsampling + Weight 87.6 78.0 82.5 79.7 92.5
BioClinicalBERT No Handling 0.0 0.0 0.0 0.0 77.3
BioClinicalBERT Class Weight 85.6 77.0 81.1 78.6 91.8
BioClinicalBERT Upsampling 78.3 85.0 81.5 83.6 91.3
BioClinicalBERT Upsampling + Weight 78.3 85.0 81.5 83.6 91.3
5.1 Data Splitting and Patient-Level
Separation
To support robust and realistic evaluation, we split
the dataset at the patient-level, ensuring that all re-
ports from a given patient were assigned to only one
of the training, validation, or test sets. This approach
prevents information leakage and simulates deploy-
ment in real-world settings where models are applied
to previously unseen patients.
Patients were randomly assigned to training, val-
idation, and test sets with proportions of 60%, 20%,
and 20%, respectively to maintain consistent cancer
prevalence across these splits. To address imbalance
in the training set, we applied random oversampling
to the cancer-positive cases, achieving a near balanced
distribution, as described in Section 4.2. The valida-
tion and test sets were left unmodified to provide un-
biased estimates of model performance.
5.2 Evaluation Metrics
Model performance was evaluated using standard
classification metrics: precision, recall, F1-score, F2-
score, accuracy, and the receiver operating character-
istic (ROC) curve. Given the critical clinical impor-
tance of correctly identifying cancer-positive cases,
particular emphasis was placed on recall (sensitivity)
to minimise false negatives and their associated risks.
While the F1-score balances precision and recall,
we also report the F2-score, which places greater
weight on recall. This offers a more clinically mean-
ingful evaluation metric in scenarios where missing
positive cases is especially detrimental. Metrics were
reported for the cancer-positive class. ROC curves
were plotted to visualise the trade-off between sen-
sitivity and specificity across decision thresholds. All
metrics were computed on the held-out test set, using
the checkpoint with the highest F1-score on the vali-
dation set (Section 4.3).
6 RESULTS AND DISCUSSION
We evaluated the performance of BERT and BioClin-
icalBERT models on the binary classification task of
detecting upper GI cancer from free text clinical re-
ports. As described in Section 5, our experiments
assessed the impact of different imbalance-handling
strategies class weighting, minority class upsampling,
and their combination on model performance.
Figure 1: Confusion matrix
for BERT on the test set
with upsampling.
Figure 2: Confusion matrix
for BioClinicalBERT on the
test set with upsampling.
6.1 Effectiveness of Imbalance
Mitigation Strategies
Table 1 summarises the impact of different class im-
balance strategies on test set performance. Metrics are
reported for the cancer-positive class, with the best-
performing models highlighted in bold. Without any
mitigation, both BERT and BioClinicalBERT failed
to identify any cancer-positive cases, resulting in F1
and F2-scores of 0.0%. Despite relatively high accu-
racy of 77.3%, this reflects the severe class imbalance
and underscores the inadequacy of accuracy alone as
a performance metric (Section 4.2).
Applying class weighting substantially improved
detection. In the test set, which included 200 cancer-
positive and 682 cancer-negative cases, BERT cor-
rectly identified 156 cancer-positive cases (TP) with
44 false negatives (FN), and 660 true negatives (TN)
From Free Text to Upper Gastrointestinal Cancer Diagnosis: Fine-Tuning Language Models on Endoscopy and Histology Narratives
505
versus 22 false positives (FP) (see confusion matrix in
Figure 1). BioClinicalBERT achieved 170 TP, 30 FN,
635 TN, and 47 FP (see Figure 2). These results high-
light the effectiveness of penalising misclassification
of the minority class.
Further improvements were achieved using ran-
dom minority class upsampling. BERT with upsam-
pling alone achieved 156 TP, 44 FN, 660 TN, and 22
FP, while BioClinicalBERT attained 170 TP, 30 FN,
635 TN, and 47 FP. This confirms enhanced sensitiv-
ity, which is critical in clinical contexts where min-
imising false negatives is a priority.
Combining class weighting with upsampling did
not yield additional gains, suggesting that upsampling
sufficiently mitigates imbalance here. Overall, Bio-
ClinicalBERT with upsampling provided the best re-
call and F2-score, representing meaningful improve-
ment in detecting cancer-positive cases.
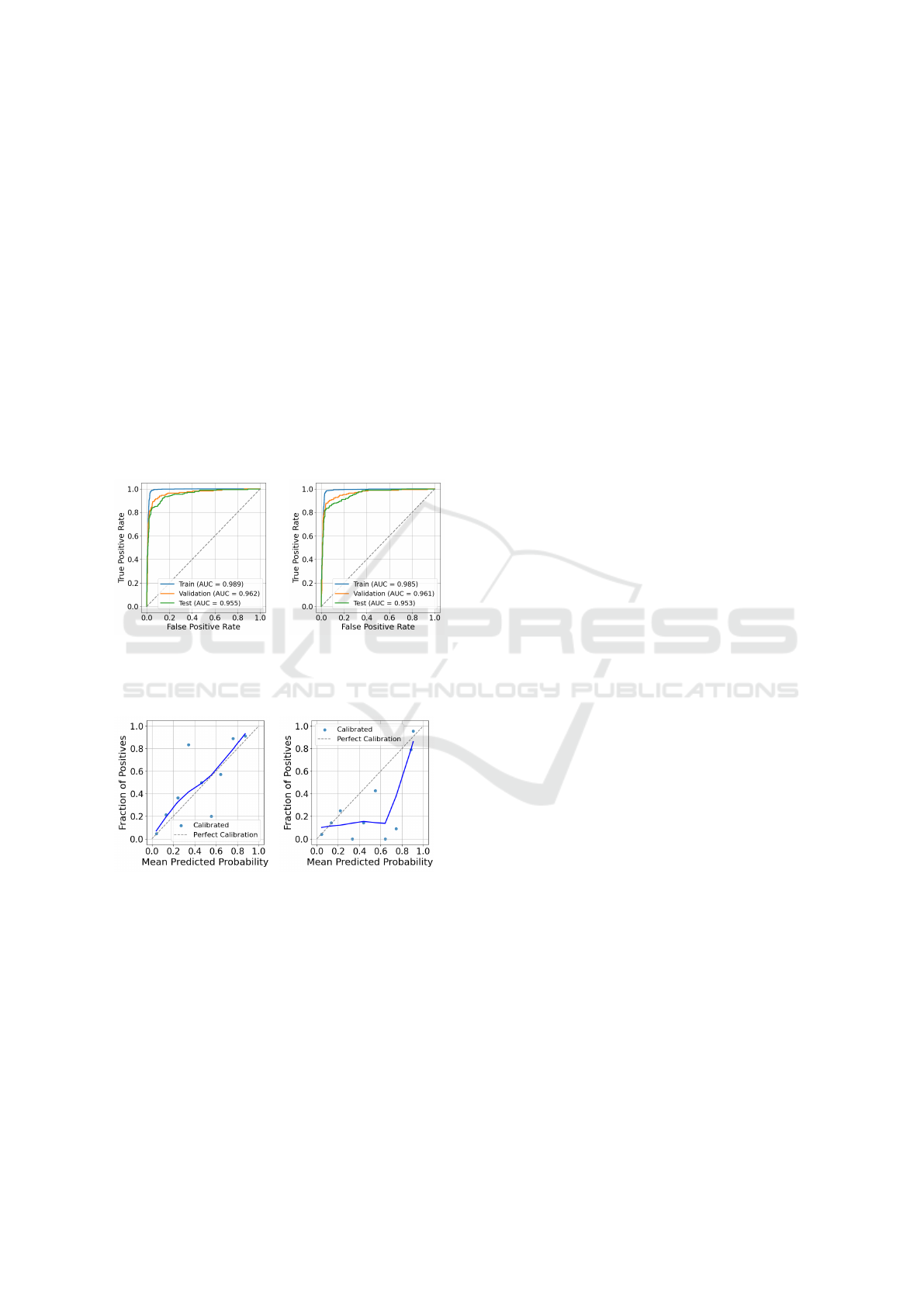
Figure 3: ROC curve for
BERT trained with upsam-
pling.
Figure 4: ROC curve for
BioClinicalBERT trained
with upsampling.
Figure 5: Calibration curve
for BERT trained with up-
sampling.
Figure 6: Calibration
curve for BioClinicalBERT
trained with upsampling.
6.2 Model Performance Evaluation
The top-performing models (BERT and BioClinical-
BERT trained with upsampling) were further evalu-
ated using ROC and calibration curves. Figures 4
and 3 present the ROC curves across the training,
validation, and test sets. BioClinicalBERT achieved
AUC scores of 0.985 (training), 0.961 (validation),
and 0.953 (test), while BERT reached slightly higher
AUCs of 0.989, 0.962, and 0.955 on the same splits.
Although differences are small, both models demon-
strated strong discriminative ability and consistent
performance across datasets.
The modest drop in AUC from training to test sets
suggests limited overfitting. However, AUC alone
cannot fully capture calibration or real-world relia-
bility. Therefore, we also assessed model calibration
separately to provide a more complete picture of per-
formance.
6.3 Calibration Analysis
Figures 5 and 6 show smoothed (lowess) calibration
curves for BERT and BioClinicalBERT, respectively,
on the test set. Both models demonstrate broadly
acceptable calibration outside the mid-probability
range, but notable miscalibration is evident in the
middle.
For example, BERT underestimates risk at pre-
dicted probabilities around 0.35 (observed positive
rate ≈ 0.85) and overestimates around 0.55 (observed
≈ 0.2). BioClinicalBERT shows a different pattern,
with strong overestimation around 0.35 (observed ≈
0.0) and 0.65 (observed ≈ 0.0), while being closer
to the diagonal near 0.45 (observed ≈ 0.15) and 0.75
(observed ≈ 0.1).
These deviations are concentrated in the
midrange, while calibration is better preserved
at lower predicted probabilities. Given that clinical
decision thresholds for cancer referral typically fall
below 20%, such midrange miscalibration is unlikely
to have major clinical impact. Overall, BERT and
BioClinicalBERT both exhibit midrange calibration
issues, with neither model demonstrating consistently
superior probability estimates across this range.
6.4 Model Comparison and Clinical
Implications
While overall F1-scores were similar, BioClinical-
BERT consistently showed higher recall and F2-score
than BERT, both critical in this clinical setting where
minimising missed cancer diagnoses is paramount.
Although BERT had slightly better precision, Bio-
ClinicalBERT’s superior recall and F2-score indicate
a better balance favoring sensitivity, which aligns with
the priority of reducing false negatives. These re-
sults reinforce the value of domain-specific pretrain-
ing (Section 4), as BioClinicalBERT appears more ef-
fective at capturing cues in clinical narratives. The
higher F2-score highlights its greater effectiveness for
cancer detection, reflecting the clinical importance of
prioritising sensitivity over precision.
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
506

Figure 7: True Positive Example: SHAP explanation for a cancer-positive case. Key terms (red) strongly influenced the
prediction.
Figure 8: False Negative Example: SHAP explanation for a cancer case missed by the model. Low or negative SHAP terms
led to misclassification.
6.5 Model Interpretability
To support interpretability and transparency, we ap-
plied SHAP (Lundberg and Lee 2017) to generate
post hoc attribution scores, quantifying the contribu-
tion of individual words to each prediction. We exam-
ined several randomly selected representative exam-
ples classified by BioClinicalBERT from both cancer-
positive and cancer-negative cases. As endoscopy re-
ports comprise the majority of the dataset, most exam-
ples are drawn from endoscopy narratives, although
some attributions also reflect histology report contri-
butions when biopsy data were available. This indi-
cates that the model can effectively make predictions
from endoscopy reports alone, even in the absence
of biopsy information, which is encouraging for real-
world clinical applicability.
Figure 7 shows a correctly classified cancer-
positive example. The model heavily weighted terms
such as “malignant-appearing tumour at the goj”,
“scope passes with slight resistance”, and “ulcerat-
ing infiltrating lesion along the lesser curve”. Proce-
dural terms like “biopsies taken”, “staging ct organ-
ised”, and “mdt referral with documented concerns”
also contributed positively. These align with clini-
cally meaningful indicators of malignancy, suggest-
ing the model relies on appropriate and interpretable
language cues.
Figure 8 illustrates a false negative example,
where the model failed to identify a cancer case. Cer-
tain terms, including “chronic gastric ulcer evident”,
“ulcer cardia bordering lesser curve”, “sessile polyp
edge”, and “biopsied await histology”, appeared with
low or negative SHAP values despite their potential
clinical relevance. These features may represent pre-
malignant or malignant pathology but were under-
weighted by the model, contributing to misclassifi-
cation. While the model generally distinguishes ma-
lignant from benign language effectively, borderline
findings require greater contextual understanding or
additional diagnostic input to avoid missed cases.
7 CONCLUSIONS
This study presents a comprehensive approach for
detecting upper GI cancer from routine clinical free
text reports, from data preparation to model inter-
pretation. We developed a high-quality, temporally
aligned, patient-level dataset by linking endoscopy
and histology reports with registry-confirmed cancer
outcomes, enabling supervised learning on real-world
clinical text. Comparing general purpose BERT and
domain specific BioClinicalBERT, we found that ad-
dressing class imbalance was crucial: models with-
out handling failed to detect cancer-positive cases,
while random upsampling consistently improved per-
formance, with no added benefit from combining it
with class weighting. Both models performed well,
but BioClinicalBERT achieved higher recall (85%)
and fewer false negatives, highlighting the value of
domain-specific pretraining for capturing subtle clin-
ical language cues. Calibration analysis confirmed
predicted probabilities were well-aligned with ob-
served outcomes, particularly at low-risk thresholds
relevant for clinical decision-making. SHAP analysis
provided token-level interpretability, showing predic-
tions relied on clinically meaningful language. Over-
all, our pipeline from curated dataset construction
through interpretable, calibrated modelling with class
From Free Text to Upper Gastrointestinal Cancer Diagnosis: Fine-Tuning Language Models on Endoscopy and Histology Narratives
507

imbalance mitigation offers a robust, clinically rele-
vant solution for upper GI cancer detection. Although
limited to a single hospital and showing some mid-
range probability miscalibration, the models demon-
strate strong clinical potential. Future work could
expand the dataset to cover more years, apply cali-
bration correction, and incorporate structured clinical
data to further improve sensitivity and robustness.
Ethical Considerations
The study was approved by the NHS Trust and Uni-
versity ethics committees (REC 22/PR/1559). All pa-
tient data were pseudonymised prior to analysis to en-
sure confidentiality.
ACKNOWLEDGEMENTS
This work used the ADA High Performance Comput-
ing cluster (HPC) at the University of East Anglia.
We thank the HPC support team for their assistance.
REFERENCES
Alexandre, L., Tsilegeridis-Legeris, T., and Lam, S. (2022).
Clinical and endoscopic characteristics associated with
post-endoscopy upper gastrointestinal cancers: a sys-
tematic review and meta-analysis. Gastroenterology,
162(4):1123–1135.
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H., Jin,
D., Naumann, T., and McDermott, M. (2019). Pub-
licly available clinical BERT embeddings. arXiv preprint
arXiv:1904.03323.
Beaton, D. R., Sharp, L., Lu, L., Trudgill, N. J., Thoufeeq,
M., Nicholson, B. D., Rogers, P., Docherty, J., Jenkins,
A., Morris, A. J., R
¨
osch, T., and Rutter, M. D. (2024).
Diagnostic yield from symptomatic gastroscopy in the
uk. Gut, 73(9):1421–1430.
Beg, S., Ragunath, K., Wyman, A., Banks, M., Markar,
S., Hawkey, C., Sanders, D., M
¨
onkem
¨
uller, K., Kaye,
P., and Fothergill, L. (2017). Quality standards in up-
per gastrointestinal endoscopy: a position statement of
the british society of gastroenterology (BSG) and asso-
ciation of upper gastrointestinal surgeons of great britain
and ireland (AUGIS). Gut, 66(11):1886–1899.
Cancer Research UK (2024a). Survival for oesophageal
cancer. https://www.cancerresearchuk.org/about-cancer/
oesophageal-cancer/survival, accessed: 2025-07-20.
Cancer Research UK (2024b). Survival for stomach
cancer. https://www.cancerresearchuk.org/about-cancer/
stomach-cancer/survival, accessed: 2025-07-20.
Cancer Research UK (2025). Common cancers compared.
https://www.cancerresearchuk.org/health-professional/
cancer-statistics/survival/common-cancers-compared,
accessed: 2025-07-20.
Cheng, J. (2022). Neural network assisted pathology case
identification. J. Pathol. Inform., 13:100008.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. In Proceedings of
NAACL-HLT 2019, pages 4171–4186.
Iyer, P. G., Sachdeva, K., Leggett, C. L., Willis, B. C.,
and Rubin, D. L. (2023). Development of electronic
health record-based machine learning models to predict
barrett’s esophagus and esophageal adenocarcinoma risk.
Clin. Transl. Gastroenterol., 14(10):e00637.
Johnson, J. M. and Khoshgoftaar, T. M. (2019). Survey on
deep learning with class imbalance. J. Big Data, 6(1):1–
54.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., and
Kang, J. (2020). BioBERT: a pre-trained biomedical lan-
guage representation model for biomedical text mining.
Bioinformatics, 36(4):1234–1240.
Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Doll
´
ar, P.
(2017). Focal loss for dense object detection. In Pro-
ceedings of ICCV 2017, pages 2980–2988.
Loshchilov, I. and Hutter, F. (2017). Decoupled weight de-
cay regularization. arXiv preprint arXiv:1711.05101.
Lundberg, S. M. and Lee, S.-I. (2017). A unified approach
to interpreting model predictions. Adv. Neural Inf. Pro-
cess. Syst., 30.
Niculescu-Mizil, A. and Caruana, R. (2005). Predicting
good probabilities with supervised learning. In Proceed-
ings of ICML 2005, pages 625–632.
Oliwa, T., Maron, S. B., Chase, L. M., Fiehn, O., and
Altman, R. B. (2019). Obtaining knowledge in pathol-
ogy reports through a natural language processing ap-
proach with classification, named-entity recognition, and
relation-extraction heuristics. JCO Clin. Cancer Inform.,
3:1–8.
Pan, J., Ding, S., Yang, S., Li, G., and Liu, X. (2020).
Endoscopy report mining for intelligent gastric cancer
screening. Expert Syst., 37(3):e12504.
Si, Y., Wang, J., Roberts, K., and Xu, H. (2022). Bench-
marking transformers on clinical notes classification. J.
Biomed. Inform., 127:104008.
Syed, S., Angel, A. J., Syeda, H. B., Jackson, T., and
Patel, R. (2022). The h-ANN model: comprehensive
colonoscopy concept compilation using combined con-
textual embeddings. In Proceedings of BIOSTEC 2022,
volume 5, page 189.
Wang, Z., Zheng, X., Zhang, J., and Zhang, M. (2024).
Three-branch bert-based text classification network for
gastroscopy diagnosis text. Int. J. Crowd Sci., 8(1):56–
63.
Wani, S., Yadlapati, R., Singh, S., Sawas, T., and Katzka,
D. A. (2022). Post-endoscopy esophageal neoplasia in
barrett’s esophagus: consensus statements from an in-
ternational expert panel. Gastroenterology, 162(2):366–
372.
Wolf, T., Debut, L., Sanh, V., Chaumond, J., Delangue,
C., Moi, A., Cistac, P., Rault, T., Louf, R., Funtowicz,
M., and Brew, J. (2020). Transformers: State-of-the-art
natural language processing. In Proceedings of EMNLP
2020: System Demonstrations, pages 38–45.
World Health Organization (WHO) (2025). Cancer. https:
//www.who.int/news-room/fact-sheets/detail/cancer, ac-
cessed: 2025-07-20.
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
508
