
Uncertainty Estimation and Calibration of a Few-Shot Transfer
Learning Model for Lettuce Phenotyping
Rusith Chamara Hathurusinghe Dewage
1,2 a
, Habib Ullah
1 b
, Muhammad Salman Siddiqui
1 c
,
Rakibul Islam
2 d
and Fadi-Al Machot
1 e
1
Faculty of Science and Technology, Norwegian University of Life Sciences, P.O. Box 5003, NO-1432 ,
˚
As, Norway
2
Photosynthetic AS, Sjøskogenveien 5, 1407 Vinterbro, Norway
Keywords:
Few-Shot Learning, Transfer Learning, Uncertainty Estimation, Calibration.
Abstract:
Computer vision-assisted automatic plant phenotyping in controlled environment agriculture (CEA) remains a
significant challenge due to the scarcity of labeled data from growing conditions. In this work, we investigate
few-shot transfer learning for estimating the maximum width of lettuce from cropped and segmented images
exhibiting non-uniform spatial distribution. The dataset presents additional complexity as images are cap-
tured using a wide-angle, off-center camera. We systematically investigate backbone architectures (ResNet,
EfficientNet, MobileNet, DenseNet, and Vision Transformer) and perform various data augmentation strate-
gies and regression head designs to identify optimal configurations under few-shot conditions. To enhance
predictive reliability, we employ post-hoc uncertainty estimation using Monte Carlo (MC) dropout and con-
formal prediction, and further evaluate model calibration to analyze alignment between predicted uncertainties
and empirical errors. Our best model, based on Vision Transformer Huge with 14×14 patch size (ViT-H/14),
achieved a root mean square error (RMSE) of 14.34 mm on the test set. For uncertainty estimation, MC
dropout achieved a miscalibration area of 0.19, an average prediction interval width of 27.89 mm, and an
empirical coverage of 73% at the nominal 90% confidence level. Our results highlight the importance of back-
bone selection, augmentation, and head architecture on model generalization and reliability. This study offers
practical guidelines for developing robust, uncertainty-aware few-shot models for plant phenotyping, enabling
more trustworthy deployment in CEA applications.
1 INTRODUCTION
Deep learning is widely used in many domains, fa-
cilitating automated analysis of complex visual data.
In controlled environment agriculture (CEA), deep
learning models have shown promise in tasks such
as crop monitoring, yield estimation, and precision
agriculture (Wang et al., 2022b; Mokhtar et al., 2022;
Zhang and Li, 2022). Nevertheless, the success of
these applications often depends on good quality,
large, and labeled datasets. Collecting useful data and
labeling it appropriately requires time and human re-
sources. Few-shot learning offers an alternative train-
a
https://orcid.org/0009-0004-6144-7807
b
https://orcid.org/0000-0002-2434-0849
c
https://orcid.org/0000-0002-6003-5286
d
https://orcid.org/0000-0002-7174-9225
e
https://orcid.org/0000-0002-1239-9261
ing approach by enabling models to learn from a lim-
ited number of samples. Despite their strong predic-
tive powers, deep learning models need an assessment
of their uncertainty to ensure reliability in real-world
applications. Without appropriate uncertainty estima-
tion, decisions based on predictive models may be un-
reliable, particularly in areas such as CEA, where un-
reliable predictions can result in huge financial losses.
In the cases of insufficient labeled data, few-shot
learning enables training a model with minimal sam-
ples by utilizing pre-trained deep-learning architec-
tures. The key objectives of this research are to ap-
ply few-shot transfer learning to predict the maximum
width of lettuce from a limited number of segmented
images, quantify uncertainty in the model’s predic-
tions on the test dataset, and evaluate model calibra-
tion. To achieve these objectives, we utilized a dataset
of lettuce images captured at various growth stages in
a controlled environment. The dataset also included
388
Dewage, R. C. H., Ullah, H., Siddiqui, M. S., Islam, R. and Machot, F.-A.
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping.
DOI: 10.5220/0013743600004000
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2025) - Volume 1: KDIR, pages 388-397
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

ground-truth measurements of the maximum lettuce
width.
This study enhances the domain of deep learning
uncertainty estimation in agricultural applications by
ensuring that deep learning models generate predic-
tions that are both accurate and reliable.
2 RELATED WORK
The success of deep learning is mainly driven by the
availability of extensive datasets, enhanced process-
ing capabilities, and improvements in training tech-
niques. Nonetheless, deep learning models frequently
encounter difficulties when faced with limited train-
ing data, and they cannot fundamentally quantify un-
certainty in their predictions, hence constraining their
use in essential fields such as healthcare and agricul-
ture.
Few-shot learning (FSL) has emerged as a pow-
erful approach in agriculture to address the challenge
of limited labeled data. Ruan et al. applied meta-
learning with hyperspectral imaging to detect drought
and freeze stress in tomatoes using as few as eight tar-
get domain samples, achieving superior performance
over traditional methods (Ruan et al., 2023). Lager-
gren et al. used FSL with convolutional neural net-
works (CNNs) to segment leaf morphology and vena-
tion traits from high-resolution field images, enabling
efficient phenotyping and genetic analysis with min-
imal annotation (Lagergren et al., 2023). FSL has
also shown promise in classification tasks and im-
proving model robustness. Belissent et al. combined
transfer and zero-shot learning for weed classifica-
tion, showing ResNet50’s effectiveness on the Toma-
toWeeds dataset and potential for identifying unseen
species (Belissent et al., 2024). Wang et al. reviewed
FSL techniques such as Siamese networks, prototyp-
ical networks, and GANs for plant disease and pest
recognition, presenting their high accuracy with lim-
ited data (Wang et al., 2022a). Luo et al. high-
lighted the importance of uncertainty estimation and
model calibration in CEA, suggesting FSL combined
with post-training techniques can enhance real-world
decision-making (Luo et al., 2023).
A significant advancement of FSL is few-shot
transfer learning (FSTL), which has demonstrated
considerable performance in plant phenotyping and
other fields. Research has shown that fine-tuning
CNNs using limited plant datasets can substantially
enhance accuracy and efficiency relative to training
from the ground up (Ojo and Zahid, 2022; Yang
et al., 2022). The recent paper by Hossen et al. re-
views the use of transfer learning (TL) in agricul-
ture, addressing data scarcity in the field. Recent ad-
vances in agricultural applications have demonstrated
the effectiveness of TL using various foundation mod-
els. Classical CNNs such as VGG16, ResNet50/101,
AlexNet, and InceptionV3 have been widely adopted
for tasks like plant species recognition, disease de-
tection, pest classification, and seedling identifica-
tion, often achieving high accuracy even with limited
or complex datasets. Lightweight models like Mo-
bileNetV2/V3, SqueezeNet, and EfficientNetB4 of-
fer improved efficiency and are particularly suited for
real-time or resource-constrained applications. Hy-
brid methods, including combinations with SVMs,
GANs, bilinear CNNs, and XGBoost, further en-
hance performance. Emerging architectures such
as DenseNet121, EfficientDet, YOLOv5, DenseY-
OLOv4, Swin Transformer, and Xception have shown
promise in specialized tasks like crop-weed detection,
plant growth prediction, and nutrient deficiency anal-
ysis. These studies consistently report high F1-scores
and accuracy, underscoring TL’s critical role in devel-
oping scalable and effective models for smart agricul-
ture (Hossen et al., 2025).
Although FSL has successfully enhanced model
generalization, uncertainty quantification continues to
pose a significant barrier, as deep learning models
generally do not offer confidence intervals for their
predictions. Uncertainty in deep learning is mainly
of two types: aleatoric, arising from inherent data
noise and irreducible by more data, and epistemic,
stemming from limited model knowledge that can be
reduced with additional data. While classification
models estimate uncertainty via predicted class prob-
abilities, regression models often lack inherent un-
certainty quantification (H
¨
ullermeier and Waegeman,
2021). Bayesian Neural Networks address epistemic
uncertainty by modeling weight distributions, but are
computationally challenging, leading to approximate
methods such as Monte Carlo dropout, which uses
dropout at inference to mimic posterior sampling, and
conformal prediction, which provides formal predic-
tion intervals (Kendall and Gal, 2017).
Recent work in few-shot learning has emphasized
improving uncertainty estimation and calibration to
enhance model reliability under limited data condi-
tions. Chang et al. proposed BMLPUC, which com-
bines calibrated uncertainty with adaptive training
for accurate Remaining Useful Life (RUL) prediction
(Chang and Lin, 2025). Similarly, Ding et al. in-
troduced BA-PML, a probabilistic few-shot learning
approach leveraging Bayesian Seq2Sep modeling and
episodic training for improved uncertainty in machin-
ery prognostics (Ding et al., 2023).
Other approaches integrate novel uncertainty-
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping
389

aware mechanisms into classification and vision-
language models. He et al. developed CLUR, us-
ing contrastive learning and pseudo uncertainty scores
for effective uncertainty estimation in few-shot text
classification (He et al., 2023). Morales et al. pro-
posed BayesAdapter, a Bayesian extension to CLIP
adapters that captures predictive distributions and en-
hances calibration in vision-language tasks (Morales-
´
Alvarez et al., 2024). Park et al. introduced meta-
XB, improving conformal prediction by reducing pre-
diction set size while ensuring strong calibration via
adaptive non-conformal scoring (Park et al., 2023).
Iwata et al. presented a meta-learning strategy for cal-
ibrating deep kernel Gaussian Processes, using task-
shared encoders and GMMs to achieve efficient and
well-calibrated few-shot regression (Iwata and Kuma-
gai, 2023).
3 METHODOLOGY
3.1 Data Collection and Preprocessing
The hydroponics lab, shown in Figure 2, served as
the primary experimental setup for this study. The
facility contains two independent chambers (zones),
which allow the simultaneous execution of two dif-
ferent plant growth protocols. These chambers reg-
ulate key environmental parameters, including light
intensity, water temperature, air temperature, nutri-
ent concentration, pH levels, carbon dioxide (CO
2
)
concentration, and relative humidity. A sensor sys-
tem continuously monitors and adjusts these condi-
tions to maintain predefined target values. Through-
out the plant growth cycle, the system systematically
collects all sensor measurements. It captures plant
images using a wide-angle camera in the top corner
of each chamber and stores them in Firebase Realtime
Database and Storage.
We collected images from an experiment con-
ducted in the hydroponics lab, where we maintained
a high carbon dioxide concentration in Zone 1 and a
low concentration in Zone 2 while keeping all other
variables nearly constant. This variation in CO
2
lev-
els led to different lettuce growth rates between the
two chambers. We manually measured the maximum
width of the lettuce on the 11
th
, 13
th
, 16
th
, and 19
th
days of growth. After retrieving images from these
days, we cropped and annotated individual plants that
were not occluded using the VGG Image Annota-
tor (Dutta and Zisserman, 2019). We then created
masked images by setting background pixel values to
zero.
The dataset included 72 masked images of lettuce
Table 1: Means and standard deviations computed from the
ImageNet dataset used to normalize the pixel values in each
channel.
Channel Mean Standard Deviation
R 0.485 0.229
G 0.456 0.224
B 0.406 0.225
as shown in Figure 3. The distribution of maximum
lettuce widths are shown in Figure 4.
The images were first padded to achieve a square
shape, ensuring that the longer dimension, either
width or height, determined the final size. Follow-
ing this, they were resized to 224 × 224 pixels. To
standardize pixel intensity values, the images were
rescaled by dividing each channel’s pixel values by
255. Subsequently, normalization was performed us-
ing the mean and standard deviation values computed
from the ImageNet dataset (Deng et al., 2009), as pre-
sented in Table 1, for each channel.
The dataset was divided into three bins based on
the maximum width of the lettuce samples. Subse-
quently, it was split into training, validation, and test
sets in a 60:20:20 ratio using stratified sampling based
on the bin assignments, resulting in 42, 15, and 15
samples, respectively. The training and validation sets
were then used to select the backbone network, tune
data augmentation strategies, and optimize the model
head architecture.
3.2 Model Architecture
The overall model architecture is illustrated in Figure
1. Let x ∈ R
224×224×3
denote an input image and y ∈
R the corresponding maximum lettuce width.
We first extract a feature vector z using a pre-
trained Vision Transformer (ViT-H/14) backbone
f (·; θ
f
):
z = f (x; θ
f
), z ∈ R
d
(1)
where θ
f
are the frozen backbone parameters and d is
the feature dimension.
The regression head comprises two fully-
connected layers with ReLU activation:
h
1
= ReLU (W
1
z + b
1
) (2)
ˆy = ReLU (W
2
h
1
+ b
2
) (3)
where W
1
∈ R
512×d
, b
1
∈ R
512
, W
2
∈ R
32×512
, b
2
∈
R
32
are trainable parameters.
Notation: ReLU(·) is the rectified linear unit acti-
vation function.
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
390
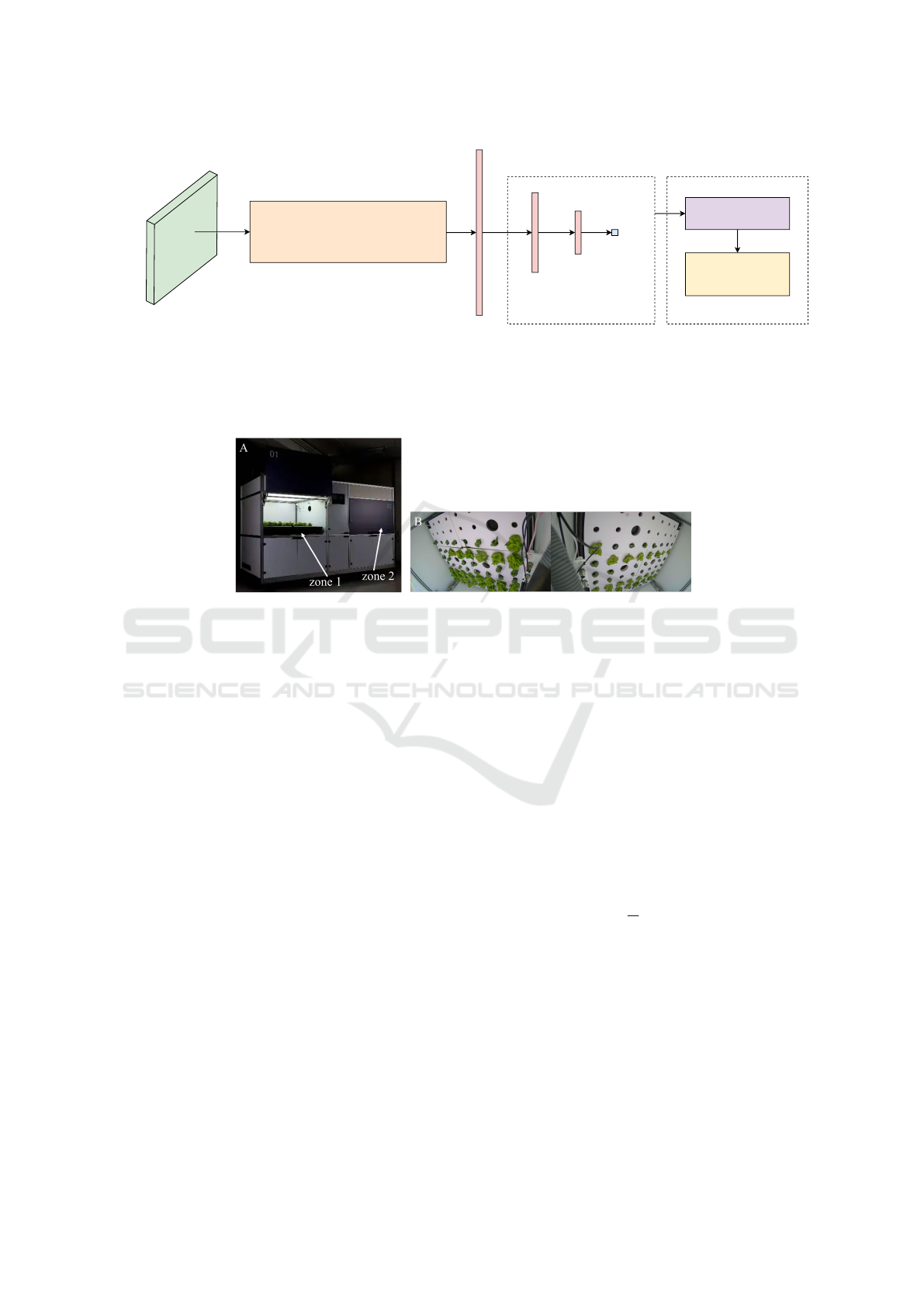
Vision Transformer (ViT-H/14)
224
224
3
1280
512
32
1
1
1
1
1
Input: Normalized
Image (224x224x3)
Backbone Network
Embedding
Dense
Layer 1
Dense
Layer 2
Maximum
width
Regression Head
Monte Carlo Dropout
Conformal Prediction
Calibration Assessment
Prediction Interval
Calibration Curve
Uncertainty Estimation
Figure 1: Architecture of the proposed model. It takes a normalized segmented lettuce image as input and extracts features
using a Vision Transformer (ViT-Huge) with a 14×14 patch size. The extracted features are passed through two dense layers
with ReLU activation to predict the maximum lettuce width. Uncertainty estimation is performed using Monte Carlo dropout
and conformal prediction, resulting in prediction intervals. Model calibration is assessed via calibration curves.
Figure 2: Hydroponics lab exterior view (A). The lab has two independent chambers (zone 1 and zone 2) that enable simul-
taneous execution of different plant growth protocols. Hydroponics lab interior view (B). Plant images are captured using a
wide-angle off-center camera in each zone.
3.3 Training Procedure
We began by evaluating various pre-trained neural
network architectures to serve as feature extractors.
Specifically, we evaluated ResNet50, ResNet101,
EfficientNetB0, MobileNetV2, DenseNet121, ViT-
B/16, and ViT-H/14. The Keras deep learning frame-
work (Chollet et al., 2015) was used to implement the
first five models, while the final two models were ac-
cessed via the PyTorch Image Models library (Wight-
man, 2019). These networks were used in a frozen
state (without updating their weights during training),
and the output features were fed into a minimal re-
gression head.
After selecting the backbone network, we inves-
tigated the effect of data augmentation on general-
ization performance. Several augmentation strategies
were evaluated, adding rotation, horizontal flip, verti-
cal flip, brightness, zoom in, zoom out, zoom in and
out, horizontal and vertical shift, and shear sequen-
tially starting from no augmentation. Each configura-
tion was evaluated while keeping the backbone frozen
and a minimal regression head. Five augmented ver-
sions were generated for each image. With the back-
bone and augmentation strategy fixed, we explored
various configurations of the model head. Specifi-
cally, we experimented with different numbers and
sizes of dense layers. Rectified Linear Unit (ReLU)
was used as the activation function.
Early stopping with a patience of 5 epochs was
employed during the hyperparameter tuning process
to avoid overfitting. The optimization was performed
using the ADAM optimizer (Kingma and Ba, 2014)
with a learning rate of 0.001 and a batch size of 8, due
to the small dataset size. The network was trained to
minimize mean squared error (MSE) over the training
set D = {(x
i
, y
i
)}
N
i=1
:
L
MSE
=
1
N
N
∑
i=1
( ˆy
i
− y
i
)
2
(4)
where ˆy
i
is given by Eq. 3. Performance was assessed
using the validation loss and R
2
score to identify the
most suitable hyperparameters.
The final model architecture employed ViT-H/14
as the backbone, followed by two fully connected hid-
den layers comprising 512 and 32 neurons, respec-
tively. The data augmentation strategy included ran-
dom rotations within a range of ±40
◦
, random bright-
ness adjustments within ±0.1, and random horizontal
flipping. The model’s performance was subsequently
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping
391
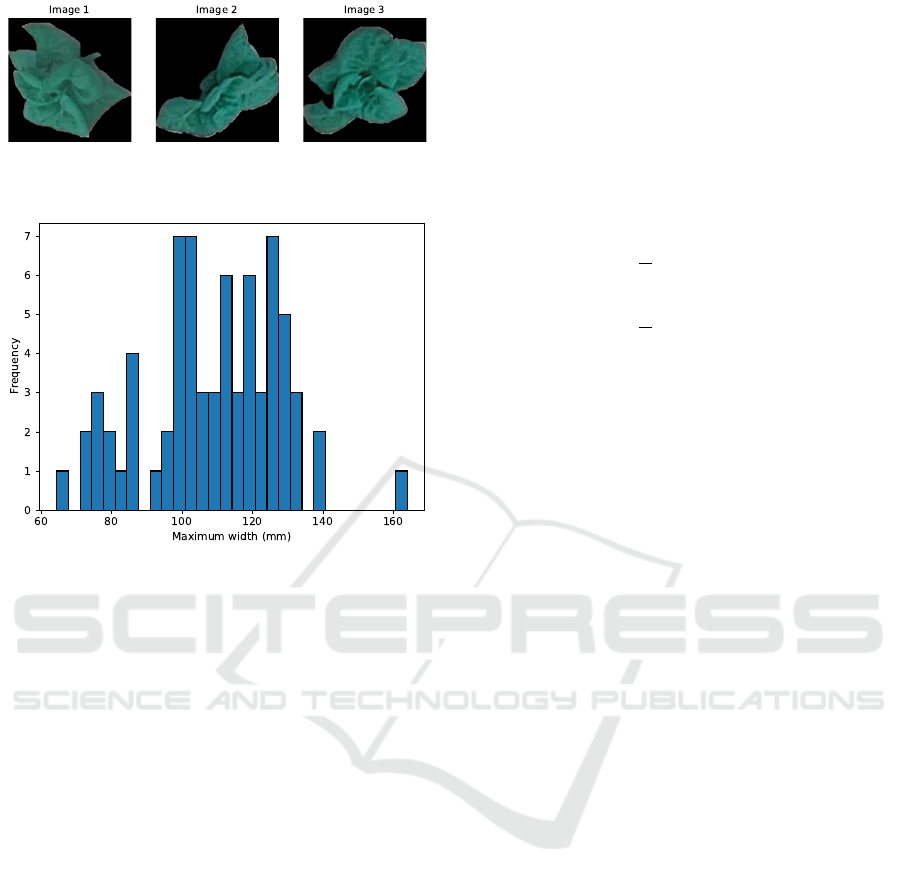
Figure 3: Few samples of masked lettuce images in the
dataset.
Figure 4: Distribution of maximum widths of lettuce in the
dataset.
evaluated on the test dataset.
3.4 Uncertainty Estimation
To quantify uncertainty using conformal prediction,
we employed the ”Naive” method, which calculates
nonconformity scores on a calibration set (Tibshirani,
2023).
Given a calibration set D
cal
, we compute absolute
residuals:
r
i
= |y
i
− ˆy
i
|, ∀(x
i
, y
i
) ∈ D
cal
(5)
For miscoverage rate α, let q
1−α
be the (1 − α)-
quantile of {r
i
}. The conformal prediction interval
for a new test sample x
new
is:
C(x
new
) = [ ˆy
new
− q
1−α
, ˆy
new
+ q
1−α
] (6)
where ˆy
new
is obtained by Eq. (3).
We computed the 95% prediction intervals using
a significance level of 0.05 and using validation set
for computing nonconformity scores. Prediction in-
tervals were computed for the test data points and pre-
dictions and their 95% prediction intervals were plot-
ted.
To estimate uncertainty using MC dropout (Gal
and Ghahramani, 2016), we modified the model head
by incorporating a dropout layers with a dropout rate
of 0.1, positioned after the hidden layers. This modi-
fied the model architecture Eq. 2 and Eq. 3 as follows,
where p denotes the dropout rate.
h
1
= Dropout (ReLU(W
1
z + b
1
), rate = p) (7)
ˆy = Dropout (ReLU (W
2
h
1
+ b
2
), rate = p) (8)
At inference, dropout remains enabled and the
model is sampled T times for each input, yielding pre-
dictions { ˆy
(t)
}
T
t=1
:
µ
ˆy
=
1
T
T
∑
t=1
ˆy
(t)
(9)
σ
2
ˆy
=
1
T
T
∑
t=1
ˆy
(t)
− µ
ˆy
2
(10)
where µ
ˆy
is the predictive mean and σ
2
ˆy
quantifies
epistemic uncertainty. We plotted the predictions
and their 95% prediction intervals for the test dataset
assuming the predictions are normally distributed
around the mean.
Calibration diagrams were plotted for the predic-
tion intervals generated by both the MC dropout and
conformal prediction methods. The results were then
compared to evaluate their performance.
4 RESULTS
The final model achieved an RMSE of 14.34 mm and
an R
2
value of 0.4464 on the test set. The average 90%
uncertainty intervals estimated by MC dropout and
conformal prediction were 27.89 mm and 27.74 mm,
respectively. Figure 5 shows the learning curve with
training and validation loss. The actual vs. predicted
maximum width of lettuce in the test set is shown in
Figure 6, where the ideal line represents perfect pre-
dictions. Figure 7 illustrates the test set predictions
along with 95% confidence intervals estimated using
the MC dropout method. Figure 8 presents the 95%
uncertainty intervals estimated using the “Naive” con-
formal prediction method for the test set samples. Fi-
nally, Figure 9 displays the calibration plot for the un-
certainties estimated by MC dropout, while Figure 10
shows the calibration plot for the uncertainties esti-
mated by the “Naive” conformal prediction method.
5 DISCUSSION
To systematically explore the influence of various
architectural and training choices on model perfor-
mance, we employed a sequential hyperparameter
tuning approach. Given the limited size of our dataset,
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
392
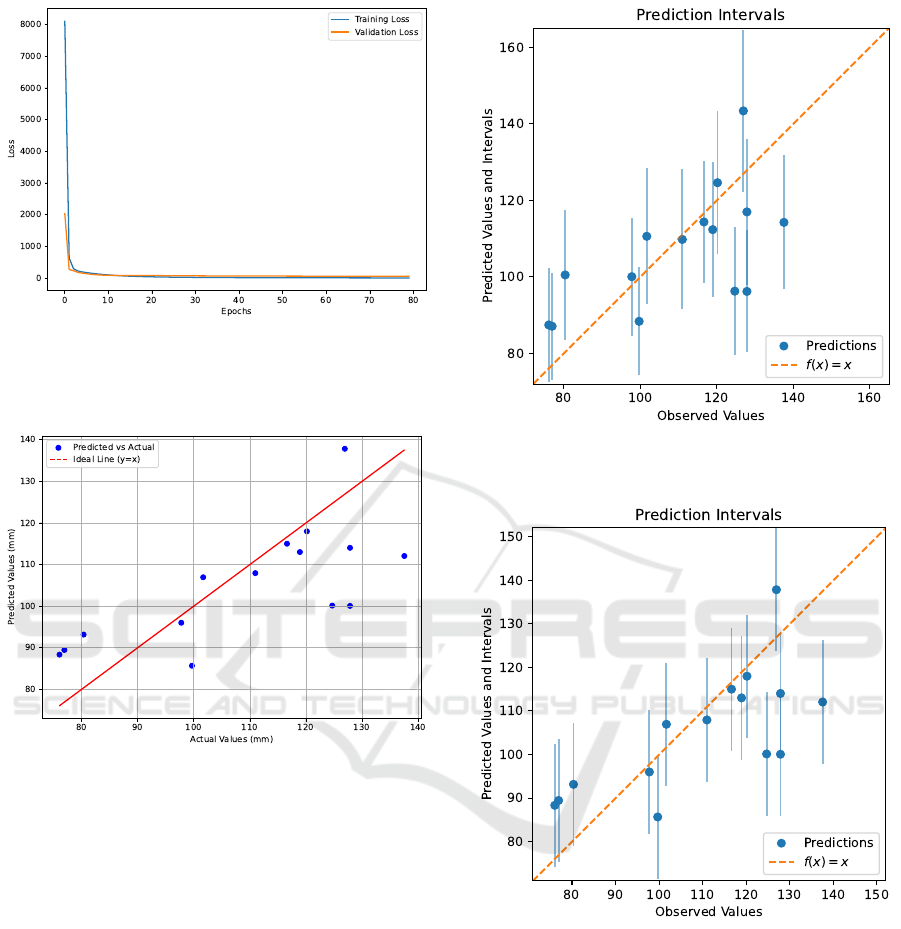
Figure 5: Training and validation loss throughout the entire
learning process. The final model was obtained at epoch 75
using early stopping with a patience of 5, restoring the best
weights.
Figure 6: Actual values vs predicted values for the test set.
this strategy allowed us to isolate and assess the in-
dividual effects of key components: the backbone
network, data augmentation techniques, and model
head architecture without introducing excessive ex-
perimental complexity or confounding variables.
Table 2 presents the validation RMSE and R
2
for the different backbone networks we evaluated.
ViT-H/14 achieved the lowest validation RMSE and
the highest R
2
, significantly outperforming the other
backbone networks. This indicates that ViT-H/14 is
highly recommended for this type of task. The fine-
tuning results of the augmentation method, presented
in Table 3, show that the quality of the training dataset
and the use of systematic sampling are crucial factors
in few-shot learning. The results also highlight that
rotation, horizontal flip, and brightness are the most
important parameters for improving the model’s gen-
eralization to real data.
Tables 4 and 5 summarize the hyperparameter tun-
ing configurations and outcomes for the model head
Figure 7: Uncertainty of predictions estimated by the MC
Dropout method for the test set.
Figure 8: Uncertainty of predictions estimated by the con-
formal prediction method for the test set.
architecture. They suggest the effect of the regression
head depth and the number of neurons in each layer
on the model’s performance.
In our current implementation, the final output
layer employs a ReLU activation. While ReLU en-
forces non-negativity, which aligns with the positive
nature of lettuce width, it may also introduce an artifi-
cial bias by truncating small predictions to zero. This
can be problematic in cases where the true value is
small but non-zero. A linear output layer could there-
fore be more appropriate, as it would preserve the
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping
393
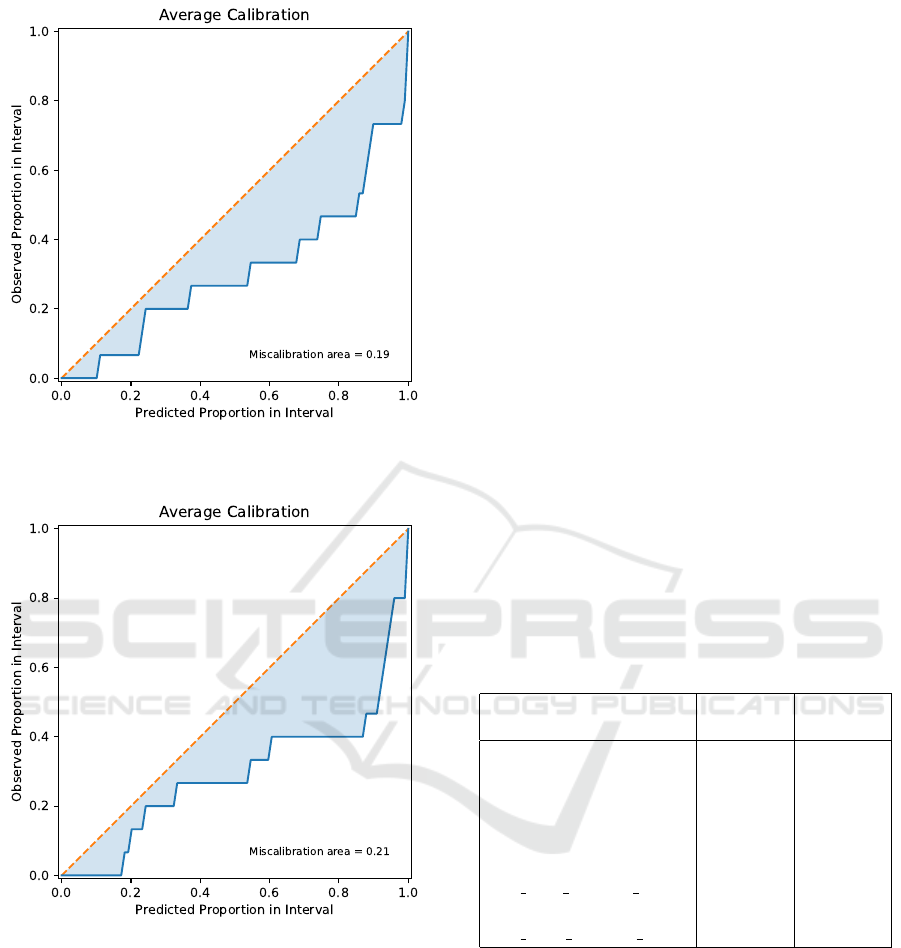
Figure 9: Calibration plot for uncertainty estimation using
the MC Dropout method.
Figure 10: Calibration plot for uncertainty estimation using
the “na
¨
ıve” conformal prediction method.
continuous nature of the regression task without im-
posing unnecessary constraints. Future work may in-
vestigate this alternative to assess whether it improves
predictive accuracy.
While the above results are based on a single ran-
dom train–validation split, we further investigated the
robustness and generalizability of these findings by
repeating the experiments across 20 distinct random
splits. This enabled a statistical evaluation of perfor-
mance differences among backbone models, augmen-
tation strategies, and head architectures. Consistent
with the single-split results, ViT-H/14 again achieved
the lowest mean RMSE and the highest mean R
2
across all splits. Since the RMSE and R
2
distri-
butions did not satisfy normality or homogeneity of
variance assumptions, we applied the Kruskal–Wallis
test to assess statistical significance. The analysis
showed that ViT-H/14 outperformed ResNet50, Effi-
cientNetB0, ResNet101, and MobileNetV2 with sta-
tistically significant differences at the 95% signifi-
cance level.
For augmentation methods, horizontal flip and
brightness yielded modest improvements compared
to the baseline, whereas rotation did not contribute
to performance gains. However, ANOVA tests on
the RMSE and R
2
distributions indicated that these
improvements were not statistically significant at the
95% significance level. Similarly, for model head
architectures, configurations with 512 and 128 neu-
rons in successive layers showed the best average per-
formance, but ANOVA suggested that these differ-
ences were also not statistically significant. Taken to-
gether, these results suggest that while augmentation
and head depth exert some influence on performance,
the choice of backbone architecture remains the most
decisive factor for generalization in this setting.
Table 2: Comparison of different backbones using valida-
tion RMSE and R
2
. The Vision Transformer Huge with a
14×14 patch size achieves the lowest validation RMSE and
the highest R
2
.
Backbone Validation
RMSE
Validation
R
2
ResNet50 20.96 -0.4730
EfficientNetB0 17.89 -0.0738
ResNet101 13.50 0.3886
MobileNetV2 15.79 0.1634
DenseNet121 14.94 0.2510
ViT
(vit base patch16 224)
15.77 0.1655
ViT
(vit huge patch14 224)
10.58 0.6244
In the transfer learning setup employed in this
study, the weights of the pretrained model were
frozen, and only a regression head was introduced at
the end of the backbone network for training. This
approach highlights the potential of backbone net-
works trained on large datasets for new tasks. A
further improvement would be fine-tuning the back-
bone network by including its weights in the training
process. Our results suggest that transformer-based
architectures offer significantly improved generaliza-
tion, with self-attention mechanisms capturing global
plant morphology more effectively than purely con-
volutional approaches.
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
394
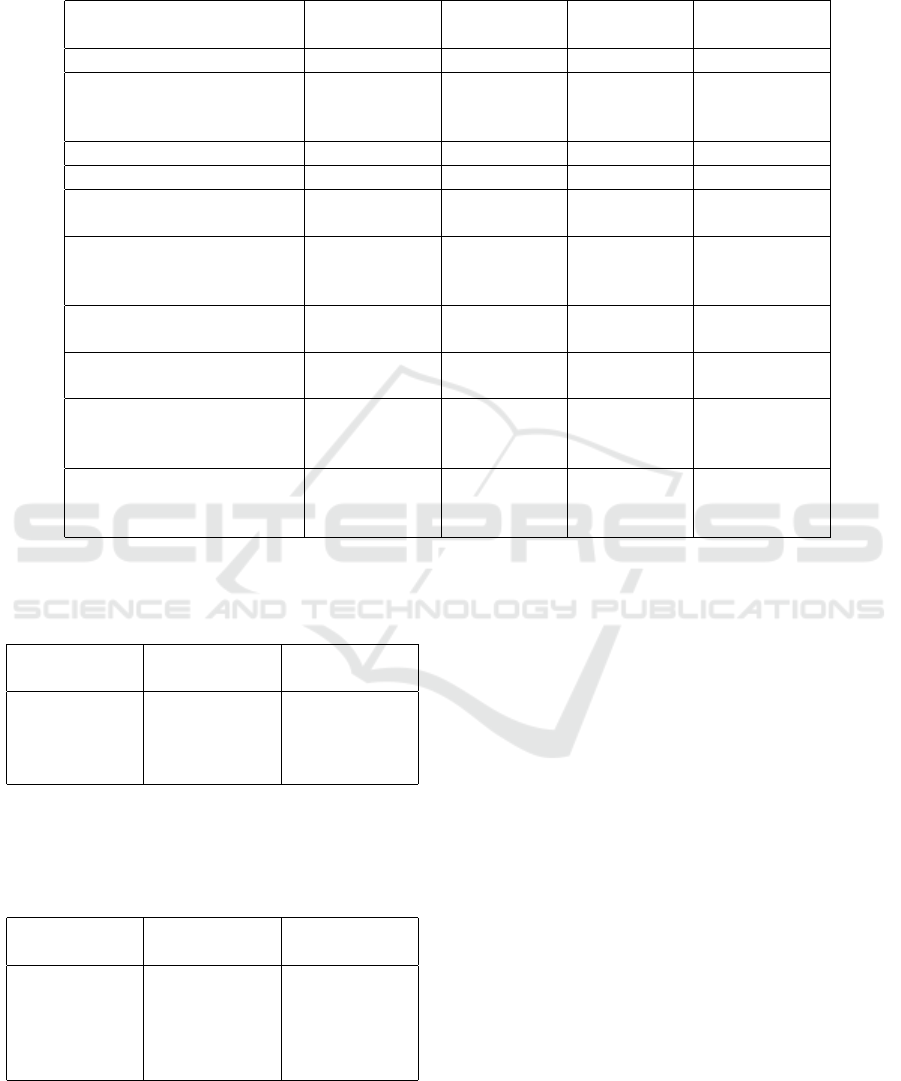
Table 3: Comparison of augmentation configurations used during hyperparameter tuning, evaluated using validation RMSE
and R
2
. Augmentations were implemented using torchvision transforms. Random rotations within a range of ±40
◦
, random
brightness adjustments within ±0.1, and random horizontal flipping each sequentially improved performance on the validation
set.
Augmentation Technique Parameter Value Validation
RMSE
Validation R
2
No augmentation - - 10.58 0.6244
RandomRotation degrees
30° 9.38 0.7051
40° 8.80 0.7401
50° 8.98 0.7293
RandomHorizontalFlip p 0.5 8.35 0.7661
RandomVerticalFlip p 0.5 9.59 0.6914
ColorJitter brightness
0.1 7.61 0.8057
0.2 7.78 0.7967
RandomAffine scale
(1, 1.1) 9.13 0.7202
(1, 1.2) 9.08 0.7232
(1, 1.3) 9.93 0.6694
RandomAffine scale
(0.9, 1) 9.63 0.6888
(0.8, 1) 10.76 0.6118
RandomAffine scale
(0.9, 1.1) 9.10 0.7225
(0.8, 1.2) 9.43 0.7020
RandomAffine translate
0.1 9.89 0.6720
0.2 9.43 0.7020
0.3 9.80 0.6782
RandomAffine shear
0.3 9.61 0.6901
10 8.64 0.7497
20 8.61 0.7516
Table 4: Effect of adding one layer. Comparison of the
number of neurons in the added layer using validation
RMSE and R
2
. Adding one layer did not result in improved
performance on the validation set.
Number of
neurons
Validation
RMSE
Validation R
2
512 7.78 0.7970
256 7.79 0.7963
128 7.86 0.7929
64 7.91 0.7903
Table 5: Effect of adding two layers. The second layer
was added after one layer with 512 neurons. Comparison
of the number of neurons in the second layer using valida-
tion RMSE and R
2
. Adding 32 neurons in the second layer
resulted in improved performance on the validation set.
Number of
neurons
Validation
RMSE
Validation R
2
256 8.31 0.7683
128 8.22 0.7736
64 8.11 0.7792
32 7.54 0.8094
16 7.88 0.7918
The learning curve (Figure 5 demonstrates a sharp
decline in both training and validation loss within the
initial epochs, indicating that the backbone network
had already learned most of the relevant information
in the input images. Consequently, the model re-
quired minimal additional learning, which it achieved
rapidly.
The dataset used in this study consisted of lettuce
samples with maximum widths ranging from 64.43
mm to 163.90 mm. Given this range, an RMSE of
14.34 mm indicated a promising model performance.
However, the R-squared value of 0.4464 suggests
that only 45% of the variance in the dependent vari-
able is explained by the model’s predictions. While
this level of accuracy may be sufficient for growth
monitoring applications, it may not be acceptable in
more complex tasks where greater precision is re-
quired. Given the constraints of few-shot learning
due to the limited number of training images, this
performance can still be considered satisfactory, em-
phasizing the applicability of few-shot transfer learn-
ing in agricultural settings. Future work should ex-
plore other few-shot learning techniques, such as pro-
totypical networks (Snell et al., ), Siamese networks
(Koch et al., 2015), Model-Agnostic Meta-Learning
(MAML) (Finn et al., 2017), and MAML++ (Anto-
niou et al., 2018), to assess their performance on this
task.
Both MC dropout and conformal prediction
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping
395

proved to be effective and easily implementable meth-
ods for uncertainty estimation. The MC dropout
method requires the inclusion of dropout layers in the
network, whereas conformal prediction does not im-
pose such a requirement. In this regard, conformal
prediction offers a more flexible and straightforward
implementation for any model.
As observed in Figure 7, the uncertainty estimated
by MC dropout remained relatively consistent across
all test dataset predictions, indicating that the model
maintains a uniform level of uncertainty across its
predictions. In contrast, the uncertainty estimated by
the ”Naive” CP method results in a fixed-width inter-
val as shown in Figure 8. There remains a potential to
explore alternative variations of conformal prediction
methods beyond the ”Naive” approach, such as split
conformal prediction, to enhance uncertainty estima-
tion.
The calibration plots for both methods given in
Figs. 9 and 10 indicate that the miscalibration area
is relatively low, suggesting that the model exhibits
some degree of calibration. However, there remains
room for further calibration improvement. The mis-
calibration area (0.19) for the Monte Carlo (MC)
dropout method was slightly smaller compared to the
conformal prediction method (0.21). MC dropout
method had empirical coverage of 73% at the nominal
90% and 95% confidence levels. In contrast, confor-
mal prediction method had empirical coverages 47%
and 80% at the nominal 90% and 95% confidence lev-
els. This further showed that the MC dropout method
resulted in better calibration in uncertainty estimation
compared to conformal prediction method in this task.
To further improve the calibration of the uncertainty
estimates produced by the MC Dropout method, iso-
tonic regression (Jiang et al., 2011) was employed us-
ing the validation set. However, this resulted in no
improvement, which could be attributed to the limited
amount of data.
6 CONCLUSION
In conclusion, the study demonstrates that our
pipeline: few-shot transfer learning, combined with
techniques such as MC dropout and conformal pre-
diction, can be effectively applied to agricultural tasks
such as lettuce growth monitoring and quantify as-
sociated uncertainties, even when trained on lim-
ited data. While the model performed promisingly
with the limited data, there are still opportunities to
improve accuracy, calibration, and uncertainty esti-
mation. Incorporating uncertainty quantification not
only improves confidence in predictions but also sup-
ports safer deployment in agricultural environments,
where decisions informed by reliable model outputs
are critical.
ACKNOWLEDGEMENTS
We would like to acknowledge the financial support
from the Research Council of Norway (project num-
ber 354125) and Photosynthetic AS. We also wish to
thank the Norwegian University of Life Sciences for
granting admission.
REFERENCES
Antoniou, A., Edwards, H., and Storkey, A. (2018). How to
train your maml. arXiv, 1810.09502.
Belissent, N., Pe
˜
na, J. M., Mes
´
ıas-Ruiz, G. A., Shawe-
Taylor, J., and P
´
erez-Ortiz, M. (2024). Transfer and
zero-shot learning for scalable weed detection and
classification in uav images. Knowledge-Based Sys-
tems, 292:111586.
Chang, L. and Lin, Y.-H. (2025). Few-shot remaining useful
life prediction based on bayesian meta-learning with
predictive uncertainty calibration. Engineering Appli-
cations of Artificial Intelligence, 142:109980.
Chollet, F. et al. (2015). Keras. https://keras.io.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). Imagenet: A large-scale hierarchical
image database. In 2009 IEEE Conference on Com-
puter Vision and Pattern Recognition, pages 248–255.
Ding, P., Jia, M., Ding, Y., Cao, Y., Zhuang, J., and Zhao,
X. (2023). Machinery probabilistic few-shot prognos-
tics considering prediction uncertainty. IEEE/ASME
Transactions on Mechatronics, 29(1):106–118.
Dutta, A. and Zisserman, A. (2019). The VIA annotation
software for images, audio and video. In Proceedings
of the 27th ACM International Conference on Multi-
media, MM ’19, New York, NY, USA. ACM.
Finn, C., Abbeel, P., and Levine, S. (2017). Model-agnostic
meta-learning for fast adaptation of deep networks. In
International conference on machine learning, pages
1126–1135. PMLR.
Gal, Y. and Ghahramani, Z. (2016). Dropout as a bayesian
approximation: Representing model uncertainty in
deep learning. In Balcan, M. F. and Weinberger,
K. Q., editors, Proceedings of The 33rd International
Conference on Machine Learning, volume 48 of Pro-
ceedings of Machine Learning Research, pages 1050–
1059, New York, New York, USA. PMLR.
He, J., Zhang, X., Lei, S., Alhamadani, A., Chen, F., Xiao,
B., and Lu, C.-T. (2023). Clur: uncertainty estima-
tion for few-shot text classification with contrastive
learning. In Proceedings of the 29th ACM SIGKDD
Conference on Knowledge Discovery and Data Min-
ing, pages 698–710.
KDIR 2025 - 17th International Conference on Knowledge Discovery and Information Retrieval
396

Hossen, M. I., Awrangjeb, M., Pan, S., and Mamun, A. A.
(2025). Transfer learning in agriculture: a review. Ar-
tificial Intelligence Review, 58(4):97.
H
¨
ullermeier, E. and Waegeman, W. (2021). Aleatoric and
epistemic uncertainty in machine learning: An intro-
duction to concepts and methods. Machine learning,
110(3):457–506.
Iwata, T. and Kumagai, A. (2023). Meta-learning to cali-
brate gaussian processes with deep kernels for regres-
sion uncertainty estimation. arXiv, 2312.07952.
Jiang, X., Osl, M., Kim, J., and Ohno-Machado, L. (2011).
Smooth isotonic regression: a new method to calibrate
predictive models. AMIA Summits on Translational
Science Proceedings, 2011:16.
Kendall, A. and Gal, Y. (2017). What uncertainties do we
need in bayesian deep learning for computer vision?
Advances in neural information processing systems,
30.
Kingma, D. P. and Ba, J. (2014). Adam: A method for
stochastic optimization. arXiv, 1412.6980.
Koch, G., Zemel, R., Salakhutdinov, R., et al. (2015).
Siamese neural networks for one-shot image recog-
nition. In ICML deep learning workshop, volume 2,
pages 1–30. Lille.
Lagergren, J., Pavicic, M., Chhetri, H. B., York, L. M., Hy-
att, D., Kainer, D., Rutter, E. M., Flores, K., Bailey-
Bale, J., Klein, M., et al. (2023). Few-shot learning
enables population-scale analysis of leaf traits in pop-
ulus trichocarpa. Plant Phenomics, 5:0072.
Luo, J., Li, B., and Leung, C. (2023). A survey of com-
puter vision technologies in urban and controlled-
environment agriculture. ACM Computing Surveys,
56(5):1–39.
Mokhtar, A., El-Ssawy, W., He, H., Al-Anasari, N., Sam-
men, S. S., Gyasi-Agyei, Y., and Abuarab, M. (2022).
Using machine learning models to predict hydroponi-
cally grown lettuce yield. Frontiers in Plant Science,
13:706042.
Morales-
´
Alvarez, P., Christodoulidis, S., Vakalopoulou, M.,
Piantanida, P., and Dolz, J. (2024). Bayesadapter: en-
hanced uncertainty estimation in clip few-shot adapta-
tion. arXiv, 2412.09718.
Ojo, M. O. and Zahid, A. (2022). Deep learning in con-
trolled environment agriculture: A review of recent
advancements, challenges and prospects.
Park, S., Cohen, K. M., and Simeone, O. (2023). Few-shot
calibration of set predictors via meta-learned cross-
validation-based conformal prediction. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
46(1):280–291.
Ruan, S., Cang, H., Chen, H., Yan, T., Tan, F., Zhang, Y.,
Duan, L., Xing, P., Guo, L., Gao, P., et al. (2023).
Hyperspectral classification of frost damage stress in
tomato plants based on few-shot learning. Agronomy,
13(9):2348.
Snell, J., Swersky, K., and Zemel, T. R. Prototypical net-
works for few-shot learning.
Tibshirani, R. (2023). Conformal prediction.
Wang, D., Cao, W., Zhang, F., Li, Z., Xu, S., and Wu, X.
(2022a). A review of deep learning in multiscale agri-
cultural sensing. Remote Sensing, 14(3):559.
Wang, X., Vladislav, Z., Viktor, O., Wu, Z., and Zhao, M.
(2022b). Online recognition and yield estimation of
tomato in plant factory based on yolov3. Scientific
Reports, 12(1):8686.
Wightman, R. (2019). Pytorch image models. https:
//github.com/rwightman/pytorch-image-models.
Yang, J., Guo, X., Li, Y., Marinello, F., Ercisli, S., and
Zhang, Z. (2022). A survey of few-shot learning
in smart agriculture: developments, applications, and
challenges.
Zhang, P. and Li, D. (2022). Yolo-volo-ls: a novel method
for variety identification of early lettuce seedlings.
Frontiers in plant science, 13:806878.
Uncertainty Estimation and Calibration of a Few-Shot Transfer Learning Model for Lettuce Phenotyping
397
