
Integrating Retrieval-Augmented Generation with the BioPortal
Annotator for Biological Sample Annotation
Andrea Riquelme-Garc
´
ıa
a
, Juan Mulero-Hern
´
andez
b
and Jesualdo Tom
´
as Fern
´
andez-Breis
c
Departamento de Inform
´
atica y Sistemas, Universidad de Murcia, CEIR Campus Mare Nostrum, IMIB-Pascual Parrilla,
Murcia, 30100, Spain
Keywords:
Large Language Models, Ontologies, Data Interoperability, Bioinformatics.
Abstract:
Integrating biological data remains a significant challenge due to heterogeneous sources, inconsistent formats,
and the evolving landscape of biomedical ontologies. Standardized annotation of biological entities with on-
tology terms is crucial for interoperability and machine-readability in line with FAIR principles. This study
compares three approaches for automatic ontology-based annotation of biomedical labels: a base GPT-4o-mini
model, a fine-tuned variant of the same model, and a Retrieval-Augmented Generation (RAG) approach. The
aim is to assess whether RAG can serve as a cost-effective alternative to fine-tuning for semantic annotation
tasks. The evaluation focuses on annotating cell lines, cell types, and anatomical structures using four on-
tologies: CLO, CL, BTO, and UBERON. The performance was measured using precision, recall, F1-score,
and error analysis. The results indicate that RAG performs best when label phrasing aligns closely with ex-
ternal sources, achieving high precision particularly with CLO (cell lines) and UBERON/BTO (anatomical
structures). The fine-tuned model performs better in cases requiring semantic inference, notably for CL and
UBERON, but struggles with lexically diverse inputs. The base model consistently underperforms. These
findings suggest that RAG is a promising and cost-effective alternative to fine-tuning. Future work will inves-
tigate ontology-aware retrieval using embeddings.
1 INTRODUCTION
The integration of biological data remains a signif-
icant challenge due to heterogeneous data sources,
inconsistent formats, and the continuous evolution
of ontologies, issues particularly pronounced in the
biomedical domain due to data that are often com-
partmentalized by specialty and presented in diverse,
non-interoperable formats (Chaudhari et al., 2024;
Mulero-Hern
´
andez and Fern
´
andez-Breis, 2022; Mor-
ris et al., 2023). Furthermore, the heterogeneity of la-
bels impedes the establishment of connections across
databases, thereby limiting the ability to construct
a comprehensive and unified view of the existing
knowledge.
Annotating biological entities with standardized
ontology terms is a critical step toward improving
data interoperability and supporting the FAIR (Find-
able, Accessible, Interoperable, Reusable) princi-
ples (Wilkinson et al., 2016). This practice enables
a
https://orcid.org/0009-0007-9516-8437
b
https://orcid.org/0000-0002-6818-3443
c
https://orcid.org/0000-0002-7558-2880
machine-readable data representation, which is es-
sential for scalable, automated analysis and effective
reuse (Bernab
´
e et al., 2023). Moreover, ontology-
based annotation reduces redundancy and establishes
semantic connections between datasets, thereby facil-
itating the construction of knowledge graphs and ad-
vancing research in areas such as precision medicine
and systems biology.
Unlike gene symbols or other well-standardized
biological entities, sample annotations frequently suf-
fer from heterogeneous, unstructured, and inconsis-
tent labeling. Samples are often described using
free-text, legacy codes, or community-specific ab-
breviations, which introduce semantic ambiguity and
impede interoperability across databases and studies
(Mulero-Hern
´
andez et al., 2024). This complexity is
exacerbated by the lack of universally adopted stan-
dards for sample annotation and the inherent vari-
ability in experimental protocols and biological con-
texts. For example, the following labels can be found
for the cell line 22Rv1:“22Rv1 delSite4 Clone22/23-
F8”, “2Rv1”, and “22rv1-arvs”. Consequently, these
challenges obstruct the seamless integration and com-
128
Riquelme-García, A., Mulero-Hernández, J. and Fernández-Breis, J. T.
Integrating Retrieval-Augmented Generation with the BioPortal Annotator for Biological Sample Annotation.
DOI: 10.5220/0013740700004000
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2025) - Volume 2: KEOD and KMIS, pages
128-135
ISBN: 978-989-758-769-6; ISSN: 2184-3228
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
parative analysis of sample-related data, despite the
pivotal role that samples play in understanding or-
ganismal function, disease states, and treatment out-
comes.
Traditional annotation tools, such as text2term,
primarily depend on surface-level lexical similarity
and struggle with sparse, ambiguous, or domain-
specific terminology, limiting their effectiveness in
real-world biomedical applications (Gonc¸alves et al.,
2024; Riquelme-Garc
´
ıa et al., 2025).
Large Language Models (LLMs) offer scalable
and modular solutions for annotating natural language
with ontology identifiers (Jahan et al., 2024). Unlike
traditional methods, LLMs leverage contextual se-
mantics for better disambiguation and alignment with
ontologies, enabling improved annotation of samples
even without direct term matches.
In previous work, we explored the use of LLMs
for automating the annotation of biological sample la-
bels to overcome the limitations of existing tools that
rely heavily on lexical similarity (Riquelme-Garc
´
ıa
et al., 2025). That effort demonstrated that fine-
tuned GPT models substantially outperform baseline
and traditional annotation tools, especially in linking
complex ontologies such as CL and UBERON. That
result reinforces the potential of LLM-based meth-
ods to resolve the semantic ambiguity and inconsis-
tency intrinsic to biological sample annotations, thus
enhancing interoperability and data reuse.
However, the fine-tuning process is both compu-
tationally intensive and economically costly, often re-
quiring substantial resources that may not be feasible
for all research settings. As an alternative, this study
explores the use of Retrieval-Augmented Generation
(RAG), a framework that enhances LLM performance
by incorporating external knowledge at inference time
rather than through parameter adjustment. In this con-
text, we leverage the BioPortal Annotator (Jonquet
et al., 2009) as an external knowledge source to guide
the annotation process, enabling the model to retrieve
and utilize relevant ontological information dynami-
cally.
Accordingly, our contribution is an approach that
reduces the dependence on fine-tuning, while main-
taining high annotation accuracy and semantic rele-
vance. This valuable tool and workflow are available
in a dedicated repository and will help researchers to
streamline and enhance the decision-making process
in the task of annotating biological entities.
2 METHODS
2.1 Dataset
For this study, we reused the dataset developed in
a previous work, which consists of 6,264 biological
sample labels collected from 27 publicly available
databases (Riquelme-Garc
´
ıa et al., 2025). These la-
bels were manually classified into three main concept
types: cell lines, cell types, and anatomical struc-
tures, based on their semantic content, although a
small subset remained unclassified due to ambiguity.
To establish a gold standard for evaluation, each label
was manually annotated with terms from four widely
adopted ontologies: Cell Line Ontology (CLO), Cell
Ontology (CL), Uber-anatomy Ontology (UBERON),
and BRENDA Tissue Ontology (BTO), all of which
are part of the OBO Foundry (Smith et al., 2007).
From the complete dataset, a subset of 1,880 labels
was randomly selected and used to evaluate the per-
formance of the RAG method (Table 1).
Table 1: Number of data per concept type in the test data
(mappings text.tsv file). CL: Cell lines, CT: Cell types, A:
Anatomical structures, No concept: label without type of
concept.
Type of concept Number of labels
CL 918
CT 696
A 208
No concept 58
Total 1880
These annotations serve as a reference for
assessing the annotation quality of the pro-
posed method, and are publicly available in the
BiosamplesRAGAnnotation repository (https:
//github.com/andreargr/BiosamplesRAGAnnotation,
“biosamples.tsv” and “mappings test.tsv” files).
2.2 OpenAI GPT Models
In the task of annotating biological sample labels
with ontology identifiers, we investigated the perfor-
mance of the GPT-4o-mini (GPT-4o-mini-2024-07-
18) model under three configurations: base, fine-
tuned, and RAG. All interactions were executed
through the OpenAI API. GPT-4o-mini was selected
due to its optimal balance between computational ef-
ficiency and task performance. The experimental re-
sults for both the base and fine-tuned configurations
were previously reported in earlier work and are in-
cluded in this present study to enable a comparative
analysis with the RAG-based approach (Riquelme-
Garc
´
ıa et al., 2025). In all configurations, prompts
Integrating Retrieval-Augmented Generation with the BioPortal Annotator for Biological Sample Annotation
129

were designed to define the role of the model, specify
the annotation task, format input/output, and enforce
constraints to ensure consistency (prompt included in
GitHub repository). This experimental framework en-
ables a rigorous evaluation of the impact of retrieval-
based augmentation on ontology-based annotation ac-
curacy.
2.3 RAG with Bioportal Annotator
RAG is an approach designed to enhance the per-
formance of LLMs by incorporating external knowl-
edge sources into the inference process. Unlike stan-
dard LLMs, which rely solely on pre-trained parame-
ters to generate responses, RAG architectures retrieve
relevant information from a curated external corpus
at query time and integrate it with the model’s in-
ternal reasoning. This allows the model to access
up-to-date, domain-specific, or otherwise unencoded
knowledge, thereby improving accuracy, contextual
relevance, and factual consistency in generated out-
puts (Ng et al., 2025).
The BioPortal Annotator is a web-based service
developed by the National Center for Biomedical On-
tology designed to facilitate the semantic annotation
of biomedical texts through the mapping of terms
to concepts drawn from an extensive repository of
biomedical ontologies (Jonquet et al., 2009). The sys-
tem operates by detecting ontology concepts within
raw English text using a highly efficient syntactic con-
cept recognition tool that leverages concept names
and synonyms, optionally enhanced by semantic ex-
pansion via hierarchical relationships such as is a as-
sertions.
In the context of ontology annotation, RAG en-
ables the model to consult structured biomedical re-
sources such as BioPortal Annotator during the an-
notation process. This mitigates the limitations as-
sociated with insufficient training data or domain-
specific terminology. In this way, RAG provides a
scalable and interpretable method for bridging gaps
between general-purpose language models and spe-
cialized knowledge domains.
In this study, the BioPortal Annotator was utilized
to generate, for each label, a list of candidate ontology
classes. Subsequently, these candidate classes were
provided to the model, which was tasked with select-
ing the most semantically appropriate class from the
candidates. The model was guided by a structured
prompt designed to ensure consistency and accuracy
in the selection process. The prompt defined a clear
task: given a label referring specifically to biological
samples, such as cell lines, cell types, or anatomical
structures, the model must identify the most suitable
ontology identifier from a specified ontology. The
prompt included representative examples of correctly
formatted identifiers obtained from BioPortal Anno-
tator and imposed strict constraints: the label must
remain unaltered, only a single identifier may be re-
turned, and no explanatory or supplementary content
is permitted. Additionally, the identifier must con-
form to a standardized format and reflect the highest
possible degree of semantic precision. This proce-
dure was systematically applied to each label across
the four ontologies selected for evaluation in the study
(see Figures 1 and 2).
2.4 Evaluation Method
The performance of the model was assessed using two
complementary approaches, following an evaluation
methodology presented and used in previous stud-
ies (Riquelme-Garc
´
ıa et al., 2025). First, the model-
generated annotations were compared against a gold
standard set of human annotations, and standard eval-
uation metrics were computed (see subsection 3.1).
Second, we compared the performance of the RAG
model with the base model and the fine-tuned model
in order to follow a rigorous evaluation of the impact
of RAG on ontology-based annotation accuracy (see
subsection 3.2).
2.4.1 Metrics for the Evaluation of the Model
The performance of the models was evaluated by clas-
sifying each prediction as true positive (TP), false
positive (FP), false negative (FN), or true negative
(TN) based on its correspondence with a gold stan-
dard set of human annotations (Table 2). A predic-
tion was considered a TP if the proposed ontological
identifier exactly matched the reference identifier or
was semantically related to it (e.g., synonymy, sub-
classing, or equivalence). This category also included
cases in which the model proposed a valid identifier
even in the absence of a corresponding annotation in
the gold standard. In contrast, FPs refer to an iden-
tifier that did not correspond to any reference anno-
tation or exhibited an invalid semantic relationship.
FNs occurred when the model failed to propose an
identifier despite the existence of a valid reference
annotation. TNs represent cases where neither the
model nor the gold standard provides an identifier for
the label.
Based on these classifications, the following stan-
dard evaluation metrics were computed:
• Precision measures the proportion of identifiers
proposed by the model that are correct. It reflects
the ability of the model to avoid false positives.
KEOD 2025 - 17th International Conference on Knowledge Engineering and Ontology Development
130
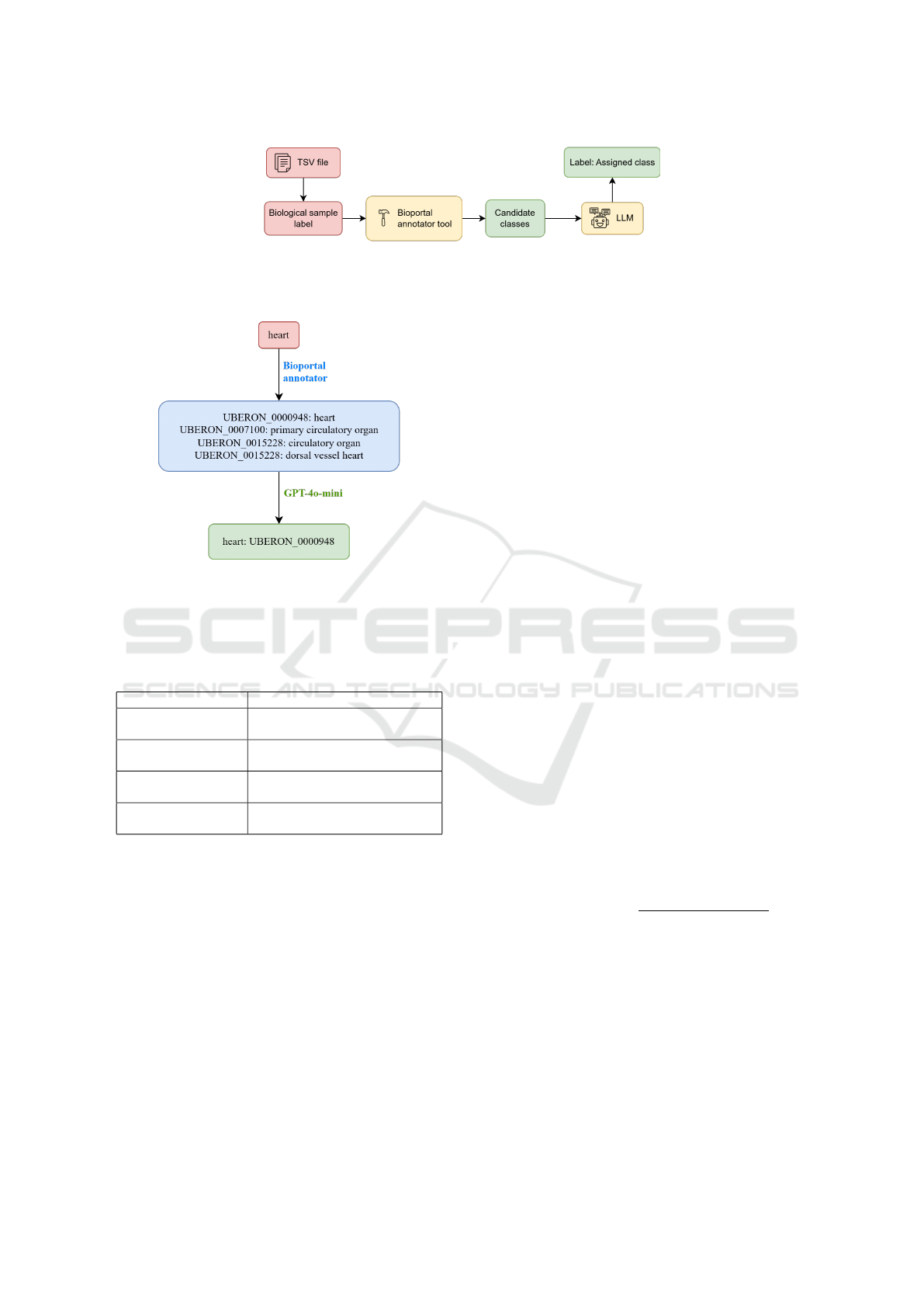
Figure 1: Annotation workflow diagram. The process starts from the extraction of the label from the TSV file, followed by the
use of the BioPortal Annotator to retrieve candidate classes. These candidates are then used to provide context to the language
model, which selects the most appropriate class. A guided example of this flow is shown in Figure 2.
Figure 2: Annotation workflow example. Red box indicates
the input of the user (biological sample), blue box repre-
sents candidate concepts proposed by BioPortal Annotator,
and green box shows the final annotation selected by LLM.
Table 2: Examples of classification outcomes: TP, FP, FN,
and TN.
Category Example
True Positive (TP) Gold standard: ovary.
Model prediction: ovary.
False Positive (FP) Gold standard: ovary.
Model prediction: prostate.
False Negative (FN) Gold standard: ovary.
Model prediction: none.
True Negative (TN) Gold standard: none.
Model prediction: none.
• Recall quantifies the proportion of relevant iden-
tifiers that were successfully retrieved by the
model. It captures the ability of the model to min-
imize false negatives.
• F1-score provides a harmonic mean of precision
and recall, offering a balanced metric that ac-
counts for both correctness and completeness of
the predictions of the model.
• Accuracy assesses the overall proportion of cor-
rect classifications (both positive and negative)
over the total number of cases. It offers a general
measure of the reliability of the model.
These metrics provide a comprehensive and inter-
pretable framework for assessing model performance,
enabling both the quantitative comparison with the
base model and the fine-tuned model.
In line with previous work, the evaluation also
considered the type of concept associated with each
label, as this determines the relevance of each ontol-
ogy. Since the ontologies under study are domain-
specific, their contribution to annotation precision
varies depending on the conceptual category. More-
over, cross-ontology inferences were taken into ac-
count when calculating precision: identifiers from one
ontology can sometimes imply related concepts in
others, though such inferences are constrained by se-
mantic granularity. Based on this reasoning, the eval-
uation considered the most informative ontologies per
label type: CLO and BTO for cell lines, CL and BTO
for cell types, UBERON and BTO for anatomical
structures, and the four ontologies equally when the
label type was not defined.
To account for the inference scenarios described,
the performance of the model was evaluated per con-
cept type and ontology, reflecting their distinct se-
mantic characteristics. In addition to standard met-
rics, a specific measure, the perfect match ratio (1),
was used to assess the ability of the model to pro-
vide comprehensive and contextually appropriate an-
notations. This metric considers whether the model
correctly predicts identifiers for all priority ontologies
associated with a given concept type. For instance, a
perfect match for a cell line label requires accurate
identifiers from both CLO and BTO. The ratio was
computed as the proportion of such cases relative to
the total number of labels of that type.
Perfect match ratio =
N of perfect matches
Total labels of type x
(1)
For labels without a defined concept type, only
precision was reported. For the remaining labels with
a specified type, the evaluation included precision, re-
call, F1-score, accuracy, and perfect match ratio.
2.4.2 Qualitative Error Analysis
To further analyze model performance, a qualita-
tive error analysis was conducted on cases where the
model-generated identifiers did not match the refer-
ence annotations. Importantly, such mismatches do
Integrating Retrieval-Augmented Generation with the BioPortal Annotator for Biological Sample Annotation
131

not necessarily imply incorrect predictions, as some
identifiers may still represent valid alternatives or
broader concepts. The analysis consisted of several
steps: retrieving the class names corresponding to
each identifier, identifying textual or semantic simi-
larities between model and reference outputs, assess-
ing whether the model contributed valid identifiers in
cases where none were provided by the human anno-
tator, and finally, identifying clear errors (FPs) pro-
duced by the model. Only the most relevant ontolo-
gies for each concept type were included in this re-
view. This process enables a more nuanced under-
standing of the model’s behavior, distinguishing be-
tween genuinely incorrect predictions and acceptable
semantic variations.
3 RESULTS
3.1 Metrics and Qualitative Analysis for
the Evaluation of the RAG Model
The results by type of concept associated with the la-
bels are included in Table 3. The TPs in these ta-
bles are derived from three distinct scenarios: (I) the
identifier generated by the model exactly matched the
human reference identifier; (R) the proposed identi-
fier differed from the reference but maintained a valid
semantic relationship with it; and (C) the model pro-
vided a valid identifier for a label in instances where
no human-annotated reference identifier was avail-
able. FPs, by contrast, arose from two main con-
ditions: (1) instances in which the model generated
identifiers that either bear no relation to the refer-
ence or were linked by an incorrect relationship, both
considered model errors (E); and (2) cases where the
model assigned an identifier in the absence of a corre-
sponding human annotation, which were designated
as incorrect contributions (IC).
Table 3a presents the results for cell line label an-
notations. The number of identifiers proposed by the
model that exactly match the human reference iden-
tifiers was significantly higher for the CLO ontology.
Moreover, CLO yielded the highest number of iden-
tifiers that exhibited a valid relationship with the ref-
erence identifier (R), compared to the other ontolo-
gies. However, in cases where the model generated an
identifier despite the absence of a reference identifier,
it frequently produced incorrect outputs, with erro-
neous contributions substantially outnumbering cor-
rect ones. Although CLO demonstrated the high-
est precision among the evaluated ontologies, as il-
lustrated in Figure 3, its precision and accuracy re-
mained relatively modest at 51% and 52%, respec-
tively. These results suggest that, when using the
RAG method, the model can effectively retrieve a
suitable identifier when the label closely resembles
existing external knowledge. Nevertheless, since bi-
ological samples are often labeled with free-text de-
scriptions, discrepancies between the label and exter-
nal knowledge sources can impede accurate cell line
annotation. In the case of the CL and UBERON on-
tologies, the low precision and recall were primarily
attributable to the limitations of the BioPortal Anno-
tator. This tool conducts a literal search and is unable
to infer information from the label. For instance, if the
label is “HeLa”, the tool does not deduce that it cor-
responds to a “uterine epithelial cell”. This inability
to perform inferential mapping represents a key lim-
itation of the annotation tool and affects recall (Fig-
ure 3). Finally, the low precision and recall observed
for the BTO ontology can be attributed to its broad
scope, which encompasses identifiers for cell lines,
cell types, and anatomical structures. This wide cov-
erage introduces ambiguity, increasing the likelihood
of incorrect identifier assignments to labels.
Table 3b presents the results for cell type label an-
notations. In this case, the number of FNs and errors
(E) was notably high for the CL ontology, leading to a
low precision and recall, despite it being specifically
designed for cell type classification (Figure 3). Addi-
tionally, the model frequently resorted to the generic
class “cell” when it failed to identify a more specific
and appropriate identifier. This observation suggests
that the use of free-text descriptions in cell type labels
can complicate the annotation process. As with the
previous case, the BTO ontology exhibited low pre-
cision and recall, which can be attributed to its broad
range of identifiers.
Finally, Table 3c presents the results for anatomi-
cal structure label annotations. In this case, the con-
tributions of the model were fewer than in the pre-
vious tasks, with the majority of them being correct.
Both precision and recall were significantly higher for
the UBERON and BTO ontologies, as shown in Fig-
ure 3. This suggests that anatomical structure labels
are generally more descriptive and human-readable,
which facilitates their interpretation and annotation.
Consequently, this led to a higher perfect match ratio,
with UBERON and BTO, both ontologies specifically
focused on anatomical structures, demonstrating the
highest precision among those evaluated.
3.2 Method Performance Comparison
Figure 4 illustrates the precision of the different meth-
ods across the four ontologies using the same test
dataset. The fine-tuned GPT-4o-mini model outper-
KEOD 2025 - 17th International Conference on Knowledge Engineering and Ontology Development
132
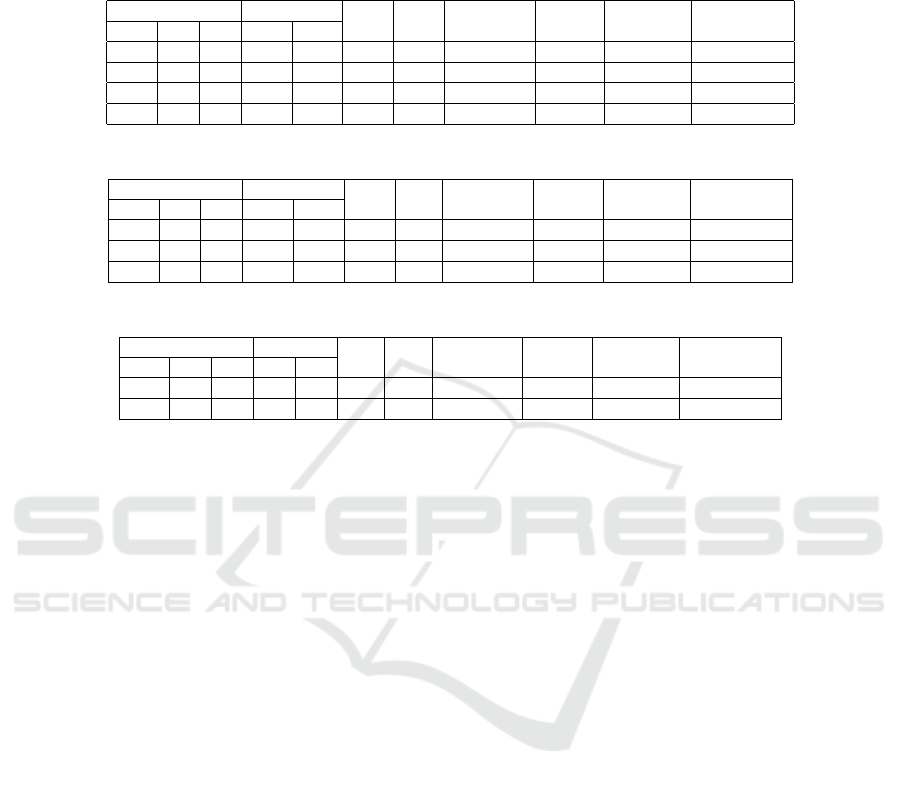
Table 3: Performance metrics of the RAG method for GPT-4o-mini model with BioPortal annotator. TP: True Positives (I:
Identical, R: valid Relation, C: Correct contribution), FP: False Positives (E: Error, IC: Incorrect Contribution), FN: False
Negatives, TN: True Negatives.
(a) Cell line concept annotation
TP FP FN TN Precision Recall F1-score Ontologies
I R C E IC
383 31 33 178 255 8 30 0.51 0.98 0.67 CLO
12 15 0 393 11 479 8 0.06 0.05 0.06 CL
77 10 0 447 19 362 3 0.16 0.19 0.17 UBERON
11 1 43 281 215 232 135 0.10 0.19 0.13 BTO
(b) Cell type concept annotation
TP FP FN TN Precision Recall F1-score Ontologies
I R C E IC
92 55 1 314 2 219 13 0.32 0.40 0.36 CL
137 27 1 392 8 131 0 0.29 0.56 0.38 UBERON
59 23 22 252 226 68 46 0.18 0.60 0.28 BTO
(c) Anatomical structure concept annotation
TP FP FN TN Precision Recall F1-score Ontologies
I R C E IC
131 37 0 40 0 0 0 0.81 1.00 0.89 UBERON
105 19 29 27 12 9 7 0.80 0.94 0.86 BTO
formed both the base model and the RAG method
for the CL and UBERON ontologies. This can be
attributed to the training of the model on domain-
specific data, enabling it to infer information from the
labels. In contrast, the RAG method demonstrated su-
perior performance for the CLO and BTO ontologies,
where the precision of the fine-tuned model was com-
paratively lower. These findings suggest that the lex-
ical diversity and the presence of alphanumeric pat-
terns in biological sample labels pose a greater chal-
lenge for the fine-tuned model in the annotation task,
meanwhile, the RAG method is more effective when
the label is sufficiently similar to external knowl-
edge sources. The base GPT-4o-mini model exhib-
ited low precision across all ontologies. Nevertheless,
the slightly higher precision observed for the CL and
UBERON ontologies suggests that the base model
may have incorporated some knowledge of these two
ontologies during its pretraining.
4 DISCUSSION
This study explored the results obtained from the au-
tomatic annotation of biomedical labels using three
different approaches: a base GPT-4o-mini model, a
fine-tuned version of the same model, and a RAG
method. The evaluation focused on three categories of
biomedical entities: cell lines, cell types, and anatom-
ical structures, across four widely used ontologies:
CLO, CL, BTO, and UBERON. By comparing the
performance of each method in terms of precision,
recall, and error distribution, this study aimed to as-
sess the capacity of models to assign appropriate on-
tology identifiers to free-text labels. The findings pro-
vide insight into the strengths and limitations of each
approach, particularly concerning the complexity and
lexical variability of the labels involved.
The performance of the RAG was evaluated using
standard metrics. The results demonstrated notably
high precision for the CLO ontology in annotating
cell line labels, as well as for the UBERON and BTO
ontologies in annotating anatomical structure labels.
These findings suggest that the RAG method is partic-
ularly effective for these types of entities and ontolo-
gies, and its application is therefore recommended for
annotating anatomical structures and cell lines using
the CLO ontology. However, the annotation of cell
line and cell type labels presented additional chal-
lenges, largely due to the lexical variation and the in-
ability of the BioPortal Annotator to infer semantic
meaning from such labels.
The comparative evaluation of the three annota-
tion methods revealed that no single approach is uni-
versally optimal across all ontologies. The fine-tuned
model demonstrated strong performance when ap-
plied to ontologies where the training data aligned
closely with the label structure, particularly in cases
where inferential reasoning is needed. However, its
performance declined when faced with labels that
contain high lexical variability or alphanumeric iden-
tifiers, as seen in certain cell line annotations. In con-
Integrating Retrieval-Augmented Generation with the BioPortal Annotator for Biological Sample Annotation
133
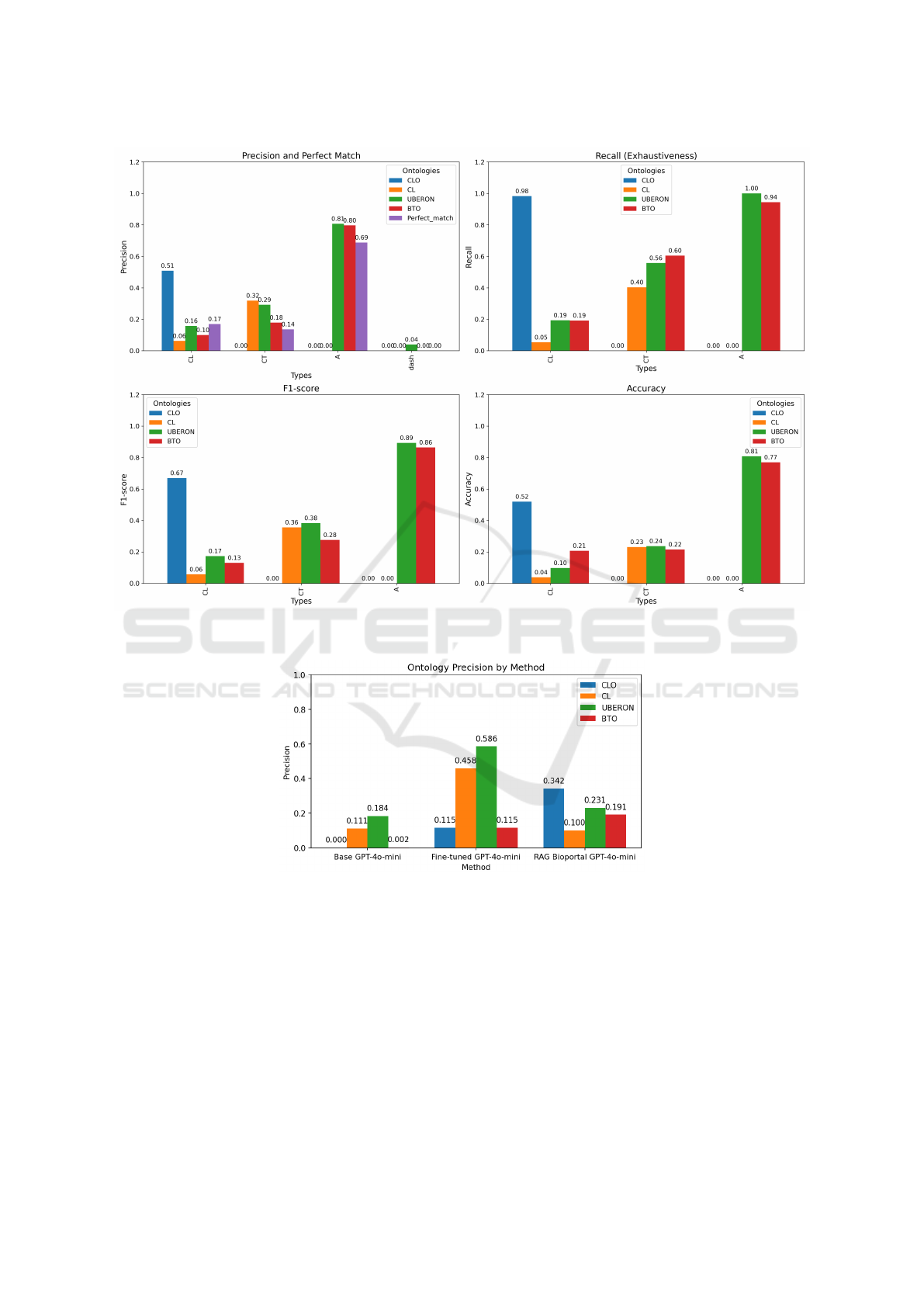
Figure 3: Performance Metrics (Precision, Recall, F1-Score, Accuracy) of GPT-4o-mini with BioPortal RAG for Biomedical
Sample Annotation by type of concept of the label.
Figure 4: Comparison of annotation performance across three methods.
trast, the RAG method proved to be more robust in
such cases, benefiting from its ability to retrieve se-
mantically similar entries from external sources. The
consistently low precision of base model confirmed
the limitations of general-purpose language models
for specialized annotation tasks without adaptation.
These findings underscore the importance of tailoring
annotation strategies to the specific characteristics of
the data and ontology involved and suggest that com-
bining retrieval-based methods with fine-tuning may
offer a promising direction for improving annotation
performance in complex biomedical domains.
A limitation of our approach is the high number
of FPs observed across the three concept types ana-
lyzed. This suggests that, in the absence of a suitable
reference identifier, the model frequently attempts to
assign an ID, often resulting in invalid contributions
that negatively impact precision across all ontologies.
This could be addressed by introducing stricter con-
straints within the prompt to prevent the generation
of inappropriate or non-existent identifiers. Since the
BioPortal Annotator cannot infer semantic informa-
KEOD 2025 - 17th International Conference on Knowledge Engineering and Ontology Development
134

tion from labels, our work was limited in its effec-
tiveness in retrieving appropriate identifiers from cer-
tain ontologies. We propose exploring alternative ap-
proaches based on the RAG framework, in particular,
replacing the BioPortal Annotator with a method that
directly interacts with the ontology structure, such
as leveraging ontology graphs or embeddings, which
may offer a more effective and flexible solution for
identifying relevant terms.
5 CONCLUSIONS
This study presented a comparative evaluation of
three methods: base GPT-4o-mini, a fine-tuned ver-
sion of the same model, and a RAG-based approach,
for the automatic annotation of biomedical labels
using four widely adopted ontologies. The results
demonstrate that the effectiveness of each method
varies depending on the ontology and the nature of the
labels. The fine-tuned model demonstrates strong per-
formance when domain-specific training supports se-
mantic inference, particularly for CL and UBERON.
Conversely, the RAG approach proves more effective
in contexts where label phrasing closely corresponds
to external knowledge sources, as observed with CLO
and BTO in relation to cell lines, and with UBERON
and BTO in the case of anatomical structures. The
limitations of using tools like BioPortal, which lack
semantic inference capabilities, highlight the need for
more flexible and ontology-aware approaches for the
RAG method. Future work will focus on improving
the integration of ontological knowledge within RAG
frameworks to enhance accuracy and generalizability
of automated annotation.
ACKNOWLEDGEMENTS
This research has been funded by MI-
CIU/AEI/10.13039/501100011033/ [grant numbers
PID2020-113723RB-C22, PID2024-155257OB-I00].
REFERENCES
Bernab
´
e, C. H., Queralt-Rosinach, N., Silva Souza, V. E.,
Bonino da Silva Santos, L. O., Mons, B., Jacobsen,
A., and Roos, M. (2023). The use of foundational on-
tologies in biomedical research. Journal of Biomedi-
cal Semantics, 14(1):21.
Chaudhari, J. K., Pant, S., Jha, R., Pathak, R. K., and Singh,
D. B. (2024). Biological big-data sources, problems
of storage, computational issues, and applications: a
comprehensive review. Knowledge and Information
Systems, pages 1–51.
Gonc¸alves, R. S., Payne, J., Tan, A., Benitez, C., Haddock,
J., and Gentleman, R. (2024). The text2term tool to
map free-text descriptions of biomedical terms to on-
tologies. Database, 2024:baae119.
Jahan, I., Laskar, M. T. R., Peng, C., and Huang, J. X.
(2024). A comprehensive evaluation of large lan-
guage models on benchmark biomedical text pro-
cessing tasks. Computers in biology and medicine,
171:108189.
Jonquet, C., Shah, N. H., Youn, C. H., Musen, M. A.,
Callendar, C., and Storey, M.-A. (2009). Ncbo an-
notator: semantic annotation of biomedical data. In
ISWC 2009-8th International Semantic Web Confer-
ence, Poster and Demo Session.
Morris, J. H., Soman, K., Akbas, R. E., Zhou, X., Smith, B.,
Meng, E. C., Huang, C. C., Cerono, G., Schenk, G.,
Rizk-Jackson, A., Harroud, A., Sanders, L., Costes,
S. V., Bharat, K., Chakraborty, A., Pico, A. R.,
Mardirossian, T., Keiser, M., Tang, A., and Baranzini,
S. E. (2023). The scalable precision medicine open
knowledge engine (spoke): A massive knowledge
graph of biomedical information. Bioinformatics,
39(2):btad080.
Mulero-Hern
´
andez, J. and Fern
´
andez-Breis, J. T. (2022).
Analysis of the landscape of human enhancer se-
quences in biological databases. Computational and
Structural Biotechnology Journal, 20:2728–2744.
Mulero-Hern
´
andez, J., Mironov, V., Mi
˜
narro-Gim
´
enez,
J. A., Kuiper, M., and Fern
´
andez-Breis, J. T. (2024).
Integration of chromosome locations and functional
aspects of enhancers and topologically associating do-
mains in knowledge graphs enables versatile queries
about gene regulation. Nucleic Acids Research,
52(15):e69–e69.
Ng, K. K. Y., Matsuba, I., and Zhang, P. C. (2025). Rag in
health care: A novel framework for improving com-
munication and decision-making by addressing llm
limitations. NEJM AI, 2(1):AIra2400380.
Riquelme-Garc
´
ıa, A., Mulero-Hern
´
andez, J., and
Fern
´
andez-Breis, J. T. (2025). Annotation of bi-
ological samples data to standard ontologies with
support from large language models. Computational
and Structural Biotechnology Journal, 27:2155–2167.
Smith, B., Ashburner, M., Rosse, C., Bard, J., Bug, W.,
Ceusters, W., Goldberg, L. J., Eilbeck, K., Ireland, A.,
Mungall, C. J., et al. (2007). The obo foundry: coor-
dinated evolution of ontologies to support biomedical
data integration. Nature biotechnology, 25(11):1251–
1255.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Apple-
ton, G., Axton, M., Baak, A., Blomberg, N., Boiten,
J.-W., da Silva Santos, L. B., Bourne, P. E., Bouwman,
J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Du-
mon, O., Edmunds, S., Evelo, C. T., Finkers, R., and
Mons, B. (2016). The fair guiding principles for sci-
entific data management and stewardship. Scientific
Data, 3(1):160018.
Integrating Retrieval-Augmented Generation with the BioPortal Annotator for Biological Sample Annotation
135
