
Deep Learning-Based Medical Image Segmentation for Brain Tumors
Yudian Pan
a
University of Toronto, Trinity College, Toronto, Canada
Keywords: Deep Learning, Segmentation, Medical Image, U-Net.
Abstract: Brain tumors present a considerable health challenge, dramatically impacting both survival and quality of life.
This study introduces a improved deep learning approach for segmenting brain tumors in MRI scans,
intending to overcome the constraints of those existing approaches. The proposed model builds upon the
conventional U-Net architecture by incorporating the Convolutional Block Attention Module (CBAM),
designed to enhance the feature extraction capabilities. By integrating both channel-wise and spatial attention
mechanisms, this approach emphasizes relevant tumor regions while preserving structural detail. Experiments
evaluations on the TCGA Brain Tumor MRI dataset confirm the remarkable advantages of our UNet+CBAM
model compared to baseline approaches, achieving a Dice coefficient of 0.936 and an IoU of 0.882. This
proposed model successfully captures tumor boundaries with high precision and provides detailed
segmentation maps that could assist clinical diagnosis. While acknowledging the challenges posed by
computational complexity, this study makes a significant contribution to the advancement of automated brain
tumor segmentation technology, which holds considerable potential for practical applications in medical
settings. Subsequent studies will prioritize the optimization of the model for real-time applications and the
enhancement of its generalizability across a range of clinical settings.
1 INTRODUCTION
Brain tumors refer to the uncontrolled growth of cells
within brain tissue and its surrounding structures,
greatly impacting both survival rates and patients’
quality of life. Their incidence is on the rise globally,
especially in developed countries. Due to the
significant differences in biological characteristics,
clinical manifestations and treatment responses, early
and accurate diagnosis and precise treatment are
always a challenge. MRI has become a major tool for
brain tumor detection and evaluation due to its
advantages in soft tissue imaging, which can present
rich tumor information through different sequences,
such as T1, T2, FLAIR, etc., but the complexity and
heterogeneity of brain tumors in images still pose a
challenge in accurate segmentation and analysis.
segmentation and analysis difficulties (Litjens & van
Ginneken, 2017).
Traditional methods (e.g., threshold segmentation,
edge detection, region growing, etc.) are often
difficult to balance accuracy and stability when
dealing with brain tumors with different
a
https://orcid.org/0009-0007-9429-7165
morphologies; machine learning can improve some of
the performance, but it relies too much on hand-
designed features, making it difficult to fully explore
high-level information (Ronneberger & Brox, 2015).
Deep learning has achieved breakthroughs in the field
of medical image analysis by leveraging the
automatic feature extraction and pattern recognition
capabilities of multi-layer neural networks recently.
Among these, U-Net and its 3D variant have largely
improved segmentation accuracy and spatial detail
capture of biomedical images by virtue of the
advantages of multi-scale feature extraction, hopping
connectivity and 3D convolution (Çiçek &
Ronneberger, 2016). Related studies, such as the
work of DeepSeg and Amin et al (Amin & Hassan,
2023), have not only excelled in automated
segmentation of brain tumors, but also promoted the
application of multi-classification and segmentation
of MRI images (Zeineldin & Burgert, 2020).
This study aims to propose an efficient and
accurate brain tumor image segmentation approach
utilizing deep learning. Firstly, advantages and
disadvantages of existing mainstream models are
systematically sorted out and compared through
340
Pan, Y.
Deep Learning-Based Medical Image Segmentation for Brain Tumors.
DOI: 10.5220/0013689700004670
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 2nd International Conference on Data Science and Engineering (ICDSE 2025), pages 340-347
ISBN: 978-989-758-765-8
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

literature research to provide a theoretical basis for
subsequent model design (Ronneberger & Brox,
2015). Secondly, by combining multi-scale feature
extraction with multi-modal information fusion, the
paper proposes an improved network structure and
training strategy to adapt to complex tumor
morphology and improve robustness, while exploring
lightweight and modular design to enhance real-time
performance (Gupta & Dayal, 2023). Again, data
augmentation, migration learning and semi-
supervised learningare helped address the issues
regarding limited labelled data, enhancing the
model’s generalization capacities (Litjens & van
Ginneken, 2017). Finally, large-scale experimental
validation is conducted on different datasets and
analysed in comparison with existing methods in
order to deeply explore its potential in clinical
applications (Isensee & Maier-Hein, 2019).
Key contributions of this research are mainly
manifested as follows:
1. Model Improvement: The number of layers and
parameters of the traditional U-Net architecture are
adjusted to enhance the recognition ability of
complex morphology of brain tumors by adding
convolutional and pooling layers.
2. Data experiments: Using the public brain tumor
MRI dataset, after data preprocessing and
enhancement, the improved U-Net is used for image
segmentation experiments.
3. Performance Comparison: The enhanced model
is compared to the original U-Net in brain tumor
segmentation, with evaluation metrics including
segmentation accuracy, Dice coefficient and
intersection ratio.
4. Discussion of results: experimental outcomes
are thoroughly analyzed, summarizing this model’s
advantages and shortcomings in terms of practical
applications, which provides a reference basis for
subsequent research.
2 RELATED WORKS
2.1 Overview of Traditional Methods
for Brain Tumor Segmentation
Early approaches to brain tumor segmentation over
the past few decades mostly depended on traditional
image processing techniques, including edge
detection, region growth, threshold segmentation, and
segmentation methods based on graph theory
. These
methods use manual features such as image grayscale
and texture to analyze the brain structure, which can
achieve a certain degree of segmentation effect in
simple cases, but when the tumor morphology is
complex or the tissue structure is more ambiguous, it
is often difficult for traditional methods to meet the
requirements of high precision. In addition,
traditional methods are more sensitive to the noise in
the image and are easily disturbed, resulting in
unstable segmentation results (Litjens & van
Ginneken, 2017). Therefore, in practical applications,
the robustness and automation level of these methods
need to be improved.
2.2 Discussion of Recent Advancements
Using Deep Learning
The advancement of deep learning technology have
triggered more researchers to begin to adopt
convolutional neural networks to address brain tumor
segmentation. Since U-Net was proposed, its
symmetric encoder-decoder structure and jump
connection design have greatly improved the
accuracy of image segmentation (Ronneberger &
Brox, 2015). To further process 3D medical images,
its variation has also been proposed and successfully
applied to volumetric data segmentation (Çiçek &
Ronneberger, 2016). Building on this, many scholars
have extensively explored the role of multi-scale
feature extraction along with attention mechanism
they have play in enhancing segmentation
performance. For example, the DeepSeg framework
developed by Zeineldin et al. achieved automatic
segmentation using FLAIR images (Zeineldin &
Burgert, 2020); Amin et al. and Gupta et al. put forth
a new network architecture for multiclass
segmentation of brain tumors (Amin & Hassan, 2023)
(Gupta & Dayal, 2023); meanwhile, Díaz-Pernas et al.
further optimized the segmentation accuracy using
multiscale convolutional neural networks. In addition,
Roy et al, Woo et al., and Fu et al. Improved the
network's ability to focus on key features by
introducing an attention module (Roy et al. 2018)
(Woo & Kweon, 2018) (Fu & Lu, 2019); while No
New-Net proposed by Isensee et al still maintains
high segmentation performance while simplifying the
network structure (Isensee & Maier-Hein, 2019).
These works show that deep learning-based methods
have achieved some huge progresses in the field of
brain tumor segmentation, demonstrating a strong
capacity to access fine lesion details.
2.3 Limitations of Existing Methods
However, despite their excellent performance in
segmentation accuracy, deep learning methods still
have some shortcomings. First, existing approaches
typically require an extensive amount of high-quality
Deep Learning-Based Medical Image Segmentation for Brain Tumors
341

labelled data, which is often scarce and expensive to
obtain in clinical practice (Litjens & van Ginneken,
2017). Second, some of the network structures are too
complex and computationally intensive, resulting in
long training time and high hardware requirements,
which is not conducive to generalization to practical
applications (Isensee & Maier-Hein, 2019). In
addition, although the attention mechanism can
improve the segmentation effect, it also increases the
parameters of the model and the training difficulty
(Roy et al. 2018; Woo & Kweon, 2018; Fu & Lu,
2019). There are also methods that are prone to miss
or missegmentation when dealing with edge details
and small-sized lesions (Zeineldin & Burgert, 2020;
Amin & Hassan, 2023; Gupta & Dayal, 2023).
Overall, how to improve this new model’s
lightweight and generalization capacities, and also
ensure the segmentation accuracy is still a difficult
problem to be solved.
2.4 Highlighting the Research Gap that
the Paper Addresses
This paper centers on improving current brain tumor
segmentation methods. Firstly, data enhancement
strategies (e.g. random horizontal flipping and
rotation) are used to extend the labelled data and
improve the model robustness (Feng & Wu, 2021).
Second, an improved U-Net network is designed to
combine multi-scale feature fusion and attention
mechanisms to enhance detail capture and maintain
lightweight (Ronneberger & Brox, 2015). The cross-
entropy loss function, Adam optimiser, and the
combination of pixel accuracy and IOU metrics are
used in training to evaluate the model, balancing
segmentation effect and computational efficiency
(Zeineldin & Burgert, 2020). In conclusion, this paper
fills the gap of brain tumor segmentation in terms of
data utilization, model design and application,
improves accuracy and efficiency, and is helpful for
clinical diagnosis (Isensee & Maier-Hein, 2019).
3 METHOD
3.1 Description of the Proposed Deep
Learning Approach
The method of deep learning proposed is mainly
developed based upon the improved U-Net
architecture, which realizes the fine segmentation of
medical images (e.g., brain MRI images) by
constructing a symmetric encoder-decoder network.
In the encoder part, a series of downsampling
modules consisting of convolution and ReLU
activation are utilized to gradually extract the multi-
scale features in the image, and at the same time, the
maximum pooling operation is used to reduce the
spatial size of the feature map; and in the decoder
section, the corresponding layers in the encoder are
fused with the up-sampled features in the decoder
through hopping connection, and then the image
resolution is recovered gradually by using the
transposed convolution, so as to retain more spatial
detail information. Additionally, to improve this new
model’s generalization ability and robust traits, data
enhancement strategies, for example, random level
flipping and random rotation, are introduced during
the training process, so as to expand the diversity of
training samples. Whole network is trained with the
cross-entropy loss function; besides the Adam
optimizer is used to continuously update the
parameters, which ultimately achieves high-precision
segmentation of the target region in medical images.
3.2 Network Architecture (U-Net)
The foundational UNet architecture is a widely
recognized and utilized model in the domain of
medical image segmentation. Introduced by
Ronneberger et al. (2015), UNet features a symmetric
encoder-decoder structure, which
comprises a
symmetric extending pathway for accurate
positioning and a contracted pathway for contextual
acquisition. The encoder path uses convolutional
layers with ReLU activation, followed by max-
pooling for downsampling, whereas the decoder path
recovers the spatial dimensions using convolutional
layers and upsampling, as seen in Table 1.
ICDSE 2025 - The International Conference on Data Science and Engineering
342
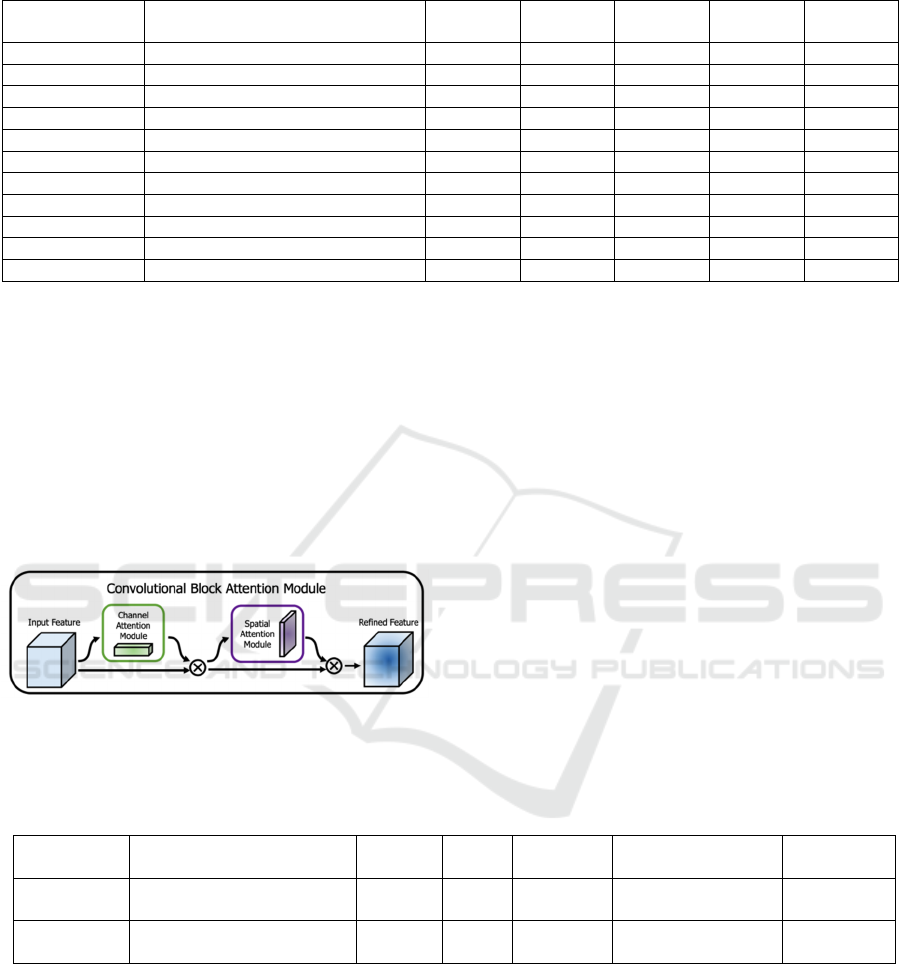
Table 1: The UNet model.
Layer Name Type Kernel
Size
Stride Padding Output
Channels
Activatio
n
Input - - - - 3 -
Downsample 1 Conv + ReLU + Conv + ReLU 3x3 1 1 64 ReLU
Downsam
p
le 2 Conv + ReLU + Conv + ReLU 3x3 1 1 128 ReLU
Downsam
p
le 3 Conv + ReLU + Conv + ReLU 3x3 1 1 256 ReLU
Downsam
p
le 4 Conv + ReLU + Conv + ReLU 3x3 1 1 512 ReLU
Downsample 5 Conv + ReLU + Conv + ReLU 3x3 1 1 1024 ReLU
Upsample Transpose Conv + ReLU 3x3 2 1 512 ReLU
Upsample 1 Conv + ReLU + Conv + ReLU 3x3 1 1 512 ReLU
U
p
sam
p
le 2 Conv + ReLU + Conv + ReLU 3x3 1 1 256 ReLU
U
p
sam
p
le 3 Conv + ReLU + Conv + ReLU 3x3 1 1 128 ReLU
Output Conv 1x1 1 0 2 Softmax
A sophisticated attention mechanism known the
Convolutional Block Attention Module (CBAM)
improves feature representation by using channel and
spatial attention in an orderly way. The Channel
Attention Module (CAM) and Spatial Attention
Module (SAM) are the two major parts of CBAM. As
illustrated in Figure 1, the spatial attention
mechanism works on “where” an essential feature is
situated, but the channel attention mechanism
concentrates on “what” is significant in a particular
feature map.
Figure 1: CAM framework (Picture credit: Original).
Mathematically, the Channel Attention Module
(CAM) can be described as formula (1):
𝑀
𝐹
=𝜎
𝑀𝐿𝑃
𝐴
𝑣𝑔𝑃𝑜𝑜𝑙
𝐹
𝑀𝐿𝑃𝑀𝑎𝑥𝑃𝑜𝑜𝑙
𝐹
(1)
The input feature map is represented by ( 𝐹), the
sigmoid function is shown by ( 𝜎), and the multi-
layer perceptron is referred to as MLP. The
operations for average-pooling and max-pooling are
denoted by AvgPool and MaxPool, respectively.
The Spatial Attention Module (SAM) can be
described as formula (2):
𝑀
𝐹
=𝜎
𝑓
×
𝐴
𝑣𝑔𝑃𝑜𝑜𝑙
𝐹
;𝑀𝑎𝑥𝑃𝑜𝑜𝑙
𝐹
(2)
Notably, (𝑓
×
) denotes a convolution operation
with a filter size of 7x7, and [;] here represents the
operation of concatenation.
Table 2: The structure of the CBAM module.
Layer Name Type Kernel
Size
Stride Padding Output Channels Activation
Channel
Attention
AdaptiveAvgPool + Conv +
ReLU + Conv
1x1 1 0 in_channels // ratio,
in_channels
Sigmoid
Spatial
Attention
Conv 7x7 or
3x3
1 3 or 1 2, 1 Sigmoid
The integration of the CBAM module into the
UNet architecture involves embedding the CBAM
module after each convolutional block in both the
downsampling and upsampling paths. Such an
integration allows the network to refine its focus on
the most informative features and regions at multiple
stages of the learning process. The CBAM module’s
structure is displayed in Table 2.
In the improved UNet+CBAM model, each
downsampling block involves convolutional layers
followed by a CBAM module, and similarly, each
upsampling block includes convolutional layers
followed by a CBAM module. Through this
connection, this model’s capacity to recognise
intricate correlations and characteristics in the
medical images becomes stronger, and this leads to an
increase in the precision of segmentation, as shown in
the data below in Table 3.
Deep Learning-Based Medical Image Segmentation for Brain Tumors
343
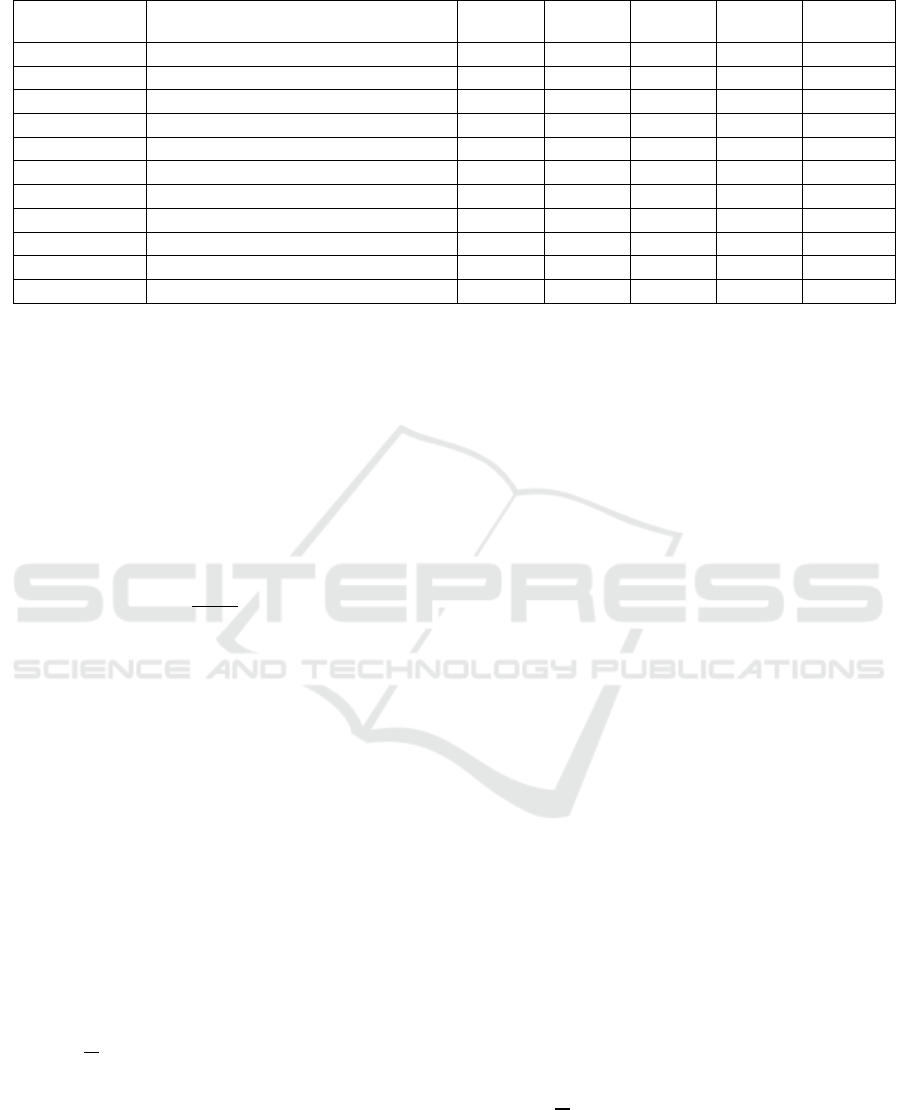
Table 3: This caption needs to be modified to justify as it includes many lines.
Layer Name Type Kernel
Size
Stride Padding Output
Channels
Activation
Input - - - - 3 -
Downsample 1 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 64 ReLU
Downsample 2 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 128 ReLU
Downsample 3 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 256 ReLU
Downsample 4 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 512 ReLU
Downsample 5 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 1024 ReLU
Upsample Transpose Conv + ReLU 3x3 2 1 512 ReLU
Upsample 1 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 512 ReLU
Upsample 2 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 256 ReLU
Upsample 3 Conv + ReLU + Conv + ReLU + CBAM 3x3 1 1 128 ReLU
Output Conv 1x1 1 0 2 Softmax
3.3 Preprocessing Techniques
Before feeding the images into the network, this work
employs multiple types of methods for preprocessing
steps to standardize the input data. Images are first
resized to a fixed dimension (e.g., 256×256). In
addition, intensity normalization is applied to adjust
the pixel values to contain zero mean and unit
variance. The normalization formula is as formula (3):
𝑥=
𝑥−𝜇
𝜎
(3)
In this equation, 𝑥 is the original pixel value, 𝜇 is
the mean, and 𝜎 is the standard deviation computed
over all pixels of the image. This normalization step
improves the convergence of the network during
training.
segmentation masks. The dataset is denoted by
formula (4):
𝐷=
𝑥
,𝑦
(4)
Where 𝑥
represents the 𝑖 th input image; 𝑦
its
related ground truth mask, with N being the total
number of samples. The training process is guided by
the cross-entropy loss function, being defined as
formula (5):
𝐿=−
1
𝑁
𝑦
𝑙𝑜𝑔 𝑙𝑜𝑔 𝑝
(5)
Notably, 𝐶denotes the number of classes, 𝑦
is
the true label for class 𝑐 (usually 0 or 1), and 𝑝
is
the predicted probability for that class. The paper
optimizes the network parameters by means of the
Adam optimizer with a learning rate typically set
to 1×10
. This combination of loss function and
optimizer has proven effective in training deep
segmentation models.
3.4 Dataset Description
The TCGA Brain Tumor MRI dataset is derived from
the TCGA (The Cancer Genome Atlas) project.
Which collects brain MRI images from real clinics
and provides the corresponding tumor segmentation.
Due to the wide range of data sources, the samples
cover different types, sizes and morphologies of brain
tumors, making this dataset an important benchmark
data in the segmentation of medical images. The
diversity of the data also contributes to this model’s
increased resilience and generalization performance.
Researchers can use this dataset to train and verify
deep learning models for better automatic
identification and fine segmentation of brain tumor
lesions.
3.5 Loss Function, Optimizer, and
Learning Rate
In this study, the paper uses the cross-entropy loss
function as the optimization objective for the
segmentation task. The cross-entropy loss measures
the discrepancy between the predicted probability
distribution and the true label distribution. Its formula
is given by equation (6):
𝐿=−
1
𝑁
𝑦
𝑙𝑜𝑔 𝑙𝑜𝑔 𝑝
(6)
Here, N denotes the total number of samples, C
represents the number of classes, 𝑦
is the true label
ICDSE 2025 - The International Conference on Data Science and Engineering
344
(either 0 or 1) for class c in the ith sample, and 𝑝
is
the probability predicted by the model for that class.
To efficiently update the model parameters, this paper
resorts to the Adam optimizer, which adaptively
adjusts learning rate during training. This paper’s
experiments reveal that the initial learning rate is
designed to 𝑏𝑒 1 × 10
and is fine-tuned based on
validation results to balance training stability and
model performance.
3.6 Evaluation Metrics for
Segmentation Performance
The Dice coefficient is used through the paper to
assess our segmentation model’s performance. The
area that overlaps between the ground truth and the
predicted segmentation zone is measured by this
statistic. Formulas (7) and (8) are applied to calculate
the dice coefficient:
𝐼𝑜𝑈 =
|
𝑃∩𝐺
|
|
𝑃∪𝐺
|
(7)
𝐷𝑖𝑐𝑒 =
2×
|
𝑃∩𝐺
|
|
𝑃
|
|
𝐺
|
(8)
The number of pixels in each set is indicated by the
formula, where P stands for the set of pixels in the
predicted segmentation and G for the set of pixels in
the ground truth mask. A greater match between the
forecast and the ground truth is demonstrated by a
larger value of the Dice coefficient, which spans from
0 to 1.
4 RESULTS
4.1 Performance Metrics Compared
with State-of-the-art Methods
Comparative tests on several popular models for the
brain tumor segmentation had been carried out in the
paper. The experiments compared the traditional
UNet base model, DeepLabV3, PSPNet, and MA-Net,
and on the basis of which the CBAM module was
embedded into UNet to form the UNet+CBAM (Full)
model. According to the experimental findings, the
IoU is 0.854 and the Dice coefficient of the UNet base
model is 0.915, respectively; the Dice coefficient of
DeepLabV3 is 0.924, and the IoU is 0.866; the Dice
coefficient of PSPNet is 0.920, and the IoU is 0.861;
the Dice coefficient of MA-Net is 0.928, and the IoU
is 0.873; while our proposed UNet+CBAM (Full)
model achieves a Dice coefficient of 0.936 and an
IoU of 0.882. From those data, it is evident that the
model’s segmentation effect has greatly enhanced
when the CBAM module is integrated. Table 4 below
summarizes the findings of the comparison
experiments:
Table 4: Main results.
Model Dice Score IoU
UNet Base 0.915 0.854
Dee
p
LabV3 0.924 0.866
PSPNet 0.920 0.861
MA-Net 0.928 0.873
UNet+CBAM (Full) 0.936 0.882
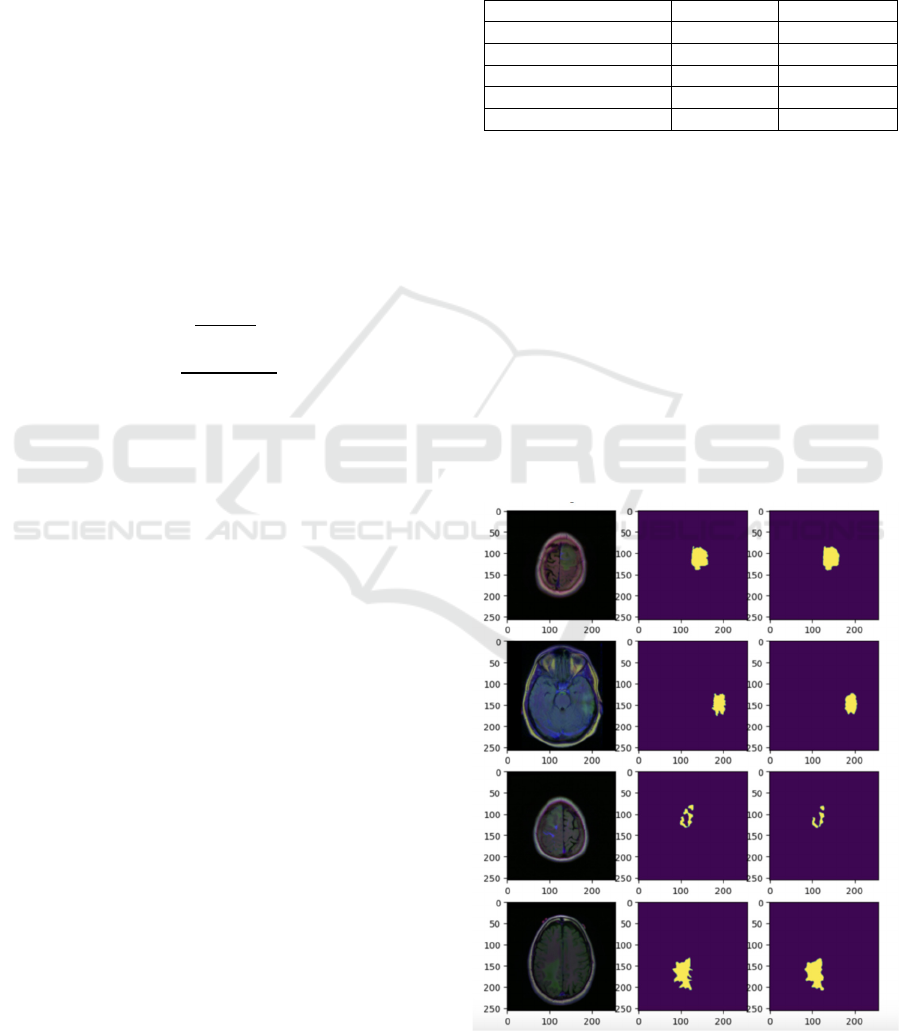
4.2 Visualizations of Segmentation
Outputs
The segmentation findings of the final model are
illustrated in this paper. The model can precisely
identify the region of the brain tumor with distinct
segmentation borders and detailed features, as shown
in Figure 2 below. Both the overall contour of the
tumor and the internal structural details are well
captured by the model and restored on the output
image. These visualization results fully demonstrate
the good performance of the model in the actual
segmentation task, as well as providing intuitive
support for subsequent clinical aid diagnosis, as
shown in Figure 2.
Figure 2: Main result (Picture credit: Original).
Deep Learning-Based Medical Image Segmentation for Brain Tumors
345

4.3 Analysis and Interpretation of
Results
From the indicators of the segmentation results, the
final model shows high accuracy and robustness. The
model’s ability to precisely identify and segment the
tumor region under a range of conditions is
demonstrated by the high degree of overlap between
the actual annotations and predicted findings shown
in the images. By observing the segmentation results,
the paper find that the model is particularly
meticulous in processing the edges of the tumor
region, and is able to effectively capture the subtle
changes of the lesion. This fully demonstrates that the
designed network structure and attention module play
an active role in feature extraction and information
fusion, making this model highly effective and
reliable in segmentation in practical applications.
4.4 Discussion of Potential limitations
Although the final model performs exceptionally well
in the segmentation task, there might still be
limitations. First, although the dataset used is highly
representative, more cases with different scanning
devices, different imaging parameters and variable
image quality may be encountered in practical
applications, which may have some impact on the
model performance. Secondly, the computational
complexity of the model is high, and although it runs
smoothly in the experimental environment, the real-
time and hardware requirements still need to be
further considered in practical clinical applications.
The adaptability and operational efficiency of the
model can be further improved in future work through
model pruning, lightweight design, and more data
enhancement means.
5 CONCLUSIONS
A brain tumor segmentation method based on
improved U-Net and attention mechanism is
proposed in this paper. Experiments show that the
new model has significant improvement in both Dice
coefficient and IoU, and the segmented tumor region
has clear edges and rich details. The method not only
reduces the workload of manual labelling, but also
provides an intuitive segmentation reference for
doctors, which has certain clinical application
potential.
Meanwhile, the study also exposed some
shortcomings. The current dataset has limited sample
sources and the model is computationally large,
which may require further optimisation in real
scenarios. In the future, the paper can try to introduce
more data, adopt semi-supervised or migration
learning methods, or design a more lightweight
network structure to boost the stability and operation
efficiency of this model.
Overall, this paper provides a new idea for
automatic brain tumor segmentation, and the paper
expect that it will play a greater role in clinical
practical applications in the future.
REFERENCES
Amin, B., Samir, R. S., Tarek, Y., Ahmed, M., Ibrahim, R.,
Ahmed, M., & Hassan, M. 2023. Brain tumor multi-
classification and segmentation in MRI images using
deep learning. arXiv preprint arXiv:2304.10039.
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., &
Ronneberger, O. 2016. 3D U-Net: Learning dense
volumetric segmentation from sparse annotation. In
International Conference on Medical Image Computing
and Computer-Assisted Intervention (424–432).
Springer, Cham.
Díaz-Pernas, F. J., Martínez-Zarzuela, M., Antón-
Rodríguez, M., & González-Ortega, D. 2024. A deep
learning approach for brain tumor classification and
segmentation using a multiscale convolutional neural
network. arXiv preprint arXiv:2402.05975.
Feng, R., Liu, X., Chen, J., Chen, D. Z., Gao, H., & Wu, J.
2021. A deep learning approach for colonoscopy
pathology WSI analysis: Accurate segmentation and
classification. IEEE Journal of Biomedical and Health
Informatics, 25(10), 3700–3708.
Fu, J., Liu, J., Tian, H., Li, Y., Bao, Y., Fang, Z., & Lu, H.
2019. Dual attention network for scene segmentation.
In Proceedings of the IEEE/CVF Conference on
Computer Vision and Pattern Recognition (3146-3154).
Gupta, A., Dixit, M., Mishra, V. K., Singh, A., & Dayal, A.
2023. Brain tumor segmentation from MRI images
using deep learning techniques. arXiv preprint
arXiv:2305.00257.
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., &
Maier-Hein, K. H. 2019. No new-net. In International
MICCAI Brainlesion Workshop (234–244). Springer,
Cham.
Kamnitsas, K., Ledig, C., Newcombe, V. F., Simpson, J. P.,
Kane, A. D., Menon, D. K., … & Glocker, B. 2017.
Efficient multi-scale 3D CNN with fully connected
CRF for accurate brain lesion segmentation. Medical
Image Analysis, 36, 61–78.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., … & van Ginneken, B.
2017. A survey on deep learning in medical image
analysis. Medical Image Analysis, 42, 60–88.
ICDSE 2025 - The International Conference on Data Science and Engineering
346

Ronneberger, O., Fischer, P., & Brox, T. 2015. U-Net:
Convolutional networks for biomedical image
segmentation. arXiv preprint arXiv:1505.04597.
Roy, A.G., Navab, N., Navab, N., et al. 2018. Concurrent
spatial and channel squeeze and excitation in fully
convolutional networks. In: Proceedings of the medical
image computing and computer-assisted intervention,
Singapore, MICCAI 2018, 421–429.
Woo, S., Park, J., Lee, J.-Y., & Kweon, I. S. 2018. CBAM:
Convolutional Block Attention Module. In:
Proceedings of the European Conference on Computer
Vision (ECCV).
Zeineldin, R. A., Karar, M. E., Coburger, J., Wirtz, C. R.,
& Burgert, O. 2020. DeepSeg: Deep neural network
framework for automatic brain tumor segmentation
using magnetic resonance FLAIR images. arXiv
preprint arXiv:2004.12333.
Deep Learning-Based Medical Image Segmentation for Brain Tumors
347
