
Comparative Analysis of Learning Strategies for Multi-Magnification
Pathological Image Classification
Yixuan Pu
a
School of Electrical and Computer Engineering, The University of Sydney, New South Wales, Australia
Keywords: Colorectal Histopathology, Multi-Channel, Stepwise Learning.
Abstract: The automatic classification of pathology images plays a crucial role in computer-aided diagnosis by
enhancing diagnostic efficiency and minimizing human error. In this paper, the Enteroscope Biopsy
Histopathological H&E Image Dataset (EBHI) is utilized to systematically compare and analyze the
performance of three strategies—Single-Magnification Training, Multi-Channel Fusion, and Stepwise
Cumulative Learning—to optimize pathology image classification. The Single-Magnification Training
strategy serves as a baseline experiment to validate the optimization effect of the model, achieving the highest
classification accuracy of 94.64% at 200× magnification. Under strict filtering conditions, Multi-Channel
Fusion achieves a peak classification accuracy of 96.06%. However, this approach remains inferior to
Stepwise Cumulative Learning. This learning strategy significantly outperforms training solely at the highest
magnification, achieving a classification accuracy of 98.27% on 400× images. This study demonstrates that
the cumulative learning strategy effectively enhances the classification performance of pathology images.
Low-magnification images contribute to improving the classification accuracy of high-magnification images,
offering new insights into multi-scale feature fusion, dynamic learning strategies, and computer-aided
pathology diagnosis. Furthermore, this study validates the applicability of the EBHI dataset in multi-
magnification pathology analysis and advances the development of intelligent pathology image analysis.
1 INTRODUCTION
Early diagnosis of colorectal cancer is crucial for
reducing its high morbidity and mortality, with
pathological image analysis remaining the gold
standard for diagnosis. Traditional pathology analysis
relies on the manual evaluation of tissue sections by
pathologists, a process that is not only time-
consuming and labor-intensive but also prone to
subjective bias. Consequently, leveraging advanced
technologies to enhance the efficiency and accuracy
of pathological image analysis has become a focal
point in contemporary medical research.
In recent years, deep learning techniques have
provided an efficient and accurate solution for the
automatic classification and detection of pathology
images by automatically extracting image features.
Current research focuses on the following three key
areas: optimizing deep learning models, improving
data preprocessing and handling class imbalance, and
integrating multi-magnification information.
a
https://orcid.org/0009-0003-9188-1252
Firstly, in terms of model optimization, numerous
studies have sought to enhance the accuracy of
pathology image classification by refining deep
learning architectures. Khan et al. (2024) proposed a
Swin Transformer-based approach that leverages the
self-attention mechanism for feature extraction in
pathology images. By incorporating normalized
preprocessing, their method significantly improves
classification accuracy. Kim et al. (2021)
systematically compared the performance of
Convolutional Neural Network (CNN), Residual
Neural Network (ResNet), and Vision Transformer
(ViT) in pathology image classification. However,
these studies primarily focused on optimizing a single
model and did not integrate multi-magnification
information.
Secondly, in terms of data preprocessing and class
imbalance handling, studies have demonstrated that
appropriate data augmentation and class balancing
strategies can enhance classification performance.
Malik et al. (2019) investigated the impact of various
222
Pu, Y.
Comparative Analysis of Learning Strategies for Multi-Magnification Pathological Image Classification.
DOI: 10.5220/0013681400004670
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 2nd International Conference on Data Science and Engineering (ICDSE 2025), pages 222-229
ISBN: 978-989-758-765-8
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

preprocessing methods on pathology image
classification and found that data augmentation
improves the model’s generalization ability. Raju and
Rao (2022) addressed the issue of class imbalance by
proposing a deep learning framework that integrates
Class-Balanced Loss with data augmentation,
enabling the model to better recognize minority class
samples. Although these approaches improve model
stability to some extent, most existing methods are
designed for single-magnification images and fail to
fully exploit the potential complementary information
across different magnifications.
Finally, in the fusion of multi-magnification
information, researchers have explored various
approaches to integrating different magnifications to
enhance the classification accuracy of pathology
images. Das et al. (2017) employed Majority Voting
Fusion by independently training a CNN model at
different magnifications and fusing classification
results from multiple perspectives at the inference
stage, thereby improving the overall classification
accuracy of full-slide images. However, this method
treats different magnifications as independent
information sources and fails to fully model the
hierarchical relationship between them, making it less
effective in simulating the gradual magnification
process that pathologists naturally follow in real-
world diagnosis.
This study aims to develop and validate an
efficient pathology image classification model based
on deep learning, explore optimization strategies for
different magnifications, and conduct an in-depth
investigation into data missing issues, magnification
combination methods, and multi-magnification
learning. The main innovations of this study include:
1) Validate and improve the single-magnification
training method and establish baseline
experiments;
2) Investigate the applicability of the 12-channel
fusion model and compare different data
imputation strategies;
3) Propose a cumulative magnification learning
strategy based on a progressive training
sequence.
2 MATERIAL AND METHODS
2.1 Dataset
The EBHI dataset, developed through a collaboration
between Northeastern University and China Medical
University Cancer Hospital, serves as a standardized
dataset for the automated classification of colorectal
cancer histopathological images (Hu et al., 2023). The
dataset comprises 5,532 electron microscope images,
categorized into five pathological groups: Normal,
Polyp, Low-grade IN, High-grade IN, and
Adenocarcinoma. Among these, the first two classes
represent non-cancerous conditions, while the latter
three exhibit pre-cancerous or malignant
characteristics to varying degrees. In this study, a
classification task was constructed based on the
distinction between benign and malignant tissue types,
following this categorization criterion.
In the data preprocessing stage, standardization
was applied to the raw data to ensure stable model
training. Size normalization was first performed to
adapt input images to the required format for deep
learning models and to maintain compatibility with
commonly used pre-trained CNN architectures
(Tellez et al., 2019). Specifically, all pathological
images were uniformly resized from 2048 × 1536 to
224 × 224 to ensure a consistent input size.
Additionally, to enhance training stability and
accelerate convergence, pixel value normalization
was conducted, scaling all pixel intensities to the
range [0,1] to minimize the impact of numerical
differences between images on model training.
Following this, all images were converted to
Tensor format to enable efficient batch processing in
PyTorch. To further enhance data diversity, data
augmentation techniques were applied to the training
set, including random horizontal flip, color jitter, and
random cropping. These augmentation strategies
were introduced to improve the model’s
generalization ability, allowing it to better adapt to
pathological images under varying conditions (Hao et
al., 2021; Tellez et al., 2019; Yuan, 2021).
In terms of data partitioning, all images were
divided into 40% for the training set, 40% for the
validation set, and 20% for the test set, ensuring that
all magnification images from the same case appeared
in only one of these subsets to prevent data leakage
(Hu et al., 2023).
Moreover, since some cases contained multiple
images at a specific magnification, a magnification
combination sampling strategy was employed. This
approach involved randomly combining different
images from the same case to expand the dataset and
enhance the robustness of the model (Hashimoto et
al., 2020; Tokunaga et al., 2019).
2.2 Methodology
This study employs Residual Network-50 (ResNet50)
as the fundamental deep-learning model for the
classification of colorectal cancer histopathological
images. To evaluate the impact of different training
Comparative Analysis of Learning Strategies for Multi-Magnification Pathological Image Classification
223

strategies, three experimental schemes were designed
and implemented: Single-Magnification Training,
Multi-Channel Fusion, and Stepwise Cumulative
Learning. A systematic analysis was conducted to
assess the classification performance of each strategy.
To ensure the comparability of experiments and
control variables, ResNet50 was consistently used in
all experiments, with modifications made to its input
layer (conv1) based on specific experimental
requirements.
ResNet50 is a deep convolutional neural network
(CNN) based on the Residual Network architecture. It
incorporates residual block structures and skip
connections to effectively mitigate the vanishing
gradient problem in deep networks (He et al., 2016).
This network has strong feature extraction
capabilities, enabling it to learn deep structural
information from pathological images. To
accommodate different input strategies, two
configurations of the ResNet50 input layer were
implemented in this study: For Single-Magnification
Training, the input channel was set to 3 channels
(standard RGB structure), ensuring that the model
learns pathological features at a single magnification.
For Multi-Channel Fusion, the input channel was
adjusted to 12 channels (4 magnifications × 3 RGB
channels), allowing the model to integrate multi-
magnification information within a single input image
and simultaneously learn structural features across
different magnifications.
To systematically investigate the impact of
different magnification levels on model classification
performance, this study designed two major
experimental schemes.
The first scheme, Single-Magnification Training,
involved independently training the model using
images at 40×, 100×, 200×, and 400× magnifications
to analyze classification performance at each
magnification level.
The second scheme, Multi-Magnification
Learning, included two distinct strategies: Multi-
Channel Fusion and Stepwise Cumulative Learning.
In the Multi-Channel Fusion experiment, 40×, 100×,
200×, and 400× magnification images were
concatenated following the RGB structure, forming a
12-channel input, which was then trained using
ResNet50 to evaluate the effect of cross-
magnification information fusion.
In the Stepwise Cumulative Learning experiment,
the model was progressively trained by sequentially
incorporating magnification information in the order
of 40× → 40×+100× → 40×+100×+200× →
40×+100×+200×+400× to examine whether gradual
learning enhances the generalization capability of the
model. At each training stage, testing was conducted
at the highest magnification level learned up to that
point (e.g., after training on 40×+100×+200×, the
final evaluation was performed on 200×) to assess
whether low-magnification information contributes to
improving classification performance at higher
magnifications.
Furthermore, considering that some cases may
lack corresponding images at certain magnifications,
three different data processing strategies were
designed in the Multi-Channel Fusion experiments to
investigate the impact of different filling methods on
model performance. These three strategies are Strict
Filtering, Black Filling, and Nearest Magnification
Filling.
2.3 Evaluation Metrics
This study employs Accuracy, Precision, Recall,
Specificity, and F1-Score as evaluation metrics to
comprehensively assess the model’s performance in
classifying Benign and Malignant tissues. Accuracy,
which measures the overall correctness of
classifications, is widely used in decision-making
models (Turing, 2009). Precision, reflecting the
reliability of malignant predictions, is a critical metric
in medical image analysis (Van Rijsbergen, 1979).
Recall evaluates the model’s ability to detect
malignant cases, while Specificity assesses its
capability to distinguish between benign and
malignant tissues (Altman & Bland, 1994). F1-Score,
as the harmonic mean of Precision and Recall, is
particularly useful for handling imbalanced datasets
(Van Rijsbergen, 1979).
3 RESULTS AND DISCUSSION
3.1 Single-Magnification Training
In this study, single-magnification training was first
conducted on pathological images at different
magnifications to validate model optimization and
investigate the impact of magnification levels on
classification performance.
Due to significant differences in tissue structural
information and cellular feature representation across
different magnifications, their performance in
classification tasks also varies. Low-magnification
images provide an overview of the tissue structure,
whereas high-magnification images reveal more
detailed cellular features. These differences influence
the classification performance at different
magnification levels. Therefore, experiments were
ICDSE 2025 - The International Conference on Data Science and Engineering
224
conducted by independently training ResNet50 at 40×,
100×, 200×, and 400× magnifications, and their
classification performance was compared.
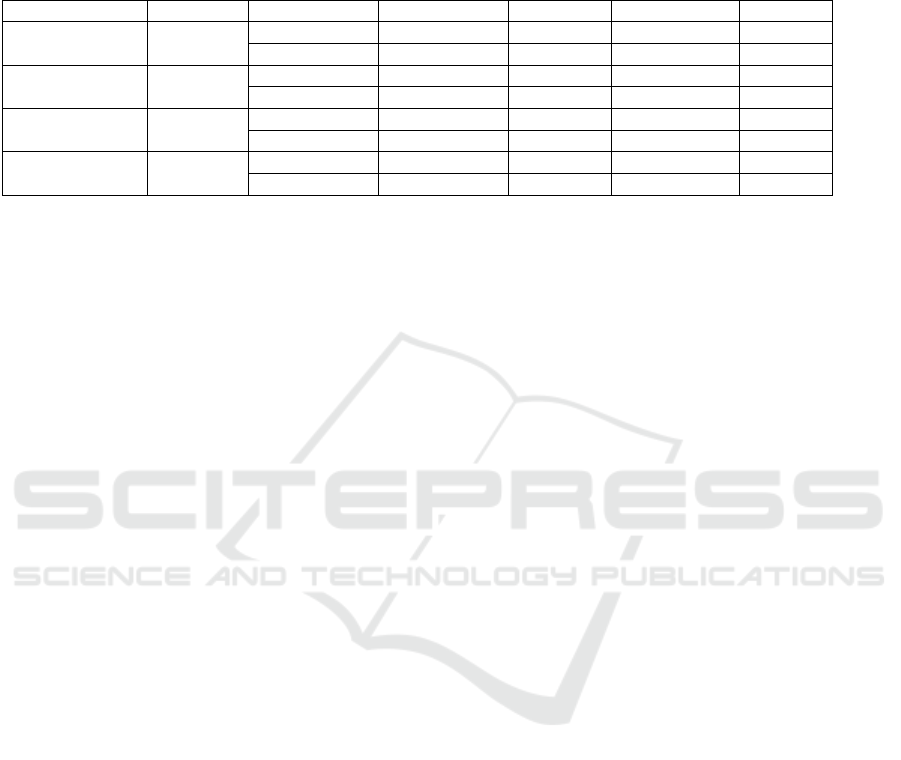
Table 1. Classification Performance of Single-Magnification Training
Ma
g
nification Accurac
y
Cate
g
or
y
Precision Recall S
p
ecificit
y
F1-score
40× 90.67
Beni
g
n 90.27 94.20 85.71 92.19
Mali
g
nant 91.30 85.71 94.20 88.42
100× 92.18
Benign 88.76 98.13 87.42 93.21
Malignant 97.88 87.42 98.13 92.35
200× 94.64
Benign 92.28 94.10 94.98 93.18
Mali
g
nant 96.19 94.98 94.10 95.58
400× 94.72
Beni
g
n 96.51 94.04 95.60 95.25
Malignant 92.55 95.60 94.04 94.05
Table 1 presents the classification performance of
the ResNet50 model at different magnifications (40×,
100×, 200×, and 400×). As shown in the table,
Classification Accuracy exhibits an overall increasing
trend with higher magnifications, reaching 94.64% at
200× and further improving to 94.72% at 400×.
Additionally, for the malignant category, both
Precision and Recall at 200× and 400× magnifications
are higher than those at lower magnifications,
indicating that high-magnification images are more
beneficial for malignant lesion detection.
This study utilizes the same EBHI dataset and
adopts ResNet50 as the baseline model, consistent
with the original study. By implementing a series of
data preprocessing and training optimization
strategies, the classification accuracy of the model has
been significantly improved. Compared to the highest
classification accuracy of 83.81% reported in the
original study using ResNet50, the optimized
strategies in this study have achieved 94.64%
accuracy at 200× magnification, demonstrating a
remarkable performance enhancement.
In the Single-Magnification Training experiment,
this study employed a data augmentation strategy to
increase data diversity and enhance the model’s
generalization capability. All pathological images
were normalized to the range [0,1] and resized to 224
× 224 for input. The data augmentation operations
included random horizontal flipping, vertical flipping,
90°, 180°, and 270° rotations, as well as color
jittering, enabling the network to develop greater
robustness to rotational transformations.
In terms of training optimization, this study
employed a dynamic learning rate adjustment method
(ReduceLROnPlateau) to adapt the learning rate at
different training stages, thereby preventing
convergence issues that may arise from a fixed
learning rate. Additionally, the Adam optimizer was
used in place of traditional Stochastic Gradient
Descent (SGD), leveraging momentum and adaptive
learning rate mechanisms to enhance training stability
and accelerate convergence.
3.2 Multi-Channel Fusion
The results of single-magnification training indicate
that images at different magnifications exhibit
varying classification performances. Among them,
high-magnification images at 200× and 400×
achieved better classification accuracy, though the
performance improvement between these two
magnifications was relatively minor. This
phenomenon suggests that relying solely on a single
magnification may not fully capture the
discriminative features of pathological images.
To address this, the study further investigates
whether the fusion of multi-magnification
information can enhance classification performance.
Compared to Single-Magnification Training, Multi-
Channel Fusion integrates information from multiple
scales, enabling the model to learn both macro-level
tissue structures and fine-grained cellular morphology
simultaneously. This approach improves
classification robustness and generalization
capability. Therefore, this study explores a Multi-
Channel Fusion strategy, in which images of the same
lesion at different magnifications are concatenated
into a 12-channel input, enhancing the model’s ability
to learn across different scales and ultimately
improving classification performance.
In the experimental design, each case contains
images at four magnifications (40×, 100×, 200×, and
400×), which are concatenated into a unified input
and fed into a modified ResNet50 model for training.
However, in the dataset, some cases lack images at
certain magnifications. To investigate the impact of
different missing data handling strategies on model
performance, this study explores the following three
Comparative Analysis of Learning Strategies for Multi-Magnification Pathological Image Classification
225
approaches:
1) Strict Filtering: Cases missing images at any
magnification are directly excluded, ensuring
that both the training and testing samples are
complete 12-channel inputs.
2) Black Filling: If an image at a specific
magnification is missing, it is replaced with a
black image (all pixel values set to 0) to maintain
input size consistency.
3) Nearest Magnification Filling: If an image at a
particular magnification is missing, it is replaced
by an image from the nearest lower
magnification. For example, if the 40× image is
missing, the 100× image is used instead; if 100×
is also missing, the 200× image is used, and so
on.
All models are trained on preprocessed multi-
channel images, and testing is also performed on
concatenated 12-channel inputs, rather than
evaluating single-magnification images separately.
By comparing the effects of these three data-filling
strategies, this study analyzes how different missing
data-handling approaches influence classification
accuracy.
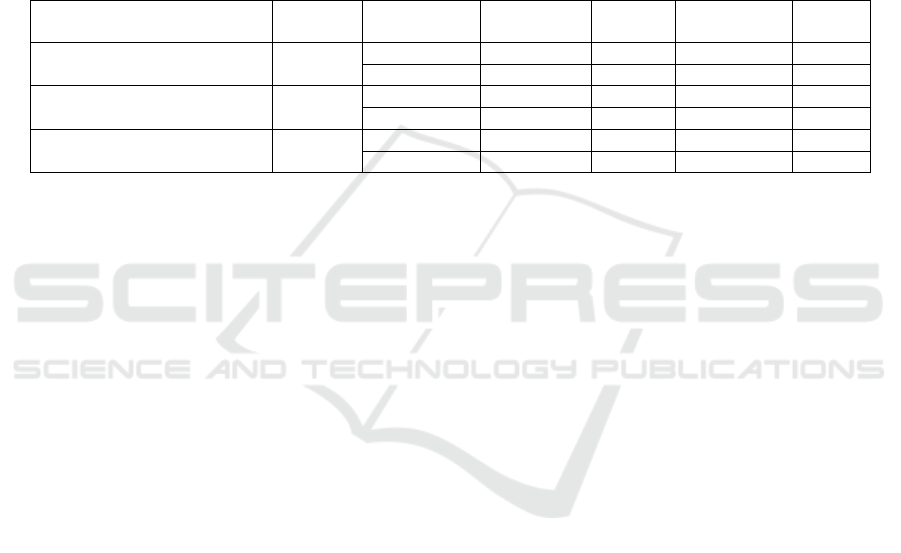
Table 2. Comparison of Classification Performance Across Multi-Channel Fusion Strategies
Strategy Accuracy Category Precision Recall Specificity F1-
score
Strict Filter 96.06
Benign 96.28 95.58 96.52 95.92
Mali
g
nant 95.86 96.52 95.58 96.19
Black Filling 95.41
Beni
g
n 94.27 96.72 94.10 95.48
Mali
g
nant 96.61 94.10 96.72 95.34
Nearest Magnification Filling 92.96
Benign 95.71 90.74 95.45 93.16
Malignant 90.21 95.45 90.74 92.76
Table 2 presents the impact of three data filling
strategies on the multi-channel fusion classification
task. The experimental results show that the Strict
Filtering strategy achieved the highest classification
accuracy of 96.06%, indicating that complete multi-
magnification information provides the most stable
feature representation, thereby enhancing the model’s
classification capability.
In contrast, when using Black Filling to replace
missing magnification images, classification
performance declined but still maintained a relatively
high accuracy. This suggests that the model can
partially adapt to the Black Filling strategy; however,
zero-value images can introduce additional noise,
leading to less stable feature representation compared
to complete data.
Nevertheless, the Black Filling strategy
outperformed the Nearest Magnification Filling
approach, suggesting that maintaining data
consistency is more beneficial than filling missing
magnifications with available images from other
magnifications.
3.3 Stepwise Cumulative Learning
Although the multi-magnification fusion strategy
improves classification performance by integrating
images from different magnifications, it does not fully
reflect the observation sequence followed by
pathologists during actual diagnosis. Typically,
pathologists begin with low-magnification images to
examine the overall tissue structure before
progressively zooming in to higher magnifications to
obtain finer details. Simply relying on multi-channel
concatenation may not fully leverage these
hierarchical features.
To address this, this study proposes a stepwise
training strategy, namely Stepwise Cumulative
Learning, in which the model is initially trained on
low-magnification images and then progressively
incorporates higher-magnification information. This
approach aims to investigate whether the gradual
accumulation of magnification information can
enhance the final classification performance. To
validate the effectiveness of stepwise learning, the
model is evaluated during the testing phase only on
the highest magnification introduced in the training
process.
During training, the model initially uses only 40×
magnification images for preliminary training,
allowing it to learn global tissue structure information
at a low magnification. Subsequently, images at 100×,
200×, and 400× magnifications are progressively
introduced, simulating a stepwise optimization
process supported by multi-scale information.
Notably, at each training stage, all case IDs must
remain consistent, ensuring that images of the same
case across different magnifications originate from
the same source. This constraint prevents the model
from merely learning single-magnification features
and instead enables it to establish robust cross-
magnification associations.
ICDSE 2025 - The International Conference on Data Science and Engineering
226
By following this design, the model not only
leverages the holistic structural information provided
by low magnifications but also gradually integrates
fine-grained details from higher magnifications,
ultimately improving classification performance.
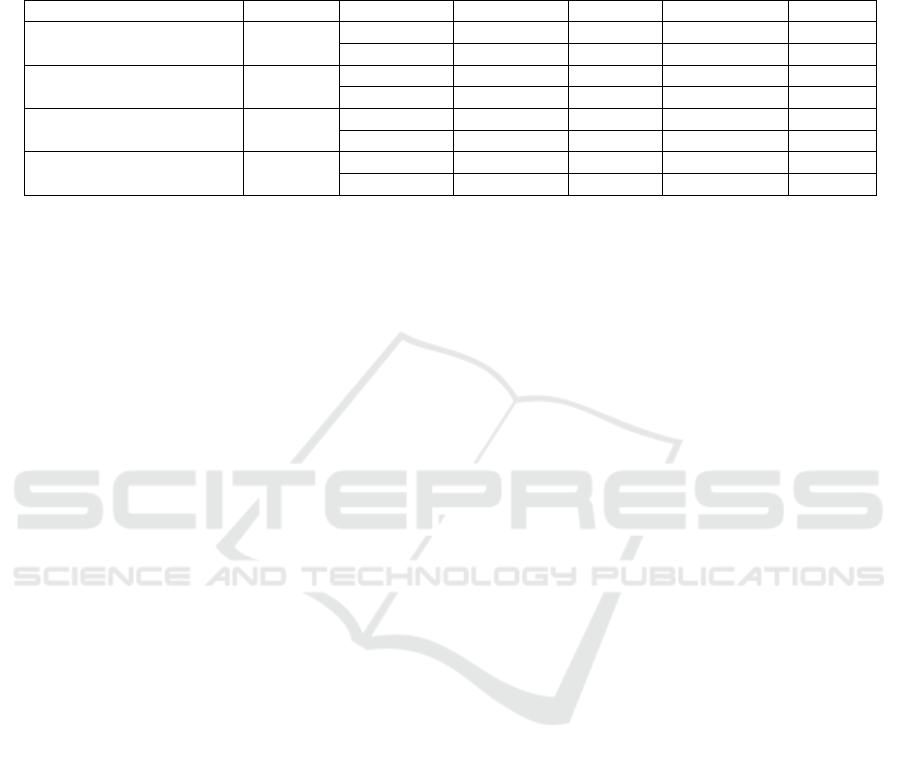
Table 3. Comparison of Classification Performance in the Stepwise Cumulative Learning Strategy
Ma
g
nification Accurac
y
Cate
g
or
y
Precision Recall S
p
ecificit
y
F1-score
40× 90.67
Beni
g
n 90.27 94.20 85.71 92.19
Mali
g
nant 91.30 85.71 94.20 88.42
40×+100× 95.43
Benign 97.45 94.26 96.91 95.83
Malignant 93.06 96.91 94.26 94.94
40×+100×+200× 96.09
Benign 95.00 95.52 96.49 95.26
Mali
g
nant 96.87 96.49 95.52 96.68
40×+100×+200×+400× 98.27
Beni
g
n 99.06 97.69 98.94 98.37
Malignant 97.39 98.94 97.69 98.16
Table 3 presents the classification performance of
the Stepwise Cumulative Learning strategy as
different magnification images are progressively
incorporated. The experiment begins with training
exclusively on 40× magnification images, followed
by the sequential addition of 100×, 200×, and 400×
magnifications, allowing the model to gradually
establish connections between global low-
magnification structural information and high-
magnification fine details.
The results demonstrate that the Stepwise
Cumulative Learning strategy consistently
outperforms single-magnification training across all
high-magnification testing tasks. For instance, in the
400× magnification task, the classification accuracy
of Single-Magnification training was 94.72%,
whereas Stepwise Cumulative Learning
(40×→100×→200×→400×) improved the accuracy
to 98.27%, achieving a 3.55% increase. Similarly, in
the 200× magnification task, Single-Magnification
training achieved an accuracy of 94.64%, while
Stepwise Cumulative Learning (40×→100×→200×)
improved the accuracy to 97.31%, representing a
2.67% increase.
These findings indicate that introducing low-
magnification information helps enhance the model’s
classification capability, and as higher magnification
information is progressively accumulated during
training, the overall model performance is further
optimized.
A more detailed analysis of classification
performance reveals that the Stepwise Cumulative
Learning strategy provides the most significant
improvement in the detection of Malignant cases. For
instance, in the 400× magnification task, the Recall
for malignant cases increased from 94.04% in Single-
Magnification training to 98.94%, representing a
4.9% improvement. This suggests that stepwise
learning helps the model better capture malignant
lesion characteristics.
In contrast, the improvement in classification
precision for Benign cases was relatively smaller. For
example, in the 400× magnification task, the
Precision increased only slightly from 96.51% to
97.39%. This discrepancy may be attributed to the
greater complexity of malignant pathological
features, where the stepwise learning strategy
provides richer hierarchical information, enabling the
model to identify malignant patterns more accurately.
Compared to the Multi-Channel Fusion strategy,
the primary advantage of Stepwise Cumulative
Learning is its alignment with the observation
sequence used by pathologists. By maintaining case
ID consistency and emphasizing the sequential
progression of magnification, this approach enables
the model to leverage low-magnification information
to refine high-magnification classification.
In summary, the Stepwise Cumulative Learning
strategy outperforms both Single-Magnification
Training and Multi-Channel Fusion across all high-
magnification testing tasks, with the most significant
improvement observed in malignant case
identification. These findings suggest that a
progressive learning approach incorporating low-
magnification information is a crucial method for
enhancing the classification accuracy of pathological
images.
3.4 Discussion
This study explores the impact of different
pathological image magnifications on classification
performance by employing three training strategies.
However, certain limitations remain, which should be
addressed in future research.
First, this study employs ResNet50 as the sole
baseline model. Although it has demonstrated strong
performance in pathological image classification, its
Comparative Analysis of Learning Strategies for Multi-Magnification Pathological Image Classification
227

reliance on local receptive fields may limit its ability
to integrate cross-magnification information
effectively. Future research could explore self-
attention-based architectures, such as ViT or Swin
Transformer, to improve global feature extraction.
Additionally, leveraging DenseNet or other feature-
reuse networks may enhance robustness, especially in
small-sample scenarios.
Second, although the EBHI dataset was used to
validate the proposed approach, further experiments
on larger and more diverse pathological datasets are
necessary to assess the generalizability and stability
of the method. Additionally, in real-world clinical
practice, pathologists rely not only on static images
but also on clinical history and lesion evolution over
time. Future research should explore Multimodal
Fusion Models, integrating multi-magnification
information with other clinical data, to enhance
diagnostic decision-making.
Overall, while this study demonstrates the
effectiveness of stepwise learning and multi-
magnification fusion, further improvements in model
selection, dataset diversity, and clinical applicability
are necessary to enhance the practical deployment of
such methods in pathology.
4 CONCLUSION
This study systematically investigates the impact of
multi-magnification information on pathological
image classification by designing and validating three
learning strategies: Single-Magnification Training,
Multi-Channel Fusion, and Stepwise Cumulative
Learning. The experiments, conducted using
ResNet50 on the EBHI dataset, demonstrate the
effectiveness of the proposed strategies in enhancing
classification performance.
The results confirm that the proposed strategies
significantly enhance classification performance. In
Single-Magnification Training, the classification
accuracy was improved from the previously reported
highest accuracy of 83.81% to 94.64% at 200×
magnification through the optimization techniques
applied in this study. Stepwise Cumulative Learning
achieved the highest accuracy among all strategies,
particularly in malignant pathology detection, where
it further improved classification accuracy to 98.27%
on 400× test images. Additionally, the study
highlights the impact of different missing
magnification image filling strategies, showing that
the Strict Filtering approach yields the best
classification performance (96.06%).
These findings suggest that progressively
incorporating low-magnification information
enhances the model’s ability to extract discriminative
features, improving overall classification accuracy.
Moreover, this study validates the suitability of the
EBHI dataset for multi-magnification learning
research, providing a useful reference for future
dataset selection.
In summary, this study presents a novel
optimization approach for multi-magnification
pathological image classification, laying the
groundwork for future advancements in intelligent
pathology image analysis.
REFERENCES
Altman, D. G., & Bland, J. M., 1994. Diagnostic tests. 1:
Sensitivity and specificity. BMJ: British Medical
Journal, 308(6943), 1552.
Das, K., Karri, S. P. K., Roy, A. G., Chatterjee, J., & Sheet,
D., 2017. Classifying histopathology whole-slides
using fusion of decisions from deep convolutional
network on a collection of random multi-views at multi-
magnification. In 2017 IEEE 14th International
Symposium on Biomedical Imaging (ISBI 2017), pp.
1024-1027. IEEE.
Hao, R., Namdar, K., Liu, L., Haider, M. A., & Khalvati, F.,
2021. A comprehensive study of data augmentation
strategies for prostate cancer detection in diffusion-
weighted MRI using convolutional neural networks.
Journal of Digital Imaging, 34, 862-876.
Hashimoto, N., Fukushima, D., Koga, R., Takagi, Y., Ko,
K., Kohno, K., et al., 2020. Multi-scale domain-
adversarial multiple-instance CNN for cancer subtype
classification with unannotated histopathological
images. In Proceedings of the IEEE/CVF conference on
computer vision and pattern recognition, pp. 3852-
3861.
He, K., Zhang, X., Ren, S., & Sun, J., 2016. Deep residual
learning for image recognition. In Proceedings of the
IEEE conference on computer vision and pattern
recognition, pp. 770-778.
Hu, W., Li, C., Rahaman, M. M., Chen, H., Liu, W., Yao,
Y., et al., 2023. EBHI: A new Enteroscope Biopsy
Histopathological H&E Image Dataset for image
classification evaluation. Physica Medica, 107, 102534.
Khan, A. A., Arslan, M., Tanzil, A., Bhatty, R. A., Khalid,
M. A. U., & Khan, A. H., 2024. Classification of colon
cancer using deep learning techniques on
histopathological images. Migration Letters, 21(S11),
449-463.
Kim, S. H., Koh, H. M., & Lee, B. D., 2021. Classification
of colorectal cancer in histological images using deep
neural networks: An investigation. Multimedia Tools
and Applications, 80(28), 35941-35953.
Malik, J., Kiranyaz, S., Kunhoth, S., Ince, T., Al-Maadeed,
S., Hamila, R., & Gabbouj, M., 2019. Colorectal cancer
ICDSE 2025 - The International Conference on Data Science and Engineering
228

diagnosis from histology images: A comparative study.
arXiv preprint arXiv:1903.11210.
Raju, M. S. N., & Rao, B. S., 2022. Colorectal multi-class
image classification using deep learning models.
Bulletin of Electrical Engineering and Informatics,
11(1), 195-200.
Tellez, D., Litjens, G., Bándi, P., Bulten, W., Bokhorst, J.
M., Ciompi, F., & Van Der Laak, J., 2019. Quantifying
the effects of data augmentation and stain color
normalization in convolutional neural networks for
computational pathology. Medical image analysis, 58,
101544.
Tokunaga, H., Teramoto, Y., Yoshizawa, A., & Bise, R.,
2019. Adaptive weighting multi-field-of-view CNN for
semantic segmentation in pathology. In Proceedings of
the IEEE/CVF conference on computer vision and
pattern recognition, pp. 12597-12606.
Turing, A. M., 2009. Computing machinery and
intelligence, pp. 23-65. Springer Netherlands.
Van Rijsbergen, C. J., 1979. Information Retrieval, 2nd edn.
Newton, MA.
Yuan, Z., 2021. Deep Learning Based Data Augmentation
for Breast Lesion Detection. Doctoral dissertation,
Carnegie Mellon University Pittsburgh, PA.
Comparative Analysis of Learning Strategies for Multi-Magnification Pathological Image Classification
229
