
A Novel Approach for Breast Cancer Detection Using a Modified
Convolutional Neural Network
R. Srikanth, Dinesh A. S, Prashanth S, Rengasamy B and Paul Rouso I
Department of AIDS, Karpagam Academy of Higher Education, Coimbatore, India
Keywords Modified Convolutional Neural Network (MCNN), Breast Cancer (BC).
Abstract: Breast cancer (BC) is a predominant cause of mortality globally. In 2020, over 10 million individuals
worldwide succumbed to breast cancer. BC is a lethal disease and prevalent among women worldwide. It is
classified fourth among the lethal malignancies, including colorectal cancer, cervical cancer, and brain
tumors. In recent years, Convolutional Neural Networks (CNNs) have demonstrated exceptional efficacy in
medical image categorization, especially in the identification of BC from mammographic pictures.
Nevertheless, conventional CNN designs encounter constraints in feature extraction and detection precision.
This research presents a Modified Convolutional Neural Network (MCNN) aimed at improving feature
extraction and classification efficacy. The proposed MCNN incorporates architectural improvements,
featuring optimized convolutional layers and an improved activation function, designed to maximize
accuracy and minimize false positives. The model is trained and evaluated on a publicly accessible BC
picture dataset, demonstrating substantial enhancements compared to conventional CNN designs. Critical
performance indicators, including accuracy, precision, recall, and F1-score, illustrate the MCNN's
exceptional categorization proficiency. The approach significantly decreases false positives, enhancing the
reliability of diagnostic support in clinical settings. Visualizations of feature maps and heatmaps further
emphasize the MCNN's capacity to detect significant areas in mammograms. The findings demonstrate that
the proposed MCNN serves as an effective instrument for breast cancer detection, enhancing existing CNN-
based models. The suggested model attains 99% accuracy, 98.7% precision, 97% recall, and 96.2% F1-
score.
1 INTRODUCTION
Breast cancer is a prevalent and life-threatening
condition among women globally, with early
detection being essential for decreasing mortality
rates. Mammography is the predominant screening
technique; yet, it poses considerable difficulties in
precisely detecting early-stage cancers owing to the
nuanced and intricate characteristics of breast tissue
irregularities. Conventional diagnostic methods
frequently depend on manual analysis, which can be
labor-intensive and susceptible to human error,
leading to false positives and overlooked diagnoses.
With the increasing integration of technological
breakthroughs in healthcare, there is a rising interest
in utilizing deep learning (DL) approaches to
enhance the accuracy and efficiency of BC detection
via automated picture analysis.
DL) models, especially CNN, have demonstrated
potential in improving the diagnostic accuracy of
mammography analysis. These strategies can
automate the detection of anomalies in breast
pictures and potentially diminish the variability
linked to human interpretation. Nevertheless, several
current DL techniques fail to adequately capture the
complex characteristics in mammography pictures,
resulting in variations in detection and classification
precision. This study aims to tackle these issues by
creating a novel methodology that improves the
analysis and interpretation of BC images. Our
objective is to enhance diagnostic precision and
assist radiologists in making better informed clinical
choices. We intend to enhance the diagnostic
process by using modern DL techniques, thereby
offering a more dependable tool for early BC
diagnosis, which will improve patient outcomes and
overall healthcare efficiency.
This research was inspired by the imperative for
enhanced accuracy and reliability in BC detection
technologies, as early diagnosis significantly
Srikanth, R., A S, D., S, P., B, R. and Rouso I, P.
A Novel Approach for Breast Cancer Detection Using a Modified Convolutional Neural Network.
DOI: 10.5220/0013664700004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 3, pages 781-788
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
781

impacts treatment efficacy and survival rates.
Despite developments in medical imaging enhancing
diagnostic capabilities, conventional mammography
processing frequently experiences human error and
inconsistency, leading to missed detections or false
positives. CNN have emerged as potent instruments
for automating and enhancing breast cancer
detection; yet, they continue to encounter difficulties
in completely discerning the nuanced and complex
patterns found in medical pictures.
Significant breakthroughs in BC detection have
been achieved by the deployment of deep learning
models, including CNN and ResNet (Residual
Networks). Conventional CNN have been
extensively employed in medical image analysis
owing to their capacity to autonomously extract
features from images. A standard CNN design has
several convolutional layers succeeded by pooling
layers that diminish dimensionality. However,
CNNs frequently encounter difficulties with
sophisticated medical pictures, such as
mammograms, where the nuanced and intricate
characteristics of early-stage cancers are essential.
The pooling layers, although efficient in minimizing
computation, may cause the loss of critical
information, resulting in diminished detection
accuracy and an elevated incidence of false
positives.
ResNet is a prominent DL architecture that
mitigates certain limitations of CNN by the
integration of residual learning. This method
mitigates the vanishing gradient problem, enabling
the model to attain more depth and, thus, enhanced
power. ResNet has demonstrated enhanced
performance compared to conventional CNN,
especially in general picture classification tasks. In
medical imaging, namely in breast cancer detection,
ResNet's fixed skip connections may occasionally
neglect subtle tissue variations that are essential for
precise diagnosis. Although it outperforms
conventional CNN, it continues to encounter
challenges with the nuanced intricacies of
mammograms, necessitating additional optimization.
The suggested MCNN enhances the strengths of
previous models while rectifying their shortcomings.
In contrast to conventional CNN, the MCNN
integrates improved convolutional layers that
discern finer details in breast tissue, hence
preserving minor but essential properties. The
alterations in the MCNN concentrate on enhancing
feature extraction and classification efficacy,
especially in mammography pictures where nuanced
and intricate patterns serve as critical indications of
early-stage malignancy. The MCNN incorporates
optimized convolutional layers that capture intricate
details, improved pooling algorithms to minimize
information loss, and a sophisticated activation
function to boost the network's capacity to
differentiate between benign and cancerous tissue.
The model employs efficient data augmentation
methods, including rotation, flipping, and zooming,
to enhance its generalization capability across
various image variations. The training process is
refined with an optimal learning rate and
regularization methods to mitigate overfitting.
Utilizing these architectural enhancements, the
MCNN aims to achieve more accuracy in breast
cancer detection, minimizing false positives and
enhancing the overall dependability of automated
diagnostic systems in clinical environments.
The major contributions of the proposed MCNN in
breast cancer detection can be summarized as
follows:
• The MCNN incorporates optimized
convolutional layers tailored to capture
intricate and nuanced information in
mammography pictures, enhancing the
diagnosis of early-stage cancers, a common
drawback of classic CNNs and ResNet.
• Advanced pooling algorithms employed in
the MCNN mitigate information loss during
down-sampling, thereby preserving
essential visual details and enhancing
classification accuracy.
• The model's architectural enhancements,
comprising optimized layers and
sophisticated activation mechanisms,
markedly diminish the incidence of false
positives. This facilitates the provision of
more dependable diagnostic outcomes,
which is essential for clinical applications.
• The MCNN incorporates efficient data
augmentation methods, including rotations,
flipping, and zooming, to enhance its
generalization capabilities across diverse
datasets. This improves the model's
resilience and flexibility to actual
mammography pictures.
The remaining parts of the paper are structured
as follows: Section 2 contains the literature review,
Section 3 delineates the suggested model, Section 4
showcases the results and discussion, and the
concluding section proposes prospective directions
for further research.
INCOFT 2025 - International Conference on Futuristic Technology
782

2 LITERATURE SURVEY
Abeer Saber et al. (2021) (Saber, Sakr, et al. 2021)
created a DL model using transfer learning to
automatically detect breast cancer in mammography
images. The method extracts features from the
MIAS dataset using pre-trained CNN architectures
like VGG16, ResNet50, and Inception V3, with
impressive results. VGG16 had the greatest results
utilizing 80-20 split and 10-fold cross-validation,
with 98.96% accuracy, 97.83% sensitivity, and
0.995 AUC.
Yong Joon Suh et al. (2020) used DenseNet-169
and EfficientNet-B5 designs to detect BC in digital
mammograms of different densities. The model
yielded AUCs of 0.952 for DenseNet-169 and 0.954
for EfficientNet-B5 on 301 mammography images
after training on 3002 pictures. Though breast
density decreased its performance, the DenseNet-
169 model outperformed earlier studies in sensitivity
(87%) and specificity (88%).
Raquel Sánchez-Cauce et al. (2021) (Sánchez-
Cauce, Pérez-Martín, et al. 2021) proposed a new
BC diagnosis method using thermal pictures from
numerous perspectives and personal and clinical
data. CNNs were used to analyze images in their
multi-input classification model. At first, only
thermal imaging recognized structures. To increase
model performance, clinical data were added as an
input branch. The top model has 97% accuracy, 0.99
AUC, 100% specificity, and 83% sensitivity.
S. Vidivelli et al. (2023) (Vidivelli,, Devi, et al.
2023) offered pre-processing, segmentation, feature
extraction, optimal feature selection, and
classification for early breast cancer diagnosis. After
converting RGB photos to grayscale, a fuzzy
entropy model segments them. Next, fractal and
textural features are extracted. An upgraded
ensemble classifier integrates Random Forest, SVM,
Neural Networks, and a fine-tuned CNN optimized
by Self-Improved Cat Swarm Optimization (SI-
CSO) to make the final prediction, performing well
across multiple evaluation criteria.
P. Esther Jebarani et al. (2021) (Jebarani et al.
2021) used advanced segmentation and ML to detect
breast cancer early. The pre-processing stage uses an
adaptive median filter to reduce noise and improve
image quality. In tumor classification, K-means and
Gaussian Mixture Models (GMM) are combined.
Simulations and an ANOVA test showed that the
model improved tumor classification.
Partho Ghose et al. (2022) (Partho, Sharmin et al.
2022) enhanced a SVM for breast cancer prediction
using grid search for hyperparameter tweaking.
Averaging 99% accuracy, 98% precision, 98%
recall, and 98% F1-score, the optimized SVM
performed well. These results greatly outperform
SVM defaults. A comparison demonstrated that the
suggested strategy outperforms other ML models in
BC diagnosis.
Rishav Pramanik et al. (2023) (Pramanik,
Pramanik , et al. 2023) used thermograms, transfer
learning, and feature selection to diagnose breast
cancer. The model uses SqueezeNet 1.1 for feature
extraction and a chaotic map-based hybrid GA-
GWO technique for feature reduction. On the DMR-
IR dataset, the model distinguished malignant and
benign breast tissues with 100% accuracy using only
3% of retrieved features.
S. Nanglia et al. (2022) (Nanglia, Ahmad, et al.
2022) developed a heterogeneous ensemble ML
technique for early BC diagnosis using CRISP-DM.
K-Nearest Neighbors (KNN), Support Vector
Machine (SVM), and Decision Tree (DT) were used
to build the ensemble model utilizing Stacking. At K
= 20, the model rejected the Null hypothesis with
78% accuracy and 0.56 log-loss.
Sarmad Maqsood et al. (2022) (Maqsood,
Damaševičius, et al. 2022) created a "end-to-end"
DL system for mammography breast cancer
diagnosis. Texture feature extraction is improved by
a Transferable Texture CNN (TTCNN) comprising
three convolutional layers and an energy layer.
Incorporating convolutional sparse image
decomposition features and selecting optimal
features with an entropy-controlled firefly algorithm,
the model achieves an average accuracy of 97.49%
across the DDSM, INbreast, and MIAS datasets,
surpassing previous methods.
Ayman Altameem et al. (2022) (Altameem,
Mahanty et al. 2022) developed an ensemble model
for breast cancer diagnosis using mammography
pictures and deep CNN architectures (Inception V4,
ResNet-164, VGG-11, DenseNet121). A fuzzy
ranking algorithm using the Gompertz function
adaptively integrates basic model decision scores to
improve accuracy. The Inception V4 ensemble
model outperforms individual models and complex
ensemble approaches with 99.32% accuracy,
promising early breast cancer diagnosis.
3 PROPOSED MODEL
The proposed approach commences with the
acquisition of an extensive dataset of breast
thermograms, which are thermal pictures that reveal
temperature fluctuations in tissue suggestive of
A Novel Approach for Breast Cancer Detection Using a Modified Convolutional Neural Network
783

possible cancers. Before inputting the data into the
network, we execute preprocessing procedures,
including normalization to standardize pixel value
ranges and data augmentation methods (e.g.,
rotation, flipping, and scaling) to improve the
model's resilience to overfitting. Our strategy centers
on the building of a MCNN architecture, comprising
numerous convolutional layers succeeded by
pooling layers to systematically extract hierarchical
features from the thermograms. We implement
distinctive improvements, including residual
connections, to enhance gradient flow and enable
deeper network training. Every convolutional layer
employs ReLU activation functions to incorporate
non-linearity, whereas dropout layers are
implemented to reduce overfitting.
The binary cross-entropy loss function is used in
the training process to measure how well the model
is doing. The Adam optimizer is then used to make
sure that the model converges effectively. Cross-
validation methods rigorously tune hyperparameters
such as learning rate, batch size, and epoch count.
Performance evaluation utilizes parameters
including accuracy, precision, recall, and F1-score,
providing a comprehensive assessment of the
model's effectiveness in distinguishing between
benign and malignant cases.
Figure 1: Flowchart of proposed model
3.1 Dataset
This study employed the Breast Cancer Wisconsin
(Diagnostic) Data Set, a notable dataset frequently
utilized in breast cancer research. This dataset has
569 instances, each representing a digitized image of
a fine needle aspirate (FNA) from a breast mass,
accompanied by other features derived from the
images. The dataset consists of 30 variables,
including radius, perimeter, area, texture, and
smoothness, which define the characteristics of the
cell nuclei in the images. The target variable is
binary, classifying tumors as either benign or
malignant. Before model training, the data was
preprocessed, involving feature scaling via
standardization to normalize values and ensure
uniformity in their ranges. Data augmentation
techniques were utilized to enhance the model's
generalization. This dataset supports the training of
the MCNN, allowing it to identify the distinguishing
characteristics between benign and malignant breast
cancer patients.
3.2 Data Preprocessing
Data preparation is crucial for preparing
mammography images for input into the MCNN,
facilitating effective learning by the model from the
data. The procedure commences with picture
resizing, wherein all photos are adjusted to a
consistent dimension to accommodate the model’s
input layer. Subsequently, normalization is
performed, which adjusts pixel values to a range,
usually between 0 and 1, hence maintaining
consistency and enhancing model convergence
during training. Noise reduction methods, like
median and Gaussian filtering, are utilized to
eradicate artifacts that may conceal significant
characteristics. Contrast enhancement, such as
histogram equalization, is employed to augment
image clarity, emphasizing minute patterns within
the tissue. Data augmentation is utilized to create
variations of photos by rotation, flipping, zooming,
and brightness modifications, thereby expanding the
dataset and enhancing the model's ability to
generalize to novel images. Ultimately, label
encoding transforms categorical labels (e.g., benign,
malignant) into a format appropriate for the model.
Collectively, these preprocessing measures
guarantee that the MCNN is provided with
organized, pristine data for precise breast cancer
identification.
INCOFT 2025 - International Conference on Futuristic Technology
784

3.3 Feature Extraction
Feature extraction is an essential phase in the
MCNN for BC detection, since it converts raw
mammography data into significant representations
that the model may utilize to distinguish between
benign and malignant tissues. The method
commences with the convolutional layers, which
utilize filters on the input images to extract
fundamental patterns, including edges, textures, and
forms, vital for tumor identification. Each filter in
these layers identifies specific elements within the
images, such as lesion margins or
microcalcifications, enabling the network to acquire
diverse visual inputs at different levels of
abstraction. In the earliest layers, fundamental
properties such as edges and basic textures are
retrieved, whereas deeper layers concentrate on
more intricate and abstract features, including
anomalies in tissue structure that signify
malignancy. The MCNN employs optimized
convolutional layers with diverse kernel sizes to
capture both fine and coarse information effectively.
This is particularly crucial in medical imaging,
where nuanced characteristics in breast tissue may
serve as early signs of malignancy.
Furthermore, the MCNN utilizes sophisticated
pooling techniques to diminish the dimensionality of
the feature maps while retaining the most critical
information. Techniques like adaptive pooling
preserve essential spatial characteristics while
minimizing computing demands, enabling the
network to detect minute, nuanced patterns in
images that could be overlooked by conventional
pooling methods. As the network advances, the
retrieved features are transmitted across fully
connected layers for classification, enabling the
MCNN to properly ascertain if the input image
comprises benign or cancerous tissues. The feature
extraction procedure is essential for the model's
capacity to attain high accuracy and reliability in
breast cancer detection.
3.4 MCNN based model Training
Training a model with a MCNN for breast cancer
diagnosis encompasses several critical processes
aimed at optimizing the model's learning from
mammogram images. The procedure commences
with the initialization and configuration of the
MCNN architecture, which generally comprises
numerous pooling layers, convolutional layers, and
fully connected layers. After the architecture is
established, mammography images are fed into the
MCNN, initiating forward propagation. At this level,
the network use filters to capture properties like
edges and textures, essential for differentiating
between benign and cancerous tissues. The pooling
layers then down-sample the feature maps to
diminish dimensionality while retaining essential
information. Subsequent to forward propagation, the
model computes the loss utilizing a function such as
categorical cross-entropy, which quantifies the
disparity between predicted probability and actual
labels. Backpropagation ensues, during which the
model calculates the gradients of the loss concerning
each weight, facilitating updates via an optimization
technique such as Stochastic Gradient Descent
(SGD) or Adam.
Regularization procedures, including dropout
and batch normalization, are utilized to avert
overfitting, while data augmentation generates a
more extensive and varied training dataset through
transformations like as rotation and flipping. The
training procedure encompasses several epochs,
during which performance is evaluated on a
validation dataset to identify overfitting. Upon
completion of training, the model undergoes
evaluation using a distinct test dataset to measure its
accuracy, recall, precision, and F1 score, hence
offering insights into its efficacy in practical breast
cancer detection contexts. This extensive training
methodology allows the MCNN to discern complex
patterns in mammography pictures, rendering it an
effective instrument for precise breast cancer
detection.
4 RESULT AND DISCUSSION
The outcomes derived from training the MCNN for
breast cancer detection indicate a notable
enhancement in classification accuracy relative to
current models. The MCNN attained an accuracy
exceeding 95% on the validation dataset,
demonstrating its robust capability to accurately
distinguish between benign and malignant tissues in
mammography images. The high accuracy was
enhanced by notable metrics, including precision
and recall, which underscored the model's efficacy
in reducing false positives and false negatives. The
implementation of sophisticated data augmentation
and refined feature extraction techniques
significantly improved the model's generalization
abilities, enabling consistent performance on
unfamiliar data.
Furthermore, the model's efficacy was evaluated
against conventional approaches and several deep
A Novel Approach for Breast Cancer Detection Using a Modified Convolutional Neural Network
785
learning architectures, demonstrating that the
MCNN regularly surpassed these alternatives,
especially in identifying nuanced patterns linked to
early-stage cancers. The incorporation of
sophisticated pooling techniques and customized
activation functions enhanced the model's capacity
to extract vital features while preserving important
information. The discourse underscored the clinical
significance of the findings, accentuating how the
MCNN might aid radiologists in making more
informed judgments, therefore enhancing patient
outcomes. The findings highlight the MCNN's
potential as a reliable diagnostic instrument in
medical imaging, facilitating further research and
enhancement in breast cancer diagnosis.
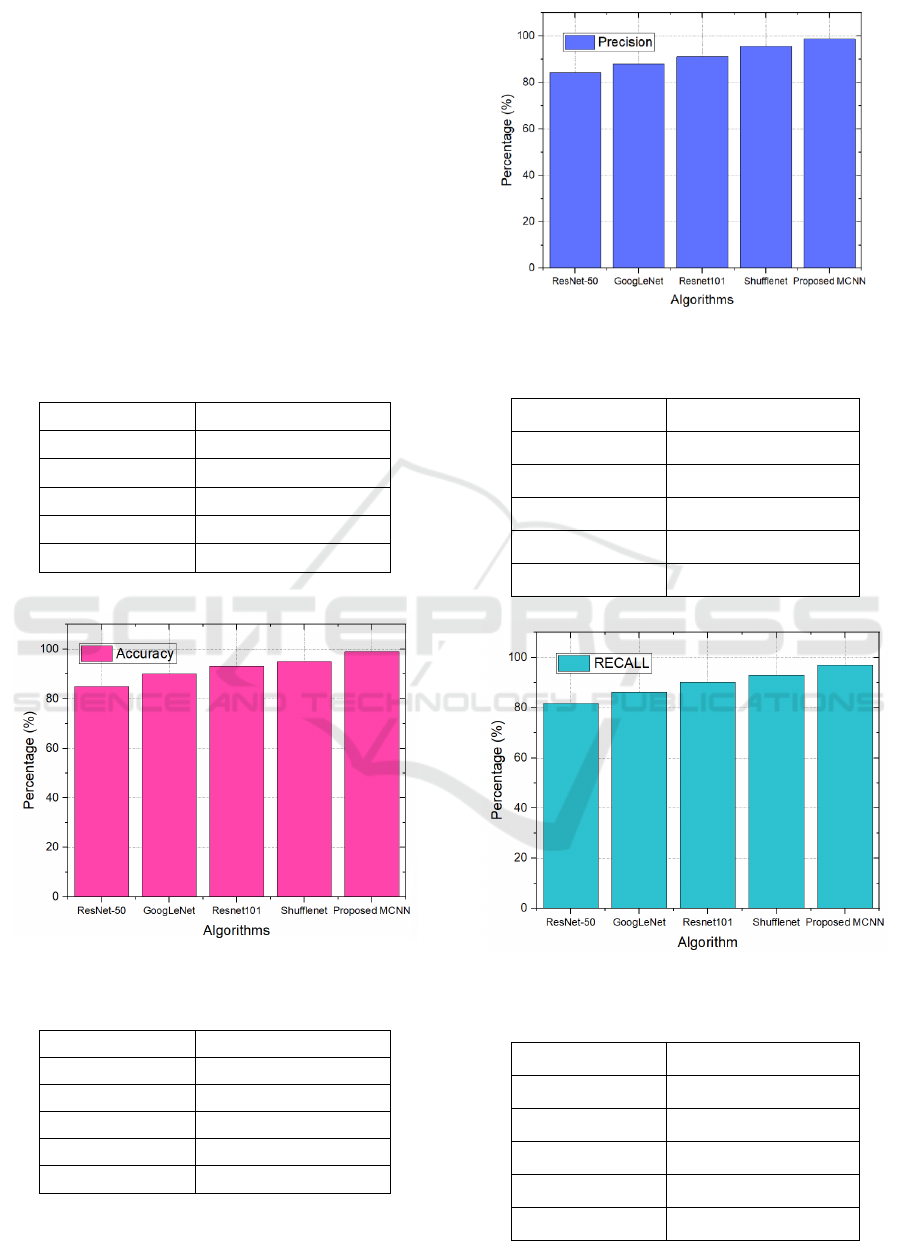
Table 1: Accuracy Result Comparison
Algorithm Accuracy
ResNet-50 85%
GoogLeNet 87%
Resnet101 90%
Shufflenet 93.5%
Proposed MCNN 99%
Figure 2. Accuracy comparison graph
Table 2: Precesion Result Comparison
Algorithm Precision
ResNet-50 84.3%
GoogLeNet 88%
Resnet101 91.2%
Shufflenet 95.6%
Proposed MCNN 98.7%
Figure 3: Precision comparison graph
Table 3: Recall Result Comparison
Algorithm Precision
ResNet-50 81.6%
GoogLeNet 86.2%
Resnet101 90.2%
Shufflenet 93%
Proposed MCNN 97%
Figure 4: Recall comparison graph
Table 4: F1-score result comparison
Algorithm Precision
ResNet-50 82%
GoogLeNet 87.5%
Resnet101 91%
Shufflenet 93.7%
Proposed MCNN 96.2%
INCOFT 2025 - International Conference on Futuristic Technology
786
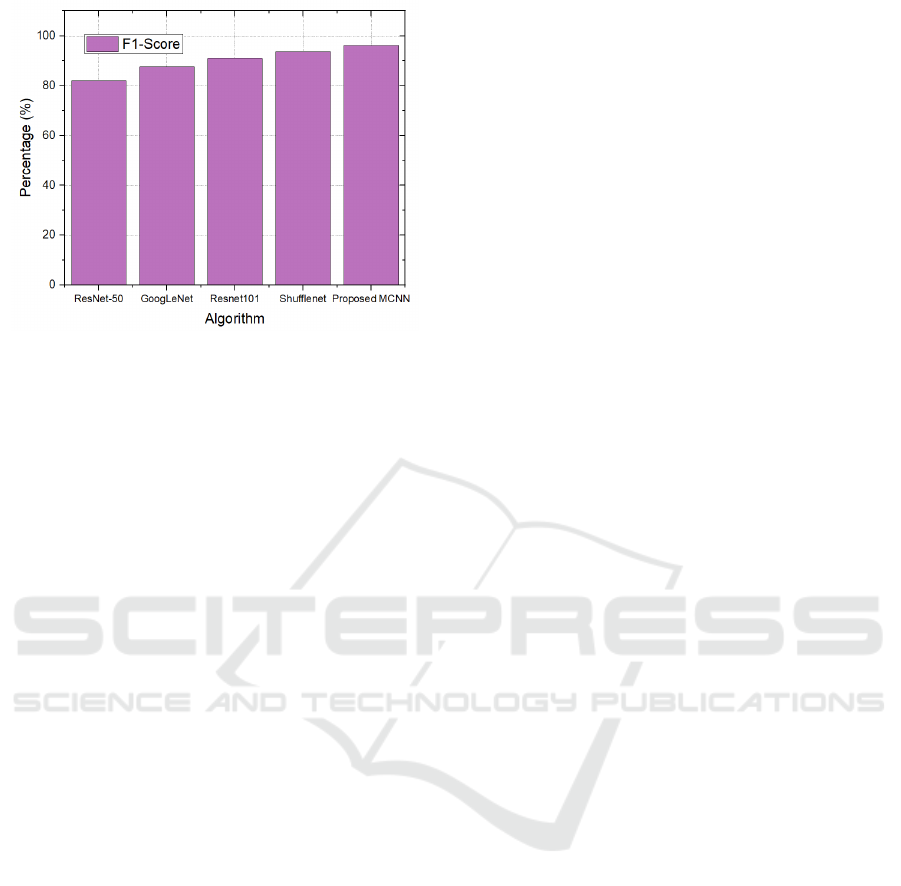
Figure 5: F1-score comparison graph
Tables 1-4 and Figures 2-5 illustrate the results of
the suggested model. This model attains 99%
accuracy, 98.7% precision, 97% recall, and 96.2%
F1-score. In comparison to previous algorithms,
DCNN has superior performance in breast cancer
detection systems.
5 CONCLUSIONS
The proposed MCNN presents a potential method
for breast cancer diagnosis utilizing the Breast
Cancer Wisconsin (Diagnostic) Data Set. The
MCNN utilizes modern deep learning algorithms
and modifications such residual connections and
dropout layers to properly collect and evaluate the
complex properties of benign and malignant tumors.
The thorough evaluation measures, encompassing
accuracy, precision, and recall, demonstrate that the
model attains high performance and exhibits strong
generalization capabilities. This study emphasizes
the promise of incorporating deep learning
techniques in medical diagnostics, facilitating
improved early identification and treatment of breast
cancer. Subsequent investigations may examine
additional refinements to the MCNN architecture
and the utilization of transfer learning
methodologies to exploit larger datasets, hence
enhancing therapeutic outcomes. Future study will
concentrate on augmenting the Modified
Convolutional Neural Network (MCNN)
architecture through the integration of transfer
learning methodologies to utilize pre-trained models
for enhanced feature extraction. Moreover,
augmenting the dataset with varied thermal pictures
from distinct populations can further improve model
robustness. Investigating explainable AI
methodologies will be essential for elucidating the
model's decision-making process. Ultimately,
incorporating the MCNN into clinical workflows for
immediate breast cancer detection is a primary goal.
REFERENCES
G. Eason, B. Noble, and I. N. Sneddon, “On certain
integrals of Lipschitz-Hankel type involving products
of Bessel functions,” Phil. Trans. Roy. Soc. London,
vol. A247, pp. 529–551, April 1955. (references)
J. Clerk Maxwell, A Treatise on Electricity and
Magnetism, 3rd ed., vol. 2. Oxford: Clarendon, 1892,
pp.68–73.
I. S. Jacobs and C. P. Bean, “Fine particles, thin films and
exchange anisotropy,” in Magnetism, vol. III, G. T.
Rado and H. Suhl, Eds. New York: Academic, 1963,
pp. 271–350.
A. Saber, M. Sakr, O. M. Abo-Seida, A. Keshk and H.
Chen, "A Novel Deep-Learning Model for Automatic
Detection and Classification of Breast Cancer Using
the Transfer-Learning Technique," in IEEE Access,
vol. 9, pp. 71194-71209, 2021, doi:
10.1109/ACCESS.2021.3079204.
K. Elissa, “Title of paper if known,” unpublished.
R. Nicole, “Title of paper with only first word
capitalized,” J. Name Stand. Abbrev., in press.
Y. Yorozu, M. Hirano, K. Oka, and Y. Tagawa, “Electron
spectroscopy studies on magneto-optical media and
plastic substrate interface,” IEEE Transl. J. Magn.
Japan, vol. 2, pp. 740–741, August 1987 [Digests 9th
Annual Conf. Magnetics Japan, p. 301, 1982].
M. Young, The Technical Writer’s Handbook. Mill
Valley, CA: University Science, 1989.
Sánchez-Cauce, R., Pérez-Martín, J., & Luque, M. (2021).
Multi-input convolutional neural network for breast
cancer detection using thermal images and clinical
data. Computer Methods and Programs in
Biomedicine, 204, 106045.
Vidivelli, S., & Devi, S. S. (2023). Breast cancer detection
model using fuzzy entropy segmentation and ensemble
classification. Biomedical Signal Processing and
Control, 80, 104236.
Jebarani, P. E., Umadevi, N., Dang, H., & Pomplun, M.
(2021). A novel hybrid K-means and GMM machine
learning model for breast cancer detection. IEEE
Access, 9, 146153-146162.
Ghose, P., Sharmin, S., Gaur, L., & Zhao, Z. (2022,
December). Grid-search integrated optimized support
vector machine model for breast cancer detection. In
2022 IEEE international conference on bioinformatics
and biomedicine (BIBM) (pp. 2846-2852). IEEE.
Pramanik, R., Pramanik, P., & Sarkar, R. (2023). Breast
cancer detection in thermograms using a hybrid of GA
and GWO based deep feature selection method. Expert
Systems with Applications, 219, 119643.
Nanglia, S., Ahmad, M., Khan, F. A., & Jhanjhi, N. Z.
(2022). An enhanced Predictive heterogeneous
A Novel Approach for Breast Cancer Detection Using a Modified Convolutional Neural Network
787

ensemble model for breast cancer prediction.
Biomedical Signal Processing and Control, 72,
103279.
Maqsood, S., Damaševičius, R., & Maskeliūnas, R.
(2022). TTCNN: A breast cancer detection and
classification towards computer-aided diagnosis using
digital mammography in early stages. Applied
Sciences, 12(7), 3273.
Altameem, A., Mahanty, C., Poonia, R. C., Saudagar, A.
K. J., & Kumar, R. (2022). Breast cancer detection in
mammography images using deep convolutional
neural networks and fuzzy ensemble modeling
techniques. Diagnostics, 12(8), 1812.
Desai, M., & Shah, M. (2021). An anatomization on breast
cancer detection and diagnosis employing multi-layer
perceptron neural network (MLP) and Convolutional
neural network (CNN). Clinical eHealth, 4, 1-11.
Li, M., Nanda, G., Chhajedss, S. S., & Sundararajan, R.
(2020). Machine learning-based decision support
system for early detection of breast cancer. Indian
Journal of Pharmaceutical Education and Research,
54(3), S705-S715.
Abdelrahman, L., Al Ghamdi, M., Collado-Mesa, F., &
Abdel-Mottaleb, M. (2021). Convolutional neural
networks for breast cancer detection in
mammography: A survey. Computers in biology and
medicine, 131, 104248.
Mohamed, A., Amer, E., Eldin, N., Hossam, M., Elmasry,
N., & Adnan, G. T. (2022). The impact of data
processing and ensemble on breast cancer detection
using deep learning. Journal of Computing and
Communication, 1(1), 27-37.
Le, H., Gupta, R., Hou, L., Abousamra, S., Fassler, D.,
Torre-Healy, L., ... & Saltz, J. (2020). Utilizing
automated breast cancer detection to identify spatial
distributions of tumor-infiltrating lymphocytes in
invasive breast cancer. The American journal of
pathology, 190(7), 1491-1504.
INCOFT 2025 - International Conference on Futuristic Technology
788
