
Leveraging Liquid Time-Constant Neural Networks for ECG
Classification: A Focus on Pre-Processing Techniques
Lisa-Maria Beneke
1
, Michell Boerger
1 a
, Philipp L
¨
ammel
1 b
, Helene Knof
1 c
,
Andrei Aleksandrov
1 d
and Nikolay Tcholtchev
1,2 e
1
Fraunhofer Institute for Open Communication Systems (FOKUS), Berlin, Germany
2
RheinMain University of Applied Sciences, Wiesbaden, Germany
fi
Keywords:
Liquid Time-Constant Neural Networks, LTC, RNN, LSTM, PTB-XL, Time Series Analysis.
Abstract:
Neural networks have become pivotal in timeseries classification due to their ability to capture complex tem-
poral relationships. This paper presents an evaluation of Liquid Time-Constant Neural Networks (LTCs), a
novel approach inspired by recurrent neural networks (RNNs) that introduces a unique mechanism to adap-
tively manage temporal dynamics through time-constant parameters. Specifically, we explore the applicability
and effectiveness of LTC in the context of classifying myocardial infarctions in electrocardiogram data by
benchmarking the performance of LTCs against RNN and LSTM models utilzing the PTB-XL dataset. More-
over, our study focuses on analyzing the impact of various pre-processing methods, including baseline wander
removal, Fourier transformation, Butterworth filtering, and a novel x-scaling method, on the efficacy of these
models. The findings provide insights into the strengths and limitations of LTCs, enhancing the understanding
of their applicability in multivariate time series classification tasks.
1 INTRODUCTION
In the field of time series classification, neural net-
works have emerged as powerful tools due to their
proficiency in capturing complex temporal relation-
ships. The ability to accurately classify tempo-
ral data can lead to significant advancements in
predictive analytics and decision-making processes.
Traditional models, such as multilayer perceptrons
(MLPs), convolutional neural networks (CNNs), re-
current neural networks (RNNs), Graph Neural Net-
works (GNNs), and Long Short-Term Memory net-
works (LSTMs) have been studied for timeseries clas-
sification tasks and have shown promising results.
(Mohammadi Foumani et al., 2024)
More recently, Liquid Time-Constant Neural Net-
works (LTCs) have emerged as a novel approach in-
spired by the principles of RNNs. LTCs introduce a
unique mechanism to adaptively manage temporal dy-
namics through their time-constant parameters. This
a
https://orcid.org/0000-0002-5741-9043
b
https://orcid.org/0000-0002-4411-0557
c
https://orcid.org/0009-0007-1364-6782
d
https://orcid.org/0000-0002-4717-4206
e
https://orcid.org/0000-0001-6821-4417
adaptability allows LTCs to model complex temporal
dependencies effectively. (Hasani et al., 2020)
However, the application of LTCs to diverse use
cases remains limited, necessitating a thorough evalu-
ation of their performance against established models
like RNNs and LSTMs. In this paper, we aim to eval-
uate LTCs on a multivariate time series benchmark-
ing dataset from the medical domain. Specifically, we
want to study their ability to classify myocardial in-
farctions (MI) in electrocardiogram (ECG) data com-
pared to RNN and LSTM models. Additionally, we
want to systematically compare the effectiveness of
various pre-processing methods for this use case. Be-
sides applying common pre-processing methods such
as baseline wander removal, Fourier transformation
and Butterworth filtering, we propose a new x-scaling
method for the task of classifying MI. By doing so, we
seek to provide insights into the strengths and limita-
tions of LTCs, contributing to the broader understand-
ing of their applicability in time series classification
tasks. In summary, our key contributions in this paper
are as follows:
• Performance Benchmarking: Comparing the
performance of Liquid Time-Constant Neural
Networks against RNNs and LSTMs for myocar-
234
Beneke, L.-M., Boerger, M., Lämmel, P., Knof, H., Aleksandrov, A., Tcholtchev and N.
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques.
DOI: 10.5220/0013648000003967
In Proceedings of the 14th International Conference on Data Science, Technology and Applications (DATA 2025), pages 234-245
ISBN: 978-989-758-758-0; ISSN: 2184-285X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
dial infarction classification using the PTB-XL
dataset.
• Analysis of Pre-processing Techniques: Ex-
amination of the effects of various signal pre-
processing methods, including baseline wander
removal, Fourier transformation, Butterworth fil-
tering, and a novel x-scaling technique on models’
performance on classifying temporal relations.
• Insights on LTC Applicability: Exploring the
applicability of LTCs in a real-world use case
while identifing the strengths and limitations of
LTCs in multivariate time series classification,
contributing valuable insights for future research
and practical applications in medical diagnostics.
The reminder of this paper is structured as follows:
Section 2 provides a background on LTCs and related
models, followed by a detailed description of the ex-
perimental setup in Section 3. Section 4 analyzes the
impact of various pre-processing methods on model
performance, while Section 5 discusses hyperparam-
eter optimization and presents the final MI detection
models. Finally, Section 6 concludes with insights
from the study and future work directions.
2 BACKGROUND AND STATE OF
THE ART
This section provides an overview of Liquid Time-
Constant Neural Networks (LTCs) and their rela-
tionship with Neural Ordinary Differential Equations
(Neural ODEs) and Continuous-Time Recurrent Neu-
ral Networks (CT-RNNs). We will outline the funda-
mental principles of these models and their distinc-
tive characteristics. In addition, we will present the
latest related work on the utilisation of ML-based ap-
proaches for MI detection.
2.1 Neural Ordinary Differential
Equations
The use of ordinary differential equations (ODEs) in
modeling dynamic systems is motivated by their abil-
ity to capture continuous-time behavior and complex
temporal relationships. Neural ODEs extend this con-
cept by incorporating neural networks into the frame-
work, allowing for flexible and expressive modeling
of system dynamics. Given a neural network function
f that is parameterized by θ (i.e. weights and biases),
the state x(t) of the modelled system at time t is de-
fined by the Ordinary Differential Equation
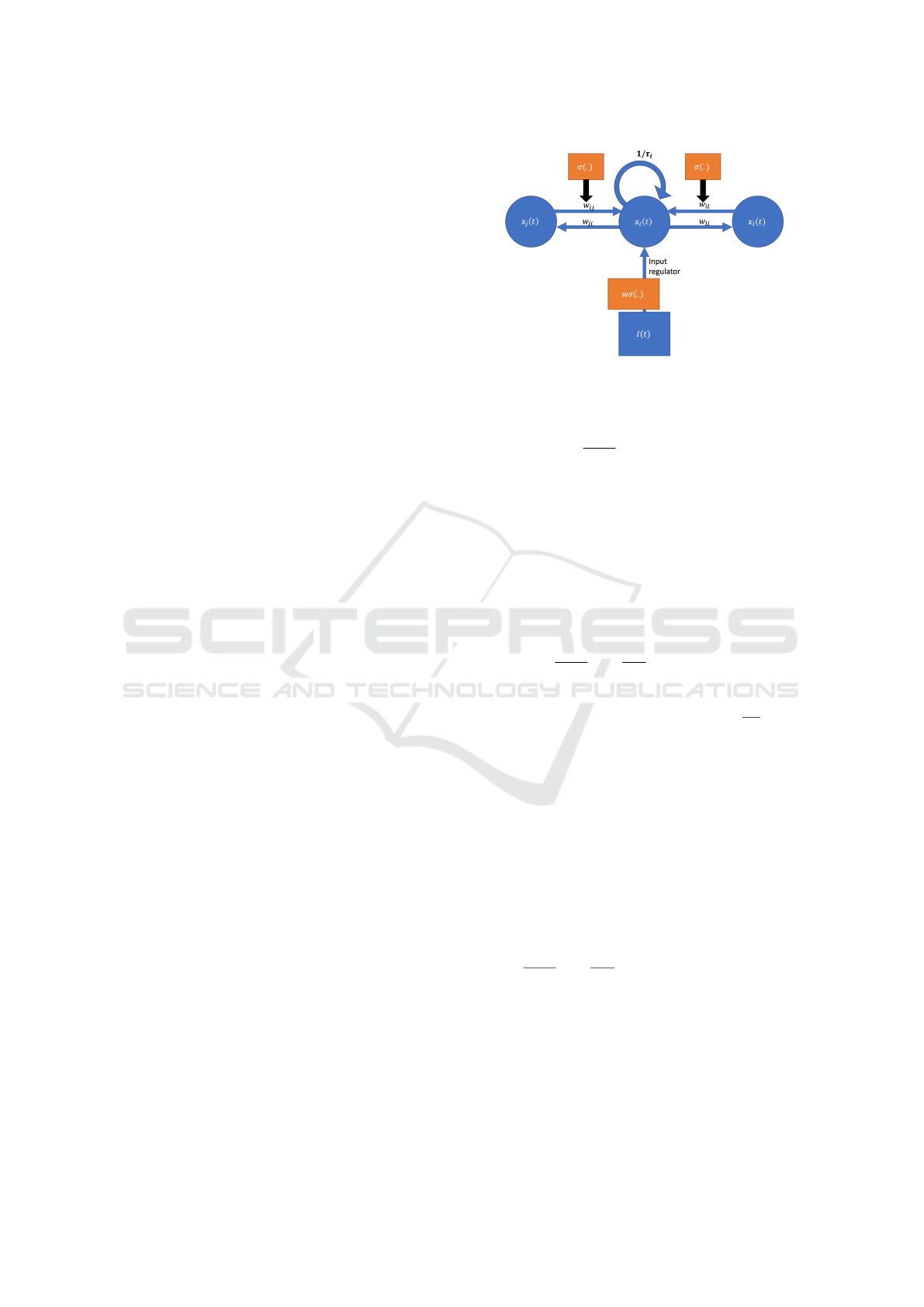
Figure 1: Illustration of a cell i ∈ [1, ..., m] of an LTC with
weights w
i j
, w
ji
, w
il
, w
li
contained in Θ, and connected neu-
rons l, j ∈ [1, ..., m], i ̸= l and i ̸= j
dx(t)
dt
= f (x(t), t, θ)
where I(t) denotes external inputs to the system
(Chen et al., 2018).
2.2 Continuous-Time Recurrent Neural
Networks
CT-RNNs also model temporal dynamics using dif-
ferential equations. The state update is given by:
dx(t)
dt
= −
x(t)
τ
+ f (x(t), t, θ)
The key difference between CT-RNNs and Neural
ODEs lies in the inclusion of the term −
x(t)
τ
, which
stabilizes the system and helps it reach an equilibrium
state with a specific time constant τ. (Funahashi and
Nakamura, 1993)
2.3 Liquid Time-Constant Neural
Networks
Hasani et al. (Hasani et al., 2020) propose yet another
formula in which the neural network not only deter-
mines the derivative of the state x but also serves as
an input-dependent varying time-constant:
dx(t)
dt
= −
x(t)
τ
+ f (x(t), t, θ) (A − x(t))
with θ and A being parameters. As described above,
CT-RNNs calculate an ODE with a time constant
τ
i
∈ R, i ∈ N for every i-th unit in the neural net-
work. Contrary to that, LTCs are utilising varying
(i.e. liquid) time-constants coupled to their hidden
state. This improves flexibility and adaption of the
network, especially on time-series prediction tasks
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
235
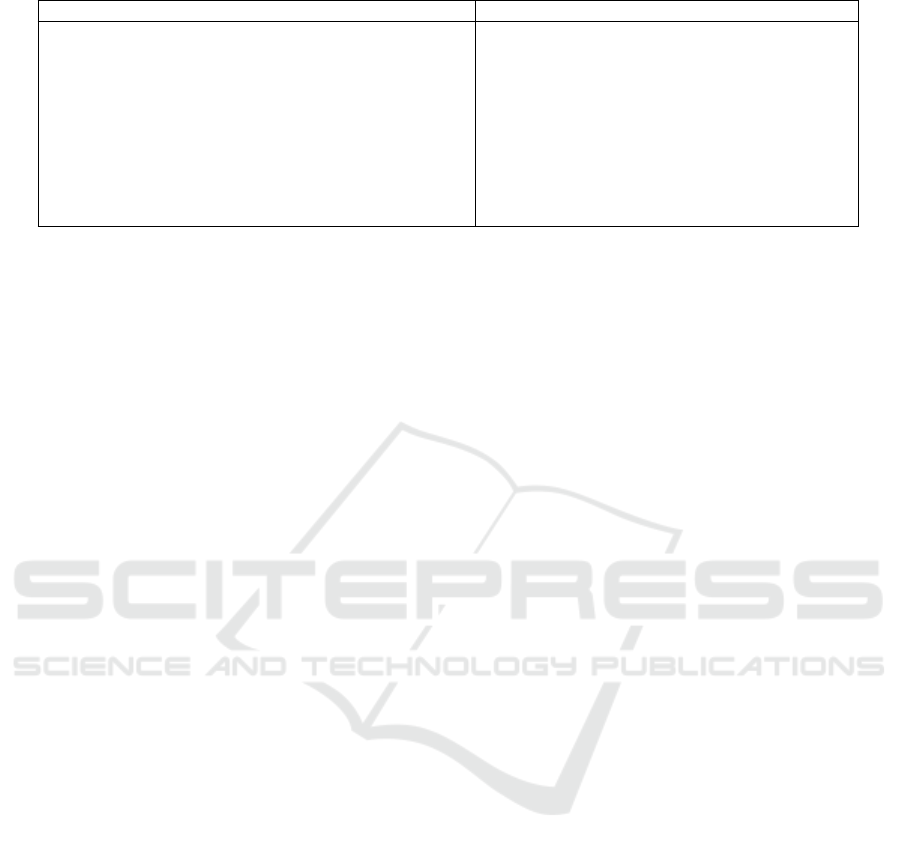
Table 1: PTB-XL based related work results.
Literature Year Classifiers Accuracy AUC
Smigiel et al. (
´
Smigiel et al., 2021) 2021 CNN 72.0% -
SincNet 73.0% -
Smigiel et al. (
´
Smigiel et al., 2021) 2021 Neural Networks 76.2% -
Pa’lczynski et al. (Pałczy
´
nski et al., 2022) 2022 Neural Networks 80.2% -
Prabhakararao et al. (Prabhakararao and Dandapat, 2022) 2022 DMSCE 84.5% -
Anand et al. (Anand et al., 2022) 2022 CNN 93.4% -
Knof et al. (Knof et al., 2024a) 2024 CNN 96.21% 98.91%
Strodthoff et al. (Strodthoff et al., 2021) 2021 LSTM - 90.7%
xresnet1d - 92.5%
inception1d - 92.5%
(Hasani et al., 2020). The general structure of a single
unit i ∈ [1, ..., m] is outlined in Figure 1.
Compared to existing algorithms, LTCs promise
to better capture the causality in data patterns over
time, while being much more robust and expressive.
They demonstrate stable and bounded behavior, offer
enhanced expressivity among neural ODEs (i.e., time-
constant neural networks), and result in better perfor-
mance on time series prediction tasks (Hasani et al.,
2020).
2.4 Related Work
In the following, we will discuss related ML-based
approaches for the detection of myocardial infarctions
based on multivariate ECG timeseries data, which are
summarized in Table 1.
The most dominant method of solving MI de-
tection of 12-lead ECG data utilises Convolutional
Neural Networks to classify between MI and non-
MI classes. (Acharya et al., 2017) (Gharaibeh and
Quwaider, 2022) (Atiea and Adel, 2022) Further-
more Segura-Saldana et al. (Segura-Salda
˜
na et al.,
2022), Xiong et. al (Xiong et al., 2022), Joloudari
et al. (Joloudari et al., 2022) and Lynn et al. (Lynn
et al., 2019) showed that Gated Recurring Units and
LSTMs are used for MI and non-MI classification
as well. Many approaches like (Singh et al., 2018)
and (Muhuri et al., 2020) utilise a multi-layer LSTM
model for accurately classifying time-series into mul-
tiple classes. Since LTCs are based on recurrent net-
works, like LSTM, these results give good insights
into the expected performance.
Hammad et al. (Hammad et al., 2022) compared
multiple models for MI detection, which a relevant
amount of is based on the PTB-XL dataset, described
in Section 3.1. As it can be seen in Table 1, Anand
et al. (Anand et al., 2022) achieved results of up to
93.4% classification accuracy.
Like others, Rai et al. (Rai and Chatterjee, 2022)
additionally utilised a combination of multiple mod-
els, for example Convolutional Neural Networks and
LSTM to further improve the results. They were
achieving detection accuracies on a combination of
multiple datasets of up to 99.89%.
Another study by Knof et al. (Knof et al., 2024a)
developed a CNN model to detect indications of my-
ocardial infarction based on ECG data. Further, they
aimed to explain and evaluate the model’s decision-
making process using explainable AI methods (Knof
et al., 2024b). They also utilised the PTB-XL dataset
and reported an MI detection performance of 96.21%
accuracy and AUC of 98.91% for their CNN model.
Strodthoff et al. (Strodthoff et al., 2021) compared
the performance of various state of the art machine
learning algorithms for the MI detection use case
based on ECG measurements as a benchmark, specif-
ically optimised for the PTB-XL data set (Wagner
et al., 2020). Unfortunately they provide their results
in AUC measurement only, which makes it hard to di-
rectly compare them to the other results in Table 1.
The comparison from Strodthoff et al. (Strodthoff
et al., 2021) focuses on different approaches of mul-
tilabel and binary classification, as well as regression
tasks. Comparing the results with our work, mostly
the results of the multilabel classification of the diag-
nostic ECG statements are relevant, since they come
close to the use case of a binary classification between
MI and non-MI diagnostic ECG statements.
Comparably to this paper, Strodthoff et al.
(Strodthoff et al., 2021) utilised both the raw ECG
signal and a feature-based approach for the classi-
fication. Interestingly they showed that many re-
sults of the image classification domain can be trans-
ferred into the time-series classification domain. They
showed that the newly proposed xresnet1d101 model
performed best in all experiments - with a 93.7% AUC
value on the classification of ECG diagnostics.
In comparison to existing work, this paper stands
out as the first to utilize LTCs for the detection of
myocardial infarctions, demonstrating their applica-
bility in a critical medical context. Additionally, it
presents a comprehensive evaluation of various pre-
processing methods tailored for noisy ECG timeseries
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
236
PTB-XL
Windowing
Encoding
Splitting
Data Preparation
Applying Signal
Pre-processing
Te ch ni qu es
Evaluate Pre-
Processing
Techniques
Final Data
Tran s f o r m a t io n :
Select & Apply
Pre-Processing
Technique( s )
on prepared
Data
Model & Hyper-
parameter
Optimization
Evaluate
Model
Build
MI Detection
Models
Balancing
Butterworth
Filtering
Baseline
Wander
Removal
Fourier Trans-
formation
X-Axis Scaling
Figure 2: Methodology followed to analyse the effect of
pre-processing techniques, and train and build MI detection
models utilzing neural ODEs.
data, highlighting the novel x-scaling approach de-
veloped specifically for this task (see Section 3.3.4).
This thorough analysis not only contributes to the
current understanding of LTCs, but also offers valu-
able insights into the optimization of pre-processing
techniques for improved classification performance in
ECG signal analysis.
3 EXPERIMENTAL SETUP
To demonstrate and validate the performance of LTCs
in real-world applications, we apply them to a medical
use case, comparing their effectiveness with RNNs
and LSTMs. Specifically, we focus on classifying
myocardial infarction (MI) using 12-lead electrocar-
diogram (ECG) data and investigate the impact of var-
ious pre-processing methods. Throughout the exper-
iments, we adhered to the methodology depicted in
Figure 2, which we will outline in the following sec-
tions.
3.1 Description of Dataset
We utilize the PTB-XL benchmarking dataset (Wag-
ner et al., 2020), which contains 21.799 ECG records
of 10 seconds length, organized into stratified train-
ing, validation, and test sets. Each ECG record is
labelled with one of five superclasses: NORM, CD,
HYP, STTC, and MI.
3.2 Data Preparation
In the following, we outline the data preparation
phase, which includes essential steps such as data en-
coding, windowing, class balancing, and splitting, all
consistently applied in our experiments.
Figure 3: Data windowing.
3.2.1 Encoding
In our use case, we focus on the detection of MI
within ECG records. Therefore, we relabel all ECG
records belonging to a different class than MI with
non-MI. Additionally, we apply one-hot encoding,
such that the models output 1 when deciding for MI
and 0 otherwise.
3.2.2 Windowing
Neural networks typically require fixed-length inputs
for class prediction. To facilitate continuous classi-
fication of time series data, we standardize the win-
dow length across our dataset by segmenting the 10-
second ECG recordings from the training set into
multiple smaller windows of fixed length, with a de-
fault length of 5 seconds. These windows may over-
lap to create additional training data and are evenly
distributed across the entire recording, with the first
window aligned to the start and the last window
aligned to the end of the data stream. Figure 3 demon-
strates how a single input data stream is divided into
four distinct windows.
3.2.3 Class Balancing
Models can be biased towards the majority class when
the training set is imbalanced (Krawczyk, 2016). In
our dataset, the number of MI-labeled recordings is
with 4.192 significantly lower than that of non-MI
recordings which is 17.607. This class imbalance is
particularly critical in medical contexts such as MI
prediction, where the minority class (i.e., MI) holds
substantial clinical significance. To address this im-
balance, we employ a data-level method that gener-
ates overlapping windows from the MI recordings, al-
lowing for the augmentation of the training set and
thereby increasing the representation of the minority
class (Krawczyk, 2016). Figure 4 illustrates how the
number of windows is increased by a factor of 1.5.
Figure 4: Balancing with overlapping windows.
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
237
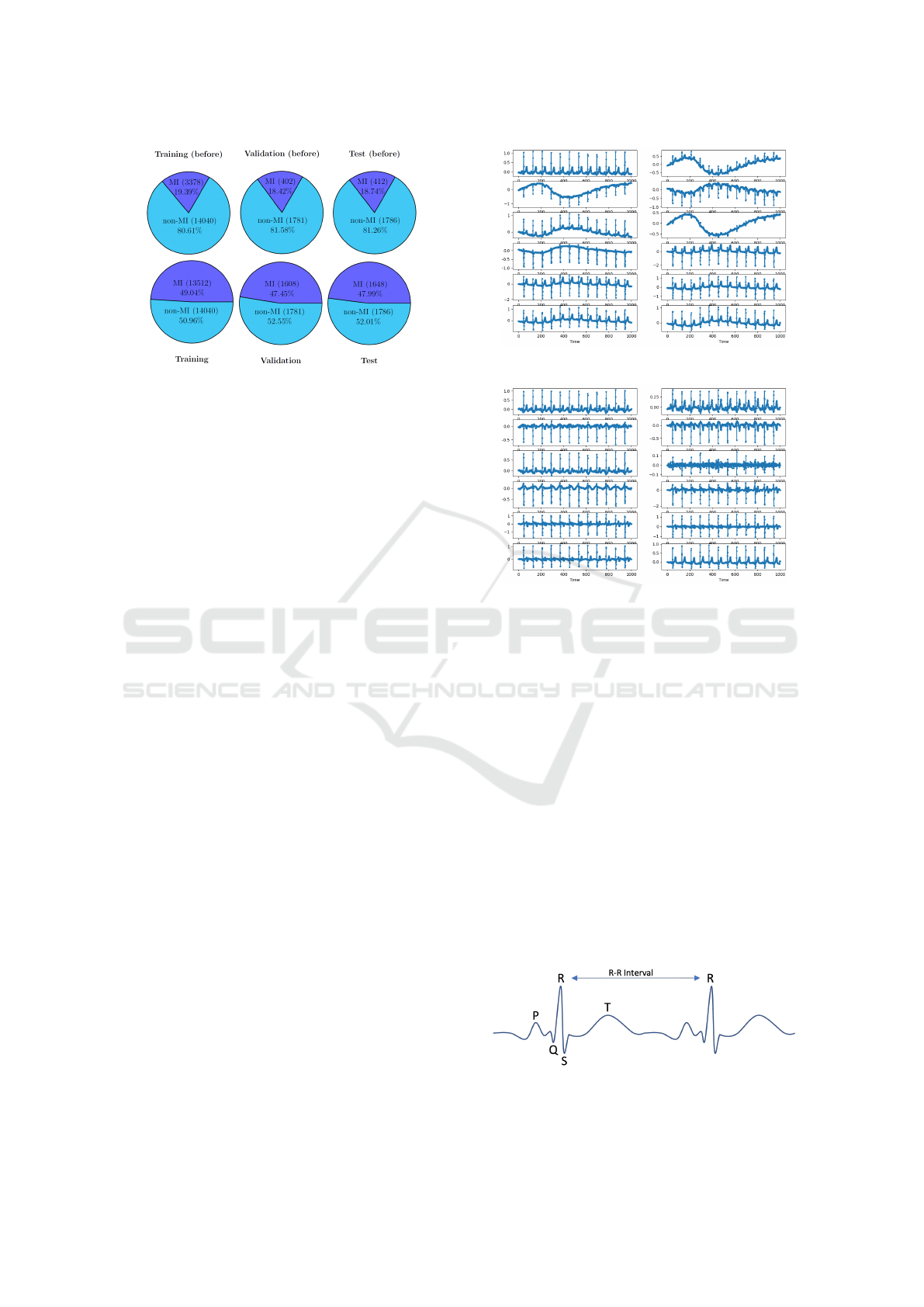
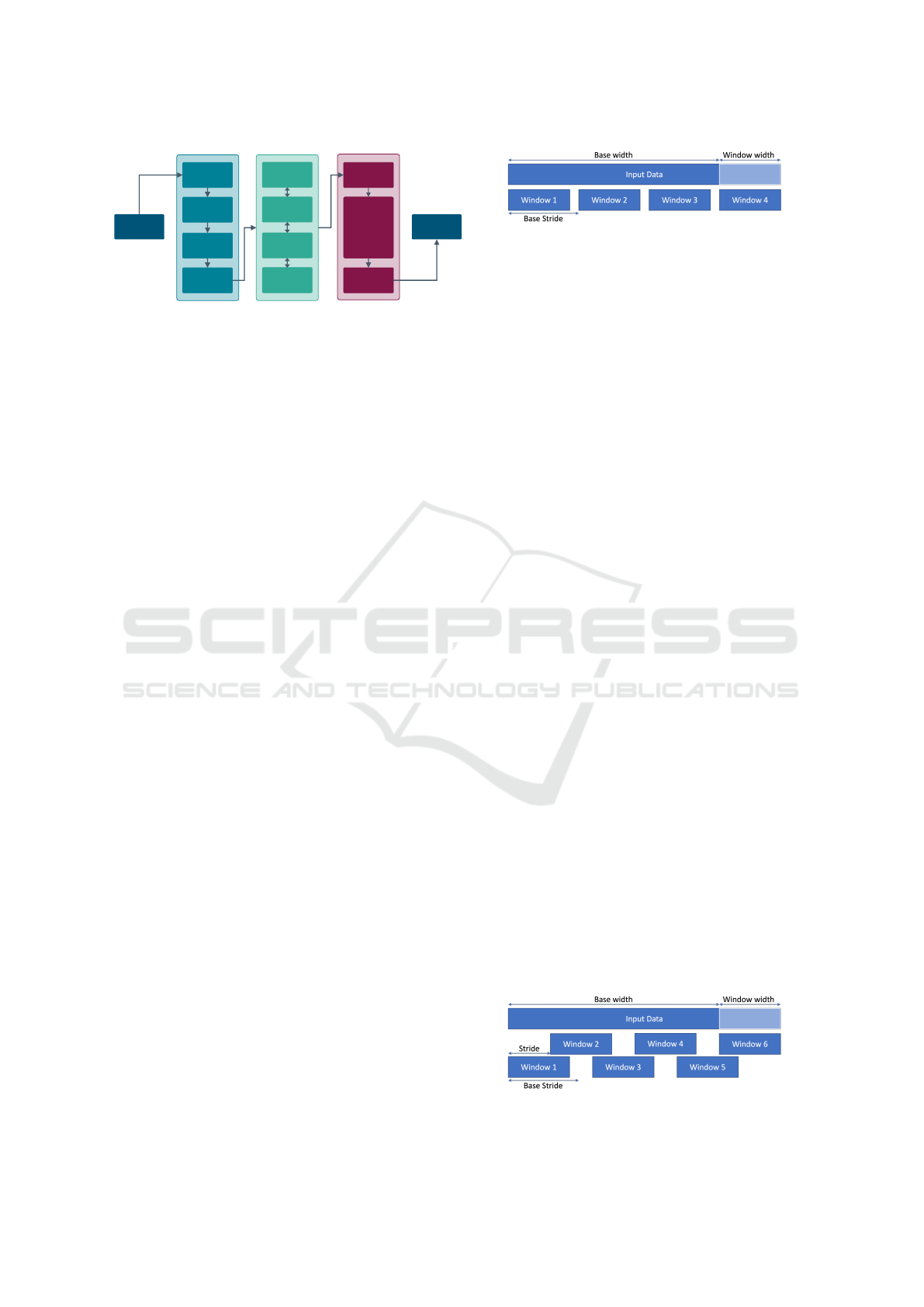
Figure 5: Distribution per class before and after balancing.
Applying this sliding-window approach to the whole
dataset, we obtain a training set of 27.552 recordings
(13.512 MI and 14.040 non-MI), a validation set of
3.389 recordings (1.608 MI and 1.781 non-MI), and
a test set of 3.434 recordings (1.648 MI and 1.786
non-MI). The distribution before and after balancing
is depicted in Figure 5.
3.3 Data Pre-Processing Techniques
Subsequently, we present pre-processing methods ap-
plied to the encoded and balanced dataset, including
baseline wander removal, x-scaling, Butterworth fil-
tering, and Fourier transformation. We will evaluate
the impact of these methods on model training by ap-
plying and analyse all possible combinations of the
selected pre-processing techniques (see Section 4).
3.3.1 Baseline Wander Removal
ECG measurements can be affected by various fac-
tors of external noise on the electrode, such as fin-
ger movement or other external influences (Sargolzaei
et al., 2009)(Degerli et al., 2021). A common source
of noise in these measurements is known as baseline
wander. This phenomenon occurs when the center of
the ECG signal deviates from a zero baseline and in-
stead follows an underlying curve. For example, the
top right lead in Figure 6 exhibits a sinusoidal pat-
tern over the first 7 seconds of data. Sargolzaei et al.
(Sargolzaei et al., 2009) proposed an algorithm to en-
hance the data quality of ECG measurements for ma-
chine learning applications. This algorithm iteratively
computes the wavelet transformation of the original
ECG signal, after which it reconstructs the baseline
of the ECG signal using an inverse wavelet transfor-
mation. Figure 6 displays the measurements of the
12 leads prior to applying the algorithm proposed by
Sargolzaei et al. (Sargolzaei et al., 2009), revealing a
noticeable wandering baseline across nearly all leads.
Figure 6: 12 ECG leads before BWR.
Figure 7: 12 ECG leads after BWR.
In contrast, Figure 7 illustrates the clarity of the signal
following the application of the algorithm.
3.3.2 R-Peak Detection
Each heartbeat exhibits several key indicator points,
as shown in Figure 8. The R-peak is typically the
most prominent amplitude in the ECG measurement,
reflecting the heart’s contraction as it pumps blood
throughout the body (S and Morris, 2002).
The Pan-Tompkins QRS-Detection algorithm
(Pan and Tompkins, 1985) identifies R-peaks in ECG
recordings. This algorithm incorporates a series of
filtering and processing steps designed for effective
noise reduction. Specifically, it employs a low-pass
and high-pass filtering technique, which is compa-
rable to the Butterworth filter described in Section
3.3.6. However, unlike the Butterworth filter, the Pan-
Tompkins noise cancellation filter is typically imple-
Figure 8: Sketch of an idealised ECG Lead-1 measurement.
Own representation based on (Singh et al., 2018).
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
238
mented in a recursive manner for any arbitrary but
fixed time-step t ∈ R.
Let f denote the ECG signal. The low-pass filter
f
low
: R → R and the high-pass filter f
high
: R → R are
defined as follows:
f
low
(x) := 2 f
low
(x −t) − f
low
(x − 2t)
+ f (x) − 2 f (x − 6t) + f (x − 12t)
f
high
(x) := 32 f (x − 16t) − f
high
(x −t)
− f (x) + f (x − 32t)
For negative time values, the functions for the
ECG signal and both filters equal zero. This approach
ensures that there is no output for time points that do
not exist in the signal.
These filters then calculate the noise-filtered sig-
nal s : R → R:
˜s(x) := f
high
( f
low
( f (x)))
s(x) :=
˜s(x)
max(| ˜s(x)|)
In the following step, the algorithm computes the
derivative of this filtered signal, and the result is
squared. This processed data stream is then aver-
aged over a moving window of 150 ms, facilitating
the identification of approximate R-peaks.
As an optimization to this algorithm, a ”climb-the-
hill” approach is applied after detecting the R-peaks
with the Pan-Tompkins algorithm. This technique
checks if an identified R-peak can adjust left or right
to increase its value. If this adjustment is possible,
the R-peak repositions until any movement in either
direction results in a decreased value of the function.
The ”climb-the-hill” optimized Pan-Tompkins
QRS-Detection algorithm demonstrates promising re-
sults, although it occasionally misidentifies T-waves
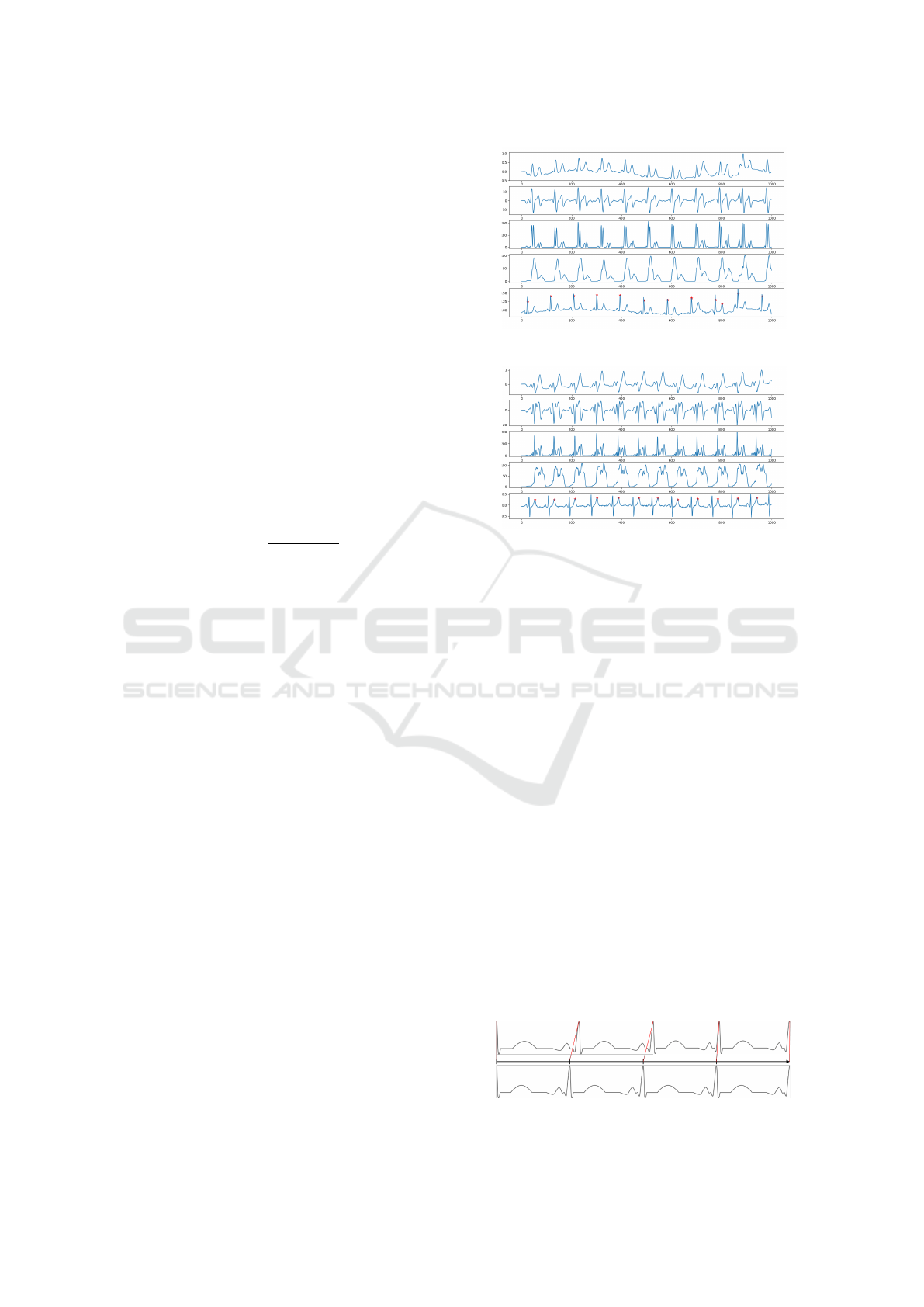
as R-peaks. For an example of accurate identification,
see Figure 9. For an instance where a T-wave is incor-
rectly identified to be an R-peak, refer to Figure 10.
Each figure illustrates data from the first four steps of
the Pan-Tompkins algorithm outlined earlier (band-
pass filter, derivative, squaring, moving average) and
marks the identified approximate R-peaks with a pink
cross on the original input data in the bottom plot.
3.3.3 Y-Axis Scaling
To normalize the data, the Scikit Learn Standard-
Scaler is used. The scaler takes all ECG recordings
from the training dataset, calculates the average value
and variance, and uses this to transform all ECGs of
the whole dataset. This generates ECG recordings
which are centered around 0 and with variance 1.
Figure 9: Correctly identified R-peaks.
Figure 10: Wrongly identified R-peaks.
3.3.4 X-Axis Scaling
We propose an x-scaling technique optimized for this
specific use case. The underlying concept of this scal-
ing addresses the issue that medical health data does
not remain within a static frequency domain; instead,
the frequency varies over time, as exemplified by the
non-constant nature of heart rate. Through x-scaling,
or time-scaling, the data is transformed into a format
that accounts for these temporal variations. Given that
this approach is not a conventional algorithm, we pro-
vide a detailed explanation of the concept and its im-
plementation, as outlined in Algorithm 1.
The x-scaler is specifically designed for ECG
recordings, as it relies on detecting and aligning heart-
beats within the observed data. To transform the
data, the algorithm iterates over all data points in
the time-series window to identify the heartbeats us-
ing the Pan-Tompkins QRS Detection algorithm (Pan
and Tompkins, 1985). Although the Pan-Tompkins
algorithm operates independently for each lead, all
12 leads in this use case correspond to measurements
from a single heart. Therefore, additional steps are
taken to determine the most likely heartbeat time
points in each recording from all 12 leads. First, the
Figure 11: X-scaling of a 4-second window.
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
239
Algorithm 1: Scaling Beats (x-axis scaling).
1: for i = 0 to len(beats) − 2 do
2: di f f erence ← beats[i + 1] − beats[i]
3: rate ←
sampling rate
di f f erence
4: for j = 0 to di f f erence − 1 do
5: k ← i ∗ sampling rate + int(round( j ∗ rate))
6: n ← i ∗ sampling rate + int(round(( j + 1) ∗
rate))
7: m ← beats[i] + j
8: if n ≥ len(records) then
9: continue
10: end if
11: # Standard case
12: if n − k = 1 then
13: scaled[k] ← records[m]
14: scaled[n] ← records[m + 1]
15: # Squashed to single index:
16: # take average value
17: else if k = n then
18: scaled[k] ←
records[m]+records[m+1]
2
19: # Stretched to three indexes:
20: # take average value as
21: # additional middle point index
22: else if n − k = 2 then
23: scaled[k] ← records[m]
24: scaled[k + 1] ←
records[m]+records[m+1]
2
25: scaled[n] ← records[m + 1]
26: end if
27: end for
28: end for
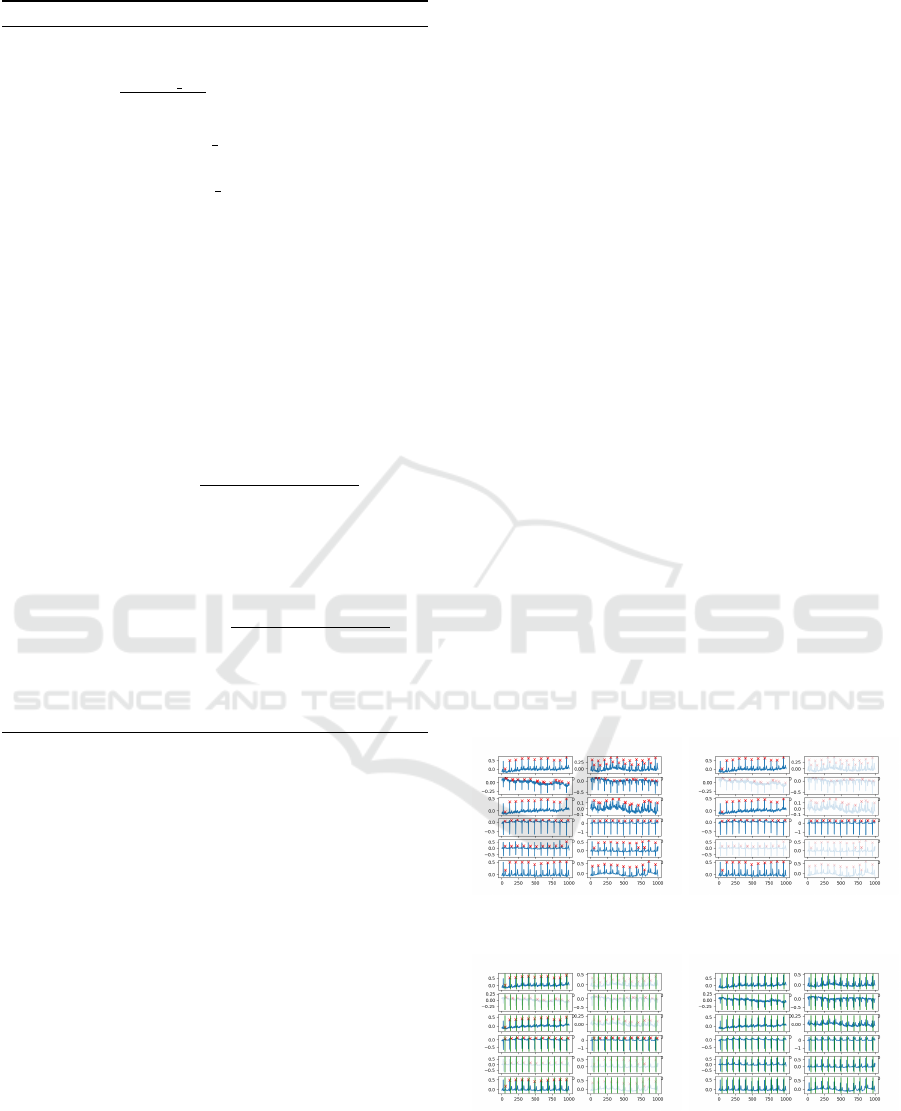
results from the Pan-Tompkins QRS detection are col-
lected, and the number of detected heartbeats per lead
is counted across the entire time window as can be
seen in Figure 12a. The dominant count of heartbeats
per lead is identified, and any leads exhibiting a differ-
ent number of beats are excluded from further analy-
sis (see Figure 12b). For the remaining selected leads,
the time points of the detected heartbeats are averaged
to establish a consensus on the overall heartbeat (see
Figure 12c). The final detected heartbeats are visual-
ized in Figure 12d.
Following this detection, the algorithm stretches
or compresses each heartbeat along the x-axis to en-
sure exactly one heartbeat is represented per ”sec-
ond”. This adjustment guarantees a constant recur-
rence rate in the data. The process for scaling a 4-
second window of irregular data to a regular pattern
is visualized in Figure 11.
As illustrated in Algorithm 1, the scaling process
begins by calculating the total number of time steps
between beat i and i + 1. The sampling rate is then
divided by this number to determine a scaling rate,
which indicates how far the points need to be com-
pressed or stretched in time. Since the data points
are stored in memory as an array, we cannot simply
move the x-axis points around; instead, we approxi-
mate their positions by rounding to the next integer
value. This approximation can introduce a timing er-
ror of up to half the sampling rate per second (e.g.,
up to 5 ms for a 100 Hz sampling rate). However,
with error rates of 0.5% or 0.1%, we consider this
error to be sufficiently small for the given use case,
outweighed by the benefits of the transformation.
After calculating the scaling rate, we iterate over
each point for each heartbeat interval. Each point is
scaled by the scaling rate and mapped onto the next
integer value. If two points project onto the same in-
dex, the average value between those points is stored
at that index. If two points project to non-adjacent in-
dexes, intermediate points are filled with a linear in-
terpolation between the two scaled points.
This procedure can be applied to a continuous
stream of data. However, a limitation is that the win-
dow lengths of each dataset might differ. To address
this, all datasets are truncated to the same length dur-
ing preprocessing, specifically to the length of the
shortest processed dataset. We expect this transforma-
tion to yield improved results, as the machine learn-
ing algorithm will focus on classifying anomalies in
the heartbeat without needing to adapt to linear trans-
formations over time. It is important to note that this
approach may result in the loss of information based
on the frequency of the heartbeat.
(a) ECG with detected R-
peaks per lead
(b) 5 selected leads of ECG
with R-peak detection
(c) 5 selected leads of ECG
with heartbeat detection
(d) ECG with heartbeat de-
tection per lead
Figure 12: ECG lead analysis.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
240

3.3.5 Fourier Transformation
A Fourier Transformation analyzes a function by ex-
tracting the amplitude and frequency of its underly-
ing structure. It generates a new function that de-
picts spikes of amplitude at corresponding frequency
points. However, this transformation does not retain
temporal shifts; thus, two functions that are identi-
cal except for their position along the x-axis yield the
same Fourier Transformation. To address the limita-
tion of identifying trends over time, the Short-Time
Fourier Transform (STFT) is employed, which di-
vides the raw input data into short time intervals as
explained in (Dey et al., 2021). A Fourier Transfor-
mation is then applied to each interval, resulting in
a two-dimensional representation that illustrates the
amplitude of various frequencies across time.
3.3.6 Butterworth Filter
The Butterworth filter is utilized to selectively filter
out frequencies outside a specified range. In this im-
plementation, a lower threshold of 20Hz and an up-
per threshold of 200Hz are chosen. These thresh-
olds are appropriate for ECG measurements, which
correspond to the rhythm of human heartbeats, typ-
ically occurring within this frequency range (Naseri
and Homaeinezhad, 2012).
3.4 Description of Models
To evaluate the effectiveness of the four pre-
processing approaches presented, we compare the
performance of an LTC against RNN and LSTM mod-
els. Each model’s architecture is minimalistic, with
a limited number of connected layers, to ensure a
clear performance comparison following various pre-
processing steps. All models accept batched time-
series input structured as a three-dimensional tensor
with the shape (batch size, number of features, num-
ber of timesteps). We set the batch size to 32, the
number of features to 12 (corresponding to the 12
leads in each ECG recording), and the number of
timesteps to 500. The input tensor is processed by a
TensorFlow base layer, where the cell type is defined
as either RNN, LSTM, or LTC. The output of this base
layer is then passed to a dense layer, producing a sin-
gle output that indicates the prediction score for My-
ocardial Infarction. For training the models, we use
the Adam optimizer with a learning rate of 0.005 for
LTCs and 0.001 for LSTMs and RNNs, following the
guidelines provided by the authors in (Hasani et al.,
2020). For each configuration of pre-processing steps,
we train all three models for 100 epochs and compare
their accuracy, precision and recall using the valida-
tion set of the PTB-XL dataset.
Afterwards, we will select the best-performing
pre-processing techniques based on the obtained re-
sults, and train and optimise final models for the de-
tection of MIs, which will be then evaluated on the
train set (compare Figure 2). This approach and the
associated models are discussed in Section 5.
4 ANALYZING THE EFFECT OF
PRE-PROCESSING
TECHNIQUES
Table 2 displays the performance metrics accuracy,
precision, and recall of the trained LTC, LSTM, and
RNN models for the four investigated pre-processing
approaches on the validation set. The pre-processing
steps are indicated by crosses (disabled) and ticks (en-
abled). We evaluate all possible combinations of the
four pre-processing steps x-scaling (XS), Butterworth
filtering (BF), Fourier transformation (BF) and base-
line wander removal (BWR). The performance met-
rics are based on the validation set after training the
models on the training set for 100 epochs.
The LSTM has an average accuracy of 0.7608,
outperforming the LTC and RNN, which have average
accuracies of 0.7319 and 0.7064, respectively. Fur-
thermore, the LSTM achieves a higher average pre-
cision of 0.7691, compared to the LTC’s 0.7034 and
the RNN’s 0.6624. In terms of average recall, the LTC
leads with 0.7616, followed by the LSTM at 0.6959
and the RNN at 0.4465. This indicates that the RNN
is not that effective as the LSTM and the LTC for
the task of detecting myocardial infarctions in 12-lead
ECG recordings. We observe the following for the
impact of the four pre-processing steps.
1) X-Scaling significantly enhances performance.
As shown in Table 2, configurations with enabled x-
scaling outperformed those with it disabled, with only
one exception. For LSTM, the x-scaling enabled con-
figurations averaged 5.28% higher in accuracy. The
RNN configurations demonstrated an even greater in-
crease of 11.47%. In contrast, the LTC configurations
showed a modest improvement of 1.98%.
2) There are no significant improvements ob-
served from enabling BWR. Indeed, it often results
in a decrease in the model’s accuracy as can be ob-
served from Table 2. When all pre-eprocessing steps
are fixed except for BWR, the LTC model shows a de-
cline in accuracy in five cases, while only two cases
exhibit an increase. Similarly, the RNN model experi-
ences a decrease in five cases and an increase in three
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
241
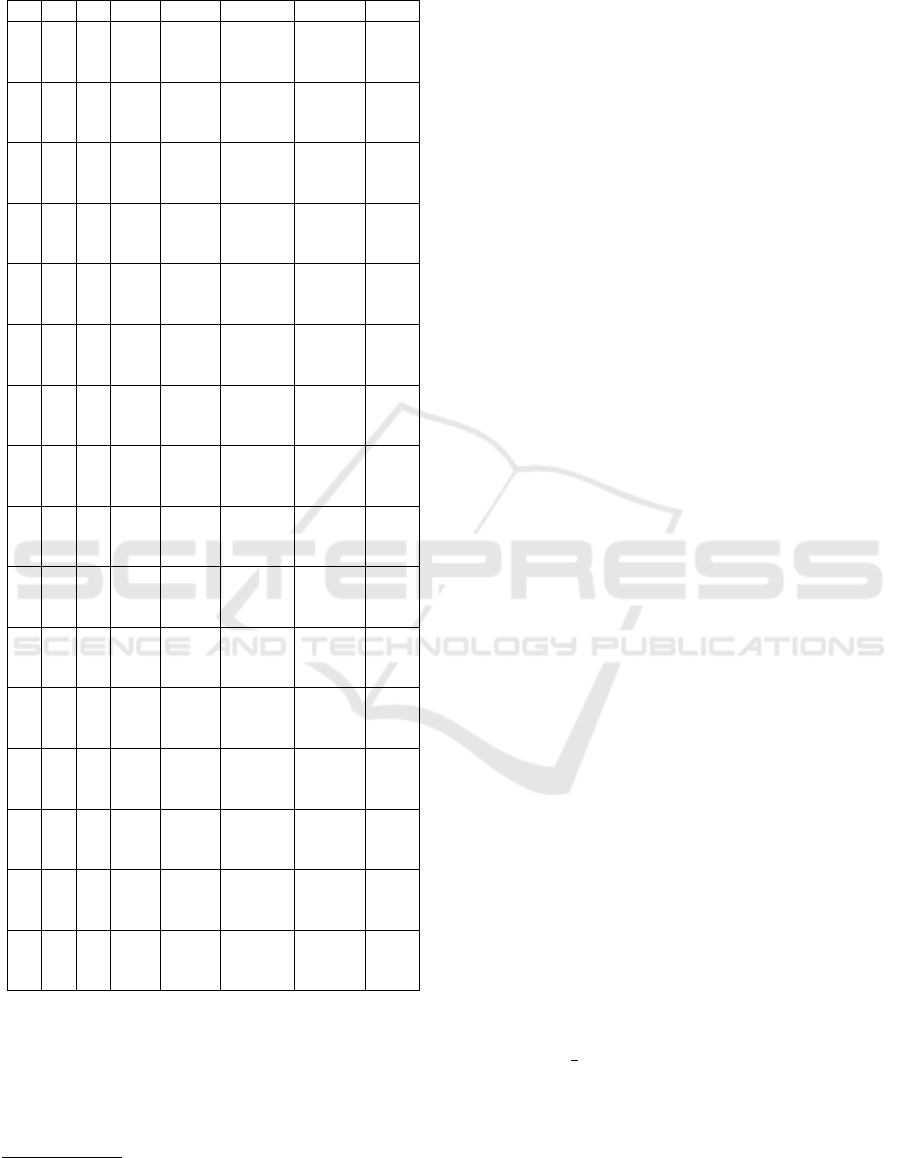
Table 2: Validation set accuracy, precision and recall for all
configurations of the investigated pre-processing methods
XS BF FT BWR Model Accuracy Precision Recall
LTC 0.790 0.819 0.649
✗ ✗ ✗ ✗ LSTM 0.732 0.730 0.714
RNN 0.626 0.539 0.420
LTC 0.756 0.762 0.725
✗ ✗ ✗ ✓ LSTM 0.780 0.764 0.797
RNN 0.650 0.630 0.623
LTC 0.743 0.715 0.791
✗ ✗ ✓ ✗ LSTM 0.739 0.745 0.718
RNN 0.680 0.589 0.675
LTC 0.780 0.751 0.817
✗ ✗ ✓ ✓ LSTM 0.750 0.774 0.660
RNN 0.672 0.656 0.612
LTC 0.739 0.715 0.785
✗ ✓ ✗ ✗ LSTM 0.724 0.737 0.630
RNN 0.649 0.587 0.377
LTC 0.715 0.671 0.784
✗ ✓ ✗ ✓ LSTM
6
- - -
RNN 0.645 0.524 0.381
LTC 0.645 0.557 0.852
✗ ✓ ✓ ✗ LSTM 0.708 0.708 0.560
RNN 0.607 0.542 0.546
LTC 0.629 0.575 0.696
✗ ✓ ✓ ✓ LSTM 0.700 0.681 0.689
RNN 0.664 0.608 0.271
LTC 0.761 0.760 0.739
✓ ✗ ✗ ✗ LSTM 0.801 0.837 0.699
RNN 0.788 0.715 0.441
LTC 0.792 0.800 0.778
✓ ✗ ✗ ✓ LSTM 0.814 0.838 0.711
RNN 0.792 0.817 0.306
LTC 0.750 0.709 0.806
✓ ✗ ✓ ✗ LSTM 0.801 0.821 0.709
RNN 0.731 0.643 0.620
LTC
7
- - -
✓ ✗ ✓ ✓ LSTM 0.801 0.825 0.730
RNN 0.771 0.768 0.348
LTC 0.773 0.693 0.863
✓ ✓ ✗ ✗ LSTM 0.769 0.779 0.724
RNN 0.777 0.750 0.545
LTC 0.636 0.576 0.802
✓ ✓ ✗ ✓ LSTM 0.775 0.785 0.693
RNN 0.757 0.770 0.302
LTC 0.742 0.700 0.793
✓ ✓ ✓ ✗ LSTM 0.764 0.751 0.711
RNN 0.767 0.768 0.353
LTC 0.730 0.750 0.545
✓ ✓ ✓ ✓ LSTM 0.754 0.761 0.694
RNN 0.728 0.693 0.326
cases
8
. In contrast, the LSTM model demonstrates
a greater number of instances where enabling BWR
enhances the model’s accuracy.
3) The use of the Butterworth Filter in the pre-
processing phase generally results in decreased accu-
racy. Specifically, there are no configurations where
8
Note that there is one case more for the RNN model
than for the LTC model due to data loss.
it improves the accuracy of the LSTM model. For the
LTC and RNN models, it only enhances performance
in one and two cases, respectively.
4) Applying the Fourier transformation also leads
to a decrease in the models’ accuracies in most cases.
This holds true for the LTC, LSTM and RNN.
The best overall performance is observed for the
LSTM model, achieving with 0.8140 the highest ac-
curacy when x-scaling and baseline wander removal
are enabled whereas Fourier transformation and But-
terworth filtering are disabled.
Based on the obtained results, we conclude that
the combination of pre-processing techniques in
which only the x-scaling and baseline wander removal
are enabled is the optimal approach for our use case
and, hence, will employ these for building the final
MI detection models as presented in the following.
5 FINAL MODEL SELECTION
AND OPTIMISATION
Having evaluated the impact of various signal pre-
processing techniques and identified the optimal com-
bination for our use case, we will now build the fi-
nal MI detection models. We will first conduct a hy-
perparameter optimization to identify the best model
configuration parameters. Subsequently, we will train
and compare the final models using the obtained opti-
mal parameters and the pre-processed data.
5.1 Hyperparameter Optimization
In the previous section, we have evaluated the im-
pact of changing the input data on the model’s per-
formance. However, additional changes to the pa-
rameters of the model itself have impact on its per-
formance. Therefore, in this section, we evaluate the
impact of changing model parameters.
The specific analysed and optimised hyperparam-
eters as well as their ranges examined are shown in
Table 3. As outlined by Hasani et al. (Hasani et al.,
2020), LTCs require a bigger learning rate than LSTM
or RNNs models. Precisely, they use 0.005 for LTCs
and 0.001 for LSTMs and RNNs, which we used as
base learning rates as well. To analyse the impact of
changing the learning rate, we scaled the base learn-
ing rate by
1
2
, 1 and 2. Additionally, the number of
units was scaled between 2 and 12 for all model types.
As before, each test-run is executed for 100 epochs
and the performance was measured using the valida-
tion split (see Section 3.1). All other configuration
parameters remain fixed as described in Section 3.4.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
242
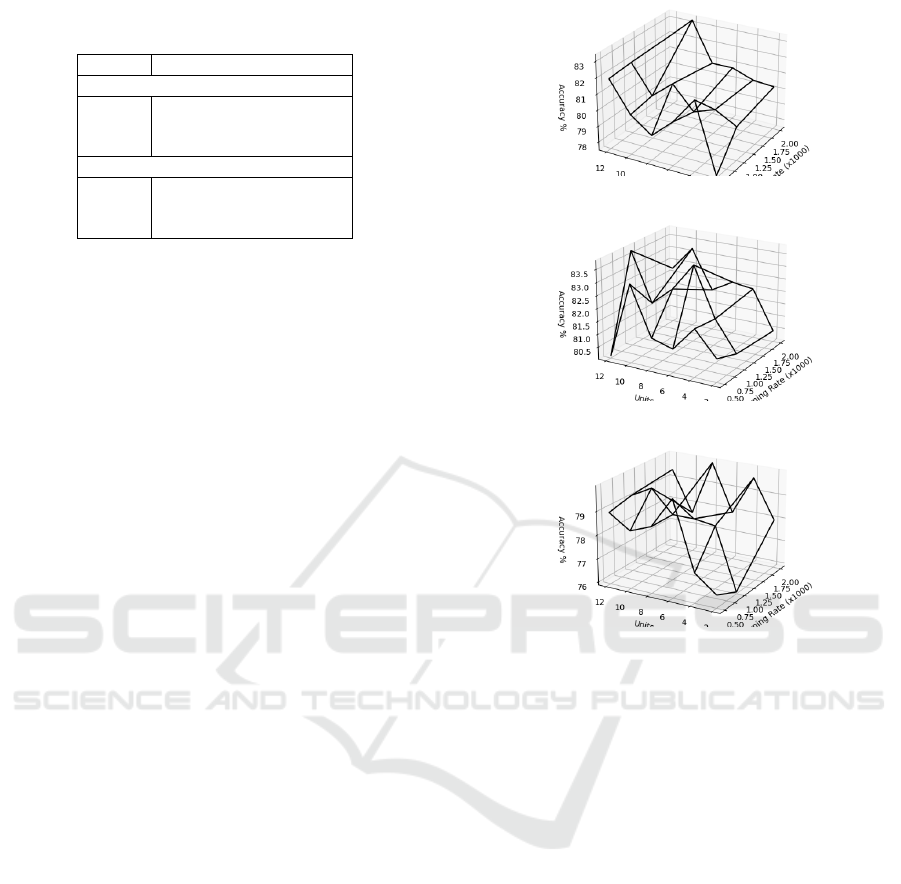
Table 3: Hyperparameter values evaluated and optimal hy-
perparameter values found for the investigated models.
Model Learning Rate Units
Evaluated
LTC 0.0025 - 0.01 2 - 12
LSTM 0.0005 - 0.002 2 - 12
RNN 0.0005 - 0.002 2 - 12
Optimal
LTC 0.01 10
LSTM 0.001 12
RNN 0.002 8
After executing all combinations, we compared
the accuracy values on the validation set for each hy-
perparameter setting. Figures 13a- 13c illustrate the
accuracy results achieved across the various investi-
gated hyperparameters for the LTC, LSTM, and RNN
model, respectively. As a result, our identified opti-
mal hyperparameters across all models are listed in
Table 3. As it can be seen, we found the optimal
settings for LSTM are 12 units and a learning rate
of 0.001. For RNN, 8 units and a learning rate of
0.002 manifest the best values. For LTC, 10 units and
a learning rate of 0.01 performed best. Therefore, we
chose these as the respective default values for the fur-
ther training of the MI detection models.
5.2 MI Detection Models
In the final step, we will use the optimal model pa-
rameters and the identified best pre-processing tech-
niques, x-scaling and BWR, to train, test, and discuss
the final MI detection models. We will perform 100-
epoch training with the optimized hyperparameters
and these techniques, while keeping all other training
and model parameters consistent with Section 3.4.
The performance results of the final models for
classifying MIs on the test set with 3434 records (see
Section 3.1) are presented in Table 4. As can be
deduced, the LSTM model shows a slightly higher
accuracy of 84.61% compared to the LTC model at
83.11%, both outperforming the RNN that manifests
70.31%. The AUC metric, which is commonly used
for measuring ECG-based performance, is a measure-
ment based on the receiver operating characteristic
curve, which measures how the true positive rate com-
pares to the false positive rate at different classifica-
tion thresholds. As we can see, the LTC has slightly
higher AUC values. Specifically, it shows an AUC
of 93.75%, whereas LSTM and RNN exhibit values
of 92.84% and 85.96%, respectively, suggesting that
the LTC model demonstrates a better trade-off be-
tween sensitivity and specificity. This can also be ob-
served when analyzing the recall and precision met-
rics. While the LTC model manifests the lowest pre-
cision at 79.92% compared to LSTM at 83.63% and
(a) LTC model
(b) LSTM model
(c) RNN model
Figure 13: Accuracy over all hyperparameter settings.
RNN at 90.57%, it demonstrates a significantly higher
recall value. The LTC achieves a recall of 89.93%,
whereas we can observe values of 84.14% for the
LSTM and a recall of only 47.73% for the RNN.
In the context of myocardial infarction detection,
precision measures how many of the predicted heart
attacks were actually heart attacks, while recall mea-
sures how many actual heart attacks were correctly
identified by the model. A model with high recall
is crucial in medical applications like MI detection,
as it minimizes the risk of missed diagnoses, as it
means fewer heart attacks are missed. The results in-
dicate that although the LTC model may incorrectly
predict some cases, it is more effective at catching ac-
tual heart attacks, leading to better patient care.
The values for true positives (TP), false positives
(FP), true negatives (TN), and false negatives (FN)
presented in Table 4 support this observation. The
RNN model shows a significant number of false neg-
atives, indicating that many instances of myocardial
infarctions are being incorrectly classified as non-MI,
which accounts for the low recall values noted ear-
lier. In contrast, both the LSTM and LTC models ex-
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
243

Table 4: Test results with optimal configurations.
Metric LTC LSTM RNN
Accuracy 0.8311 0.8461 0.7031
Precision 0.7992 0.8363 0.9057
Recall 0.8993 0.8414 0.4773
AUC 0.9375 0.9284 0.8596
TP 1611 1384 853
FP 398 267 83
TN 1253 1528 1568
FN 172 255 930
hibit lower numbers of false positives and false nega-
tives. Specifically, while the LSTM maintains a more
balanced distribution between FP and FN, the LTC
model has a higher FP count but fewer FNs. In this
context, a false negative represents an undetected MI,
which could lead to preventable death, whereas a false
positive could trigger unnecessary false alarms.
In conclusion, our results indicate that LTCs
outperform LSTMs and RNNs in the MI detection
use case. While LTCs generate more false alarms,
they also identify more actual myocardial infarctions,
making them a more reliable choice in the medical
context. Detecting true MIs is crucial, as missed di-
agnoses can severely impact patient health. Although
false positives may cause unnecessary stress for emer-
gency services, they are a lesser concern compared
to the risk of lost lives. Thus, we believe the LTC
model’s ability to reliably detect MIs justifies the
trade-off of additional false alarms, making it the
more effective choice for this critical application.
6 CONCLUSION & FUTURE
WORK
In this paper, we evaluated the performance of Liquid
Time-Constant Neural Networks in classifying my-
ocardial infarctions in ECG data, focusing on the ef-
fects of various pre-processing techniques using the
PTB-XL dataset. From our experiments, we can con-
clude the following regarding the impact of the four
pre-processing steps: The combination of applying
our novel presented x-scaling approach in combina-
tion with the Baseline Wander Removal technique
tends to improve model performance, especially for
the LSTM and the LTC models. On the other hand,
the Butterworth Filter and Fourier transformation tend
to decrease the models’ performance.
Based on the insights gained from the evalua-
tion of pre-processing techniques, we developed MI
detection models utilizing LTCs and benchmarked
their performance against LSTM and RNN models.
This comparison highlights the potential of LTCs for
real-world applications in the medical domain. Our
findings suggest that LTCs show competitive per-
formance, achieving accuracy values comparable to
LSTMs while maintaining strong recall rates. These
attributes position LTCs as a promising option for
enhancing diagnostic capabilities in medical applica-
tions, suggesting the need for further exploration.
Future work could focus on optimizing LTC ar-
chitectures and evaluating their performance across a
wider range of medical datasets to validate their ef-
fectiveness in diverse clinical scenarios. Additionally,
we intend to validate our findings by comparing them
against more complex model architectures with opti-
mized hyperparameters. We will also assess the im-
pact of x-scaling on other time series datasets, consid-
ering its promising results in improving performance.
ACKNOWLEDGMENT
This research was supported by SPATIAL project
that has received funding from the European Union’s
Horizon 2020 research and innovation programme
under grant agreement No.101021808.
REFERENCES
Acharya, U. R., Fujita, H., Oh, S. L., Hagiwara, Y., Tan,
J. H., and Adam, M. (2017). Application of deep con-
volutional neural network for automated detection of
myocardial infarction using ecg signals. Information
Sciences, 415-416:190–198.
Anand, A., Kadian, T., Shetty, M. K., and Gupta, A. (2022).
Explainable ai decision model for ecg data of cardiac
disorders. Biomedical Signal Processing and Control,
75:103584.
Atiea, M. A. and Adel, M. (2022). Transformer-based neu-
ral network for electrocardiogram classification. Inter-
national Journal of Advanced Computer Science and
Applications, 13(11).
Chen, R. T. Q., Rubanova, Y., Bettencourt, J., and Duve-
naud, D. (2018). Neural ordinary differential equa-
tions. arXiv, 5.
Degerli, A., Zabihi, M., Kiranyaz, S., Hamid, T., Mazhar,
R., Hamila, R., and Gabbouj, M. (2021). Early detec-
tion of myocardial infarction in low-quality echocar-
diography. IEEE Access, 9:34442–34453.
Dey, M., Omar, N., and Ullah, M. A. (2021). Temporal
feature-based classification into myocardial infarction
and other cvds merging cnn and bi-lstm from ecg sig-
nal. IEEE Sensors Journal, 21(19):21688–21695.
Funahashi, K. and Nakamura, Y. (1993). Approximation of
dynamical systems by continuous time recurrent neu-
ral networks. Neural Networks, 6(6):801–806.
Gharaibeh, A. and Quwaider, M. (2022). Electrocardio-
gram classification problem solving using deep learn-
ing algorithms : Fully connected neural networks. In
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
244

2022 13th International Conference on Information
and Communication Systems (ICICS), pages 281–288.
Hammad, M., Chelloug, S. A., Alkanhel, R., Prakash,
A. J., Muthanna, A., Elgendy, I. A., and Pławiak, P.
(2022). Automated detection of myocardial infarction
and heart conduction disorders based on feature selec-
tion and a deep learning model. Sensors, 22(17).
Hasani, R., Lechner, M., Amini, A., Rus, D., and Grosu, R.
(2020). Liquid time-constant networks. arXiv, 4.
Joloudari, J. H., Mojrian, S., Nodehi, I., Mashmool, A.,
Zadegan, Z. K., Shirkharkolaie, S. K., Alizadehsani,
R., Tamadon, T., Khosravi, S., Kohnehshari, M. A.,
Hassannatajjeloudari, E., Sharifrazi, D., Mosavi, A.,
Loh, H. W., Tan, R.-S., and Acharya, U. R. (2022).
Application of artificial intelligence techniques for au-
tomated detection of myocardial infarction: a review.
Physiological Measurement, 43(8):08TR01.
Knof, H., Bagave, P., Boerger, M., Tcholtchev, N., and
Ding, A. Y. (2024a). Exploring cnn and xai-based ap-
proaches for accountable mi detection in the context
of iot-enabled emergency communication systems. In
Proceedings of the 13th International Conference on
the Internet of Things, IoT ’23, page 50–57, New
York, NY, USA. Association for Computing Machin-
ery.
Knof, H., Boerger, M., and Tcholtchev, N. (2024b). Quanti-
tative evaluation of xai methods for multivariate time
series - a case study for a cnn-based mi detection
model. In Longo, L., Lapuschkin, S., and Seifert,
C., editors, Explainable Artificial Intelligence, pages
169–190, Cham. Springer Nature Switzerland.
Krawczyk, B. (2016). Learning from imbalanced data: open
challenges and future directions. Progress in Artificial
Intelligence, 5:221–232.
Lynn, H. M., Pan, S. B., and Kim, P. (2019). A deep bidi-
rectional gru network model for biometric electrocar-
diogram classification based on recurrent neural net-
works. IEEE Access, 7:145395–145405.
Mohammadi Foumani, N., Miller, L., Tan, C. W., Webb,
G. I., Forestier, G., and Salehi, M. (2024). Deep
learning for time series classification and extrinsic re-
gression: A current survey. ACM Computing Surveys,
56(9):1–45.
Muhuri, P. S., Chatterjee, P., Yuan, X., Roy, K., and Ester-
line, A. (2020). Using a long short-term memory re-
current neural network (lstm-rnn) to classify network
attacks. Information, 11(5).
Naseri, H. and Homaeinezhad, M. R. (2012). Comput-
erized quality assessment of phonocardiogram signal
measurement-acquisition parameters. Journal of Med-
ical Engineering & Technology, 36(6):308–318.
Pałczy
´
nski, K.,
´
Smigiel, S., Ledzi
´
nski, D., and Bujnowski,
B. (2022). Study of the few-shot learning for ecg clas-
sification based on the ptb-xl dataset. Sensors, 22(3).
Pan, J. and Tompkins, W. J. (1985). A real-time qrs de-
tection algorithm. IEEE Transactions on Biomedical
Engineering, BME-32(3):230–236.
Prabhakararao, E. and Dandapat, S. (2022). Multi-scale
convolutional neural network ensemble for multi-class
arrhythmia classification. IEEE Journal of Biomedical
and Health Informatics, 26(8):3802–3812.
Rai, H. M. and Chatterjee, K. (2022). Hybrid cnn-lstm deep
learning model and ensemble technique for automatic
detection of myocardial infarction using big ecg data.
Applied Intelligence, 52(5):5366–5384.
S, S. M. and Morris, F. (2002). Introduction. i-leads, rate,
rhythm, and cardiac axis. BMJ, 324(7334):415–418.
Sargolzaei, A., Faez, K., and Sargolzaei, S. (2009). A new
robust wavelet based algorithm for baseline wander-
ing cancellation in ecg signals. In 2009 IEEE Inter-
national Conference on Signal and Image Processing
Applications, pages 33–38.
Segura-Salda
˜
na, P., Britto-Bisso, F., Pacheco, D. V.,
Alvarez-Vargas, M. L., Manrique, A. L., and Nicho,
G. M. B. (2022). Automated detection of myocardial
infarction using ecg-based artificial intelligence mod-
els: a systematic review. In 2022 IEEE 16th Interna-
tional Conference on Application of Information and
Communication Technologies (AICT), pages 1–6.
Singh, S., Pandey, S. K., Pawar, U., and Janghel, R. R.
(2018). Classification of ecg arrhythmia using recur-
rent neural networks. Procedia Computer Science,
132:1290–1297.
´
Smigiel, S., Pałczy
´
nski, K., and Ledzi
´
nski, D. (2021). Deep
learning techniques in the classification of ecg sig-
nals using r-peak detection based on the ptb-xl dataset.
Sensors, 21(24).
Strodthoff, N., Wagner, P., Schaeffter, T., and Samek, W.
(2021). Deep learning for ecg analysis: Benchmarks
and insights from ptb-xl. IEEE Journal of Biomedical
and Health Informatics, 25(5):1519–1528.
Wagner, P., Strodthoff, N., Bousseljot, R.-D., Kreiseler, D.,
Lunze, F. I., Samek, W., and Schaeffter, T. (2020).
Ptb-xl, a large publicly available electrocardiography
dataset. Scientific Data, 7(1).
Xiong, P., Lee, S. M.-Y., and Chan, G. (2022). Deep learn-
ing for detecting and locating myocardial infarction
by electrocardiogram: A literature review. Frontiers
in Cardiovascular Medicine, 9.
´
Smigiel, S., Pałczy
´
nski, K., and Ledzi
´
nski, D. (2021). Ecg
signal classification using deep learning techniques
based on the ptb-xl dataset. Entropy, 23(9):1121.
Leveraging Liquid Time-Constant Neural Networks for ECG Classification: A Focus on Pre-Processing Techniques
245
