
Early Diagnosis of Ovarian Cancer by the Integration of Whole Side
Images and Deep Learning Models
Suma K V
a
, Suma P
b
, Ananya D Hedge
c
and Rakshith R
d
Dept. of Electronics and Communication, Ramaiah Institute of Technology, Affiliated to Visvesvaraya Technological
University, Belagavi-590018, Karnataka, India
Keywords: Mitosis, Histopathology, Deep Learning, Tiling
Abstract: The Ovarian cancer subtypes have been demonstrated to represent unique pathologic entities with varying
prognosis and of Ovarian cancers have been shown to be diverse pathologic entities with different treatment
outcomes and predictions. Even though pathologists are capable of performing the tissue biopsy process with
reliability, there are some challenging situations that necessitate consulting with a specialist. We propose an
automated approach for ovarian cancer classification to enhance pathologists' performance and satisfy the
need for more accurate and reproducible diagnosis. Whole Slide Images (WSIs) tiled into accessible datasets
are used in this study. For the diagnosis and prognosis of ovarian cancer, precise measurement of mitotic
activity is essential. In order to identify two forms of mitotic activity, multipolar and caterpillar mitosis that
are frequently seen in the histopathology of ovarian tumors, an average of more than 2000 tiles were taken
from each of the WSIs using GPU-optimized tiling algorithms. To detect malignant mitotic activity, this
paper's focus includes the detection and classification of the aforementioned kinds of mitosis using deep
learning architectures. Following training, YOLO-based object detection models achieved accuracies of
78.20% and 89.33%, respectively. A trained ResNet-34 model yielded 86.25%. One important factor that
makes it possible for strong deep-learning pipelines for cancer is the tiling technique, which reduces resource
usage while preserving good image quality.
1 INTRODUCTION
It is currently acknowledged that ovarian tumors are
a diverse group of multiple different histotypes rather
than a singular illness (Kussaibi et al., 2024). These
tumors vary not only at the cellular level but also in a
wide range of other ways, including aggressiveness
and how well they respond to therapy. Until recently,
all ovarian cancers had the same treatment, which
often had unsatisfactory outcomes. Depending on the
stage of the disease, this included surgery and/or
standard chemotherapy regimens (Suma et al., 2022).
The identification and classification of cancer is
among the most popular uses for automatic
histopathology image analysis.
Histopathology
images can be analyzed using nuclear and textural
features (Farahani et al., 2022). There are studies that
describe the appearance of tissue component using
a
https://orcid.org/0000-0002-6824-068X
b
https://orcid.org/ 0000-0002-9385-9468
c
https://orcid.org/0009-0003-9379-9332
d
https://orcid.org/0009-0009-5546-9600
segmentation-based characteristics. Ovarian cancer
presents significant diagnostic challenges due to its
heterogeneity across subtypes. Histopathological
analysis, relying on mitotic activity, remains central
to its assessment. However, manual quantification is
prone to variability and significant time, motivating
automated detection methods (Kasture et al., 2021).
This paper is a part of a much wider study of “Ovarian
Cancer Detection using Deep Learning Techniques”
and explores the use of tiled WSIs, obtained from
another study, part of the same wider pursuit (“A
Novel Tile-Based Methods for Identifying Ovarian
Cancer in Histopathological Images”), for training
deep learning models to identify mitotic activity,
leveraging GPU-accelerated tiling for efficient
dataset creation. We compare various state-of-the-art
models to determine the most reliable approach for
mitosis detection in ovarian cancer. The models
K V, S., P, S., D Hedge, A. and R, R.
Early Diagnosis of Ovarian Cancer by the Integration of Whole Side Images and Deep Learning Models.
DOI: 10.5220/0013621900004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 3, pages 463-469
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
463
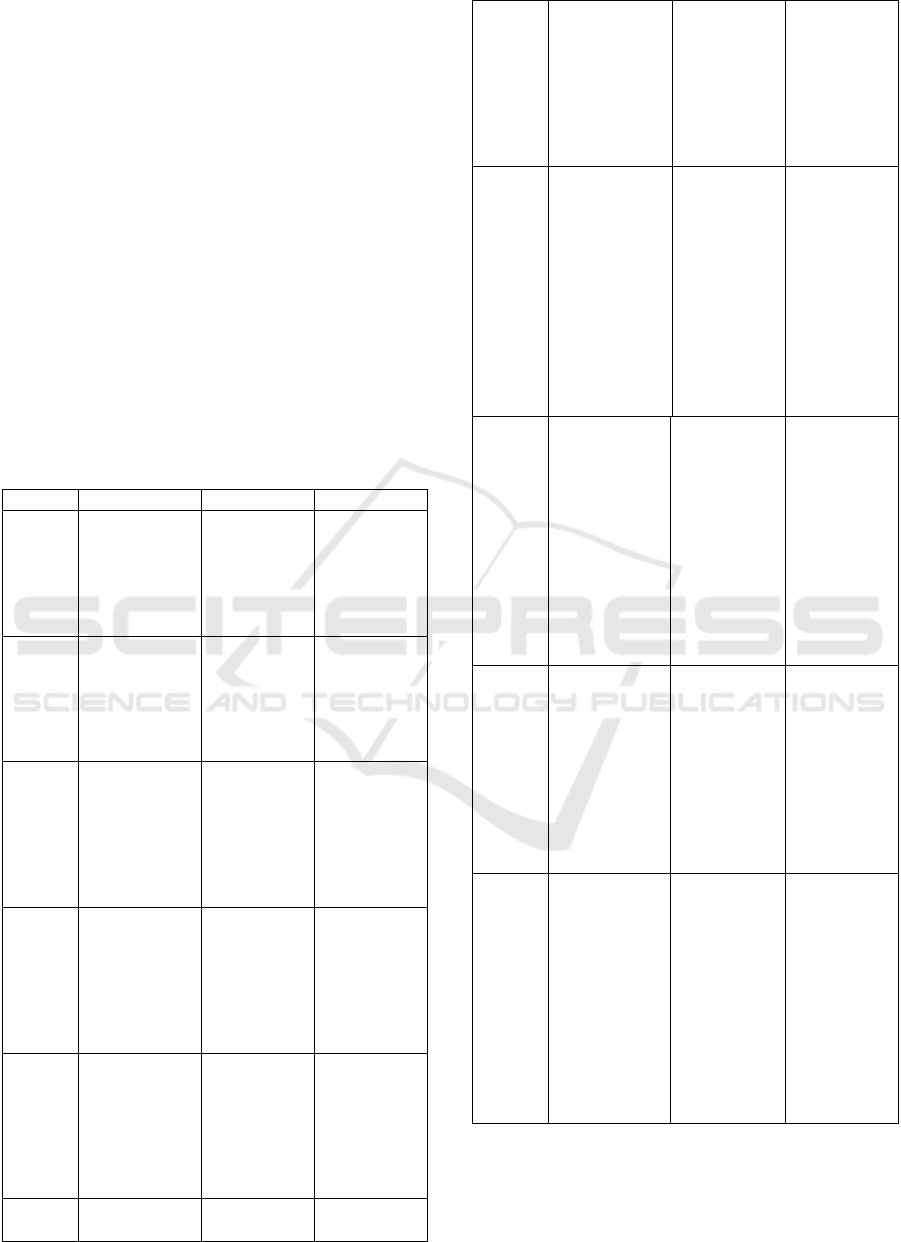
perform detection and classification for the mitosis
subtypes: Multipolar and caterpillar. As a
"translation" of pathologists' diagnostic process into
a system of computer vision that chooses
discriminative image characteristics to carry out an
automatic diagnosis, we present our suggested
automatic ovarian cancer classifier. In Figure 1, the
suggested model is summarized. Four components
comprise the design: feature extraction, machine
learning-powered categorization, picture the process
of segmentation and image pre-processing.
2 LITERATURE SURVEY
Mitotic detection in histopathology has advanced
with machine learning. Recent works emphasize
efficiency and scalability, addressing challenges like
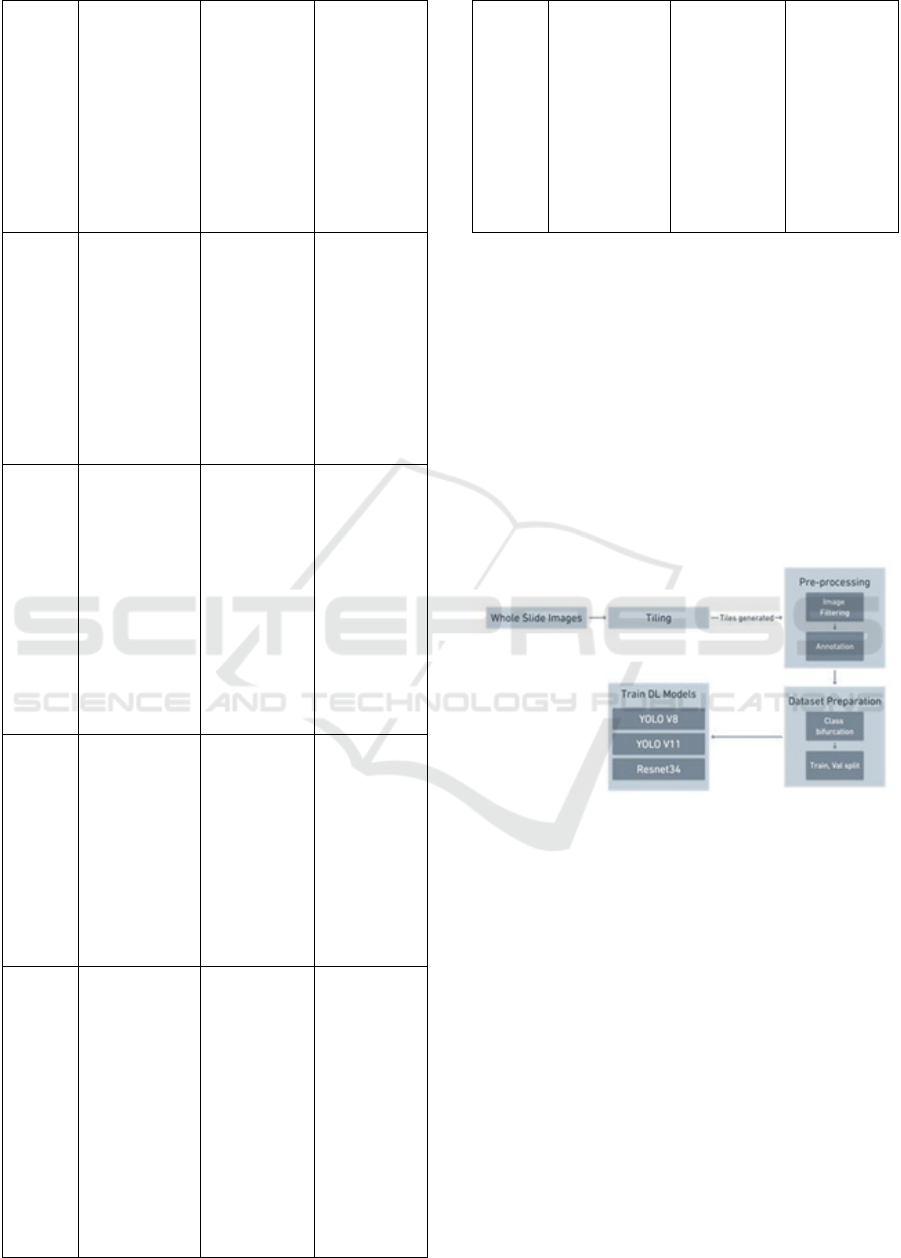
image resolution and variability as per table I.
Table 1: Study of Prior Work.
Ref. Dataset A
pp
roach Deliverables
(Farahan
i et al.,
2022)
Not explicitly
mentioned,
likely publicly
available
histopathology
datasets
Convolutiona
l Neural
Networks
(CNNs) for
histotype
classification
High
accuracy in
histotype
classification
(up to 94%).
(Kasture
et al.,
2021)
Not explicitly
mentioned,
likely publicly
available
histopathology
datasets
Convolutiona
l Neural
Networks
(CNNs)
High
accuracy in
histological
analysis.
(Cireşan
et al.,
2013)
Mitosis
Detection
dataset (breast
cancer
histology
images)
Deep
MaxPoolingC
NN with data
augmentation
High F1-
score,
winning the
ICPR 2012
mitosis
detection
contest.
(li et al.,
2019)
Publicly
available
breast cancer
datasets (ICPR,
AMIDA)
Weakly
supervised
learning with
concentric
loss function
and CNNs
(
ResNet
)
Achieves
competitive
performance
with only
image-level
labels.
(alom et
al.,
2018)
Diverse
medical image
datasets
(retinal blood
vessels, skin
lesions, lung
se
g
mentation
)
Recurrent
residual U-
Net (R2U-
Net)
architecture
for image
se
g
mentation
Improved
segmentation
accuracy
compared to
standard U-
Net.
(Mousav
i, 2023
)
99 whole-slide
ima
g
es of
First stage
detects
A two-stage
framework
canine
mammary
gland (CMG)
tumors
potential
mitotic
candidates;
second stage
classifies true
mitoses using
deep
learnin
g
.
using Mask
R-CNN and
ResNet-50
achieves an
F1 score of
76.0%.
(Aubrev
ille,
2020)
Laserendomicr
oscopy (CLE)
images from
84 patients
undergoing
surgery for oral
squamous cell
carcinoma
(OSCC)
ResNet-50
and ResNet-
101for
feature
extraction
and
classification
and Transfer
learning from
ImageNet,
Data
augmentation
High
sensitivity,
specificity,
and accuracy
for in vivo
and ex vivo
image
datasets.
(Tellez
et al.,
2018)
Three public
datasets
(MITOS-
ATYPIA-14,
ICPR 2012,
and
AMIDA13) of
breast cancer
histology
images
Two-stage
approach:
candidate
detection
using a deep
learning
model,
followed by
classification
using another
deep learning
model
High
performance
across all
three
datasets, with
an F1-score
of 0.743 on
MITOS-
ATYPIA-14.
(Bertra
m et al.,
20198)
Canine
cutaneous mast
cell tumor
(CCMCT)
dataset of
1,000 WSIs
Deep
learning-
based object
detection
(Faster R-
CNN) to
identify and
classify
mitotic
figures
Mitotic count
and spatial
distribution
of mitoses
can be used
to predict
tumor grade
and patient
outcome.
(Aksac,
2019)
Histopathologi
cal images of
papillary
thyroid
carcinoma
(PTC)
Deep
learning-
based object
detection
models
(YOLOv3,
RetinaNet) to
detect and
classify
nuclei and
mitotic
fi
g
ures
High
accuracy in
detecting
nuclei and
mitotic
figures, with
potential for
use in
automated
pathology
diagnosis and
g
radin
g
.
INCOFT 2025 - International Conference on Futuristic Technology
464
(li et al.,
2020)
Three public
datasets of
breast cancer
histology
images (ICPR
2012, ICPR
2014, and
MITOS-
ATYPIA-14)
Deep
cascaded
networks
consisting of
multiple
CNNs to
detect mitoses
in a coarse-
to-fine
manner
Improved
accuracy and
efficiency
compared to
single-stage
models, with
an F1-score
of 0.821 on
ICPR 2012.
(Chen et
al.,
2016)
Breast cancer
histology
images from
the ICPR 2012
mitosis
detection
challenge
Parallel
computation
using GPUs
to accelerate
mitosis
detection
algorithms
based on
feature
extraction and
classification
Significant
speedup
compared to
CPU-based
methods,
enabling
faster
analysis of
large
histology
images.
(Malon
et al.,
2013)
Breast cancer
histology
images from
the AMIDA13
dataset
Evaluation of
various
mitosis
detection
algorithms
based on
feature
extraction,
classification,
and deep
learning
Comparison
of different
algorithms
and
identification
of their
strengths and
weaknesses,
providing
insights for
future
algorithm
develo
p
ment
(Veta et
al.,
2015)
Breast cancer
histology
images from a
local hospital
Morphologica
l operators
and image
processing
techniques to
detect mitotic
cells
Simpler and
faster
compared to
deep
learning-
based
methods, but
might not be
as accurate.
(Paul
and
Mukherj
ee,
2013)
Various
histopathology
images,
including
breast,
prostate, and
colon cancer
Review of
different
methods for
nuclei
detection,
segmentation,
and
classification,
including
traditional
image
processing
and machine
learning
Comprehensi
ve overview
of the field
and
discussion of
various
techniques,
challenges,
and future
directions.
(Irshad
et al.,
2014)
Breast cancer
histology
images from
the ICPR 2012
mitosis
detection
challenge
Deep
cascaded
networks with
multiple
stages for
candidate
detection and
classificatio
High
accuracy in
mitosis
detection,
demonstratin
g the
effectiveness
of multi-stage
approaches.
3 METHODOLOGY
3.1 Block Diagram
Numerous techniques have been put forth to identify
nuclei in histological images. It is clear from the
results of these studies that the current approaches
work well for nuclei with consistent shapes but fall
short when the nucleus's size and shape change. A
straightforward method for categorizing mitotic
nuclei is offered in the current study. The nucleus
segmentation process is depicted in Fig. 1.
Figure. 1 Proposed model for detection of mitotic region in
whole side images
3.2 Dataset
The dataset was created using GPU-based tiling as
detailed in the referenced paper. Whole Slide Images
(WSIs) of ovarian histopathology were sourced from
the UBC-OCEAN dataset, comprising 538 WSIs
scanned at 200× magnification. Each WSI is
30,000×23,000 pixels in resolution, on average.
3.3 Pre-Processing and Dataset
Preparation
The whole slide images that make up the original
dataset are too large in file size and image dimensions
to use directly. The mitotic activity that is desired to
be represented in the images are observed only at
200x zoom of the images. This makes the division of
Early Diagnosis of Ovarian Cancer by the Integration of Whole Side Images and Deep Learning Models
465

the large images into tiles, required. Thus, the WSIs
are tiled using the Tiling algorithm proposed as a part
of the wider study. The Tiling algorithm proposed is
a novel method, that uses a custom CUDA kernel
implementing the DALI feature by Nvidia, to perform
a GPU-based tiling which utilized the GPU at its
fullest. This can be seen in table 1, where the
proposed method outperforms the existing methods.
The obtained tiles are then pre-processed, i.e. They
are filtered for observable mitotic activity and
annotated to the classes in context. These classes are
then represented in a metadata file and the dataset is
split into Training and Validation sets, Concluding
the dataset preparation.
3.4 Tiling Process
Following section describes the tiling in detail:
Tile Size: Images were divided into tiles of
1024×1024 pixels using a CUDA-accelerated tiling
algorithm.
Filtering: Empty tiles or tiles with non-relevant
regions were identified using a
thresholding
algorithm and discarded to ensure informative
datasets.
GPU Optimization: GPU acceleration via
NVIDIA DALI and CuPy minimized data transfer
between CPU and GPU, significantly reducing tiling
time and memory usage. Processing metrics included
execution time, resource utilization, and scalability.
The tiling process was benchmarked on an AMD
Ryzen 7600X CPU, NVIDIA RTX 4080 GPU, and
32 GB RAM.
Table 2: Tiling Performance
Method
Execution
Time
(min)
RAM
Usage
(GB)
Utilization
(
%
)
CPU GPU
CPU Only 28 12 67 0
GPU
Acceleration
19 10 42 14
GPU with
DALI and
CUDA
6 6 12 32
3.5 Deep Learning Models
We investigated two deep learning architectures for
mitosis detection:
YOLO-based Models: A Pretrained YOLO model
were fine-tuned for mitotic detection. The purpose of
YOLO model is to minimize the input image's
dimension to half and enables the extraction of low-
level parameters like patterns and edges. The initial
level of the YOLO Model architecture includes a
convolutional layer with 32 filters and a 3x3 kernel
size. After each convolutional layer, Batch
Normalization is applied. Pooling is not used directly
in the first layers of YOLO model. Rather, the stride-
2 convolution were used. SiLU activation, a
computationally effective method for improving the
cancer detection was applied.
ResNet: A pre trained ResNet model was
incorporated to simplify the training of the system.
In
the beginning, there is a convolutional layer with 64
filters and a 7x7 kernel size. This is the first
convolution layer and a max-pooling layer follows
next. In all situations, the stride is set to 2. The
pooling layer and the convolution layers follow in
conv2_x. Due to the way in which the residuals are
related, these layers tend to appear in pairs. Prior to
the final output layer, fully connected layers were
placed into position, and cancer variations were
categorized using ReLU activation.
Hierarchical Framework for the process of mitotic
detection :
Environment: Models were trained on NVIDIA
RTX 4080 using PyTorch and TensorFlow
frameworks. Training used cross-entropy loss for
classification and IoU loss for bounding box
predictions.
Data Augmentation: Augmentations included
rotations, flips, color jitter, and noise addition to
improve robustness.
Optimization: Learning rates and batch sizes were
optimized through grid search. Early stopping
prevented overfitting.
Evaluation Metrics: Models were assessed on
accuracy, precision, recall and F1 score.
4 RESULTS AND DISCUSSION
The models trained for the aforementioned task are
YOLO V8, YOLO V11, ResNet-34. The details
pertaining to the models and their performance are
mentioned in the upcoming section. The models were
trained on the discussed dataset, to perform detection
and classification. The classes trained in, i.e.
Caterpillar and Multipolar mitosis are well
represented and this is reflected upon inference.
4.1 Model Performance
The below table III details on the model’s
performance, followed by the confusion matrices and
the validation set of each model from Fig. 2 to 7.
INCOFT 2025 - International Conference on Futuristic Technology
466
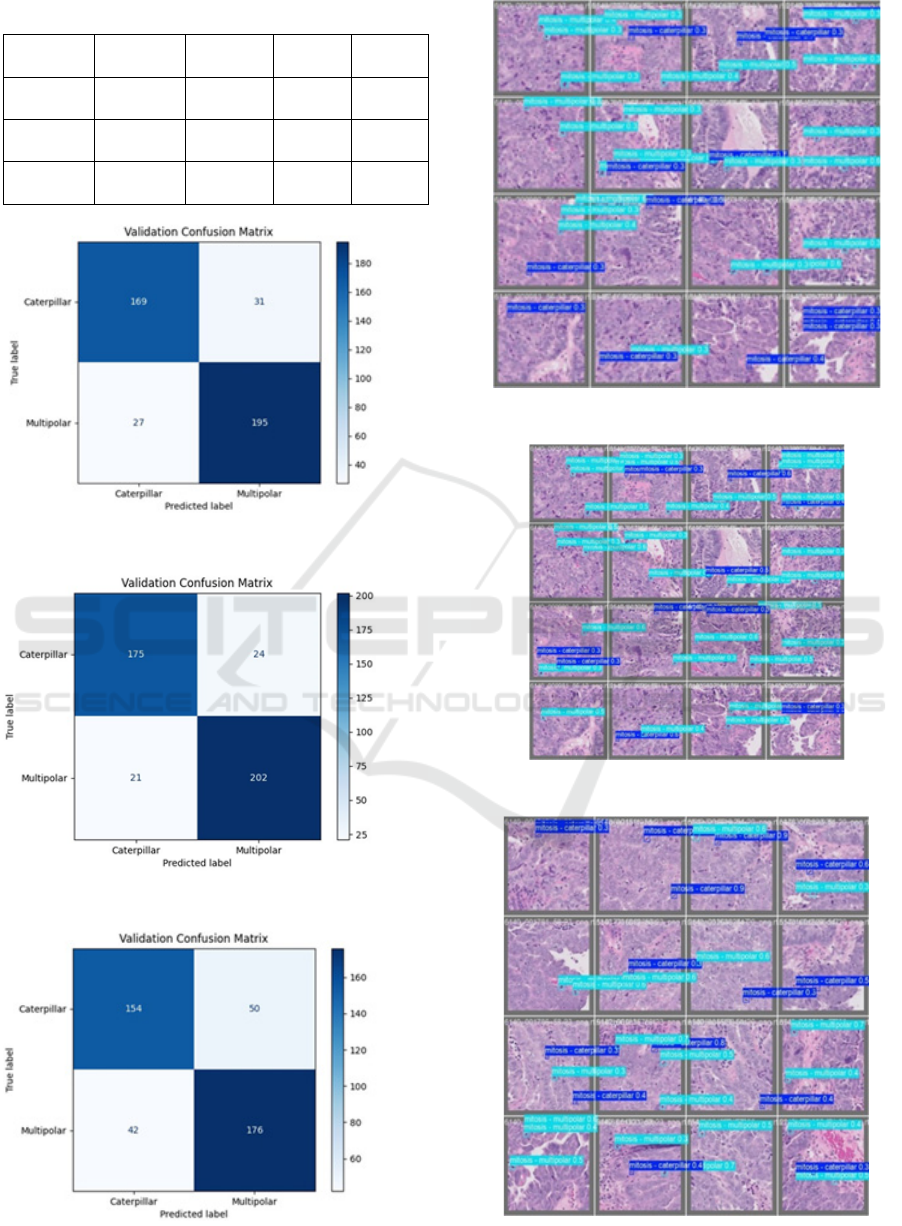
Table 3: Performance of Deep Learning Model
Figure. 2: Confusion matrix for validation of YOLO V8
Figure. 3: Confusion matrix for validation of YOLO V11
Figure. 4. Confusion matrix for validation of Resnet 34
Figure 5: Validation set inference YOLO V8
Figure 6: Validation set inference YOLO V11
Figure.7: Validation set inference Resnet34
Model Accuracy Precision Recall
F1
Score
YOLO
V8
78.20% 78.57% 75.49% 76.20%
YOLO
V11
89.33% 87.93% 89.28 88.60%
ResNet-
34
86.25% 84.50% 86.22% 85.35%
Early Diagnosis of Ovarian Cancer by the Integration of Whole Side Images and Deep Learning Models
467
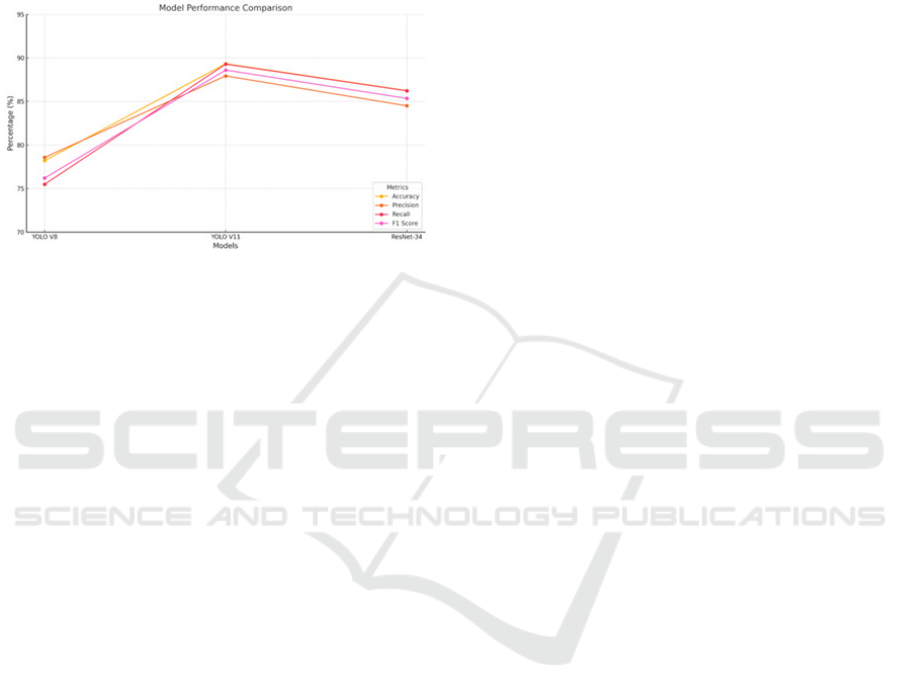
4.2 Discussion
This study successfully demonstrates the application
of deep learning models, specifically YOLOv8,
YOLOv11, and ResNet-34, for the detection and
classification of mitotic figures in ovarian cancer
histopathology images.
Figure . 8. Consolidated graph all models
By employing GPU-accelerated tiling, the
methodology overcomes the challenges associated
with analyzing large Whole Slide Images (WSIs),
enabling efficient resource utilization while
maintaining high image quality. The results indicate
that YOLOv11 achieved the highest accuracy
(89.33%) and F1-score (88.60%), outperforming both
YOLOv8 and ResNet-34. The confusion matrices
further reveal the efficacy of the models in
distinguishing between the two targeted classes:
caterpillar and multipolar mitoses. These subtypes,
known to be critical in ovarian cancer
characterization, were accurately identified.
The significance of this work lies in its potential to
provide pathologists with an automated tool that can
greatly aid in diagnosis and prognosis. The study’s
focus on the crucial mitotic activities and subtypes is
an important step in enhancing diagnostic accuracy.
5 CONCLUSION AND FUTURE
WORK
This work presents a robust deep learning-based
framework for the detection and classification of
malignant mitotic activity in ovarian cancer using
tiled WSIs. The implemented GPU-optimized tiling
and model architecture achieved high performance,
with the YOLOv11 model demonstrating superior
detection capabilities. This methodology offers a
significant contribution towards developing
automated diagnostic tools, reducing the time and
subjectivity associated with manual pathological
analysis. This work validates the use of deep learning
architectures for accurately detecting mitotic figures
and provides a strong foundation for future research
and clinical applications.
Future research will focus on expanding the
dataset to include a broader range of ovarian cancer
subtypes and exploring methods to improve the
robustness and of the models.
Contribution of authors – Suma P, Ananya D
Hedge and Rakshith R are involved in the data
analysis and paper structure. Suma K V is involved in
the comprehension and critical review of the
manuscript for conceptual substance. Each author
pledges to be accountable for every aspect of the
work.
ACKNOWLEDGMENT
The authors would like to thank Ramaiah Medical
College and Ramaiah Institute of Technology for the
logistical assistance received in completing the study.
REFERENCES
Kussaibi, H., Alibrahim, E., Alamer, E., Alhaji, G.,
Alshehab, S., Shabib, Z., Alsafwani, N. and Meneses,
R.G., 2024. Al-Powered classification of Ovarian
cancers Based on Histopathological lmages. medRxiv,
pp.2024-06.
Suma K V, C. S. Sonali, Chinmayi B S, John Kiran B,
Muhammad Easa. CNN Models Comparison for Lung
Cancer Classification using CT and PET scans, 2022
IEEE 2nd Mysore Sub Section International
Conference (MysuruCon), 2022, 16th – 17th Oct 2022,
SJCE, Mysuru, pp. 1-5, doi:
10.1109/MysuruCon55714.2022.9972704.
Farahani, H., Boschman, J., Farnell, D., Darbandsari, A.,
Zhang, A., Ahmadvand, P., Jones, S.J., Huntsman, D.,
Köbel, M., Gilks, C.B. and Singh, N., 2022. Deep
learning-based histotype diagnosis of ovarian
carcinoma whole-slide pathology images. Modern
Pathology, 35(12), pp.1983-1990.
Kasture, K.R., Sayankar, B.B. and Matte, P.N., 2021,
October. Multi-class classification of ovarian cancer
from histopathological images using deep learning-
VGG-16. In 2021 2nd Global Conference for
Advancement in Technology (GCAT) (pp. 1-6). IEEE.
Cireşan, D.C., Giusti, A., Gambardella, L.M. and
Schmidhuber, J., 2013. Mitosis detection in breast
cancer histology images with deep neural networks.
In Medical Image Computing and Computer-Assisted
Intervention–MICCAI 2013: 16th International
Conference, Nagoya, Japan, September 22-26, 2013,
INCOFT 2025 - International Conference on Futuristic Technology
468

Proceedings, Part II 16 (pp. 411-418). Springer
BerlinHeidelberg.
Li, C., Wang, X., Liu, W., Latecki, L.J., Wang, B. and
Huang, J., 2019. Weakly supervised mitosis detection
in breast histopathology images using concentric
loss. Medical image analysis, 53, pp.165-178.
Alom, M.Z., Hasan, M., Yakopcic, C., Taha, T.M. and
Asari, V.K., 2018. Recurrent residual convolutional
neural network based on u-net (r2u-net) for medical
image segmentation. arXiv preprint arXiv:1802.06955.
S. M. Mousavi , "Automated mitosis detection in
histopathology images of canine mammary gland
tumours using deep learning," Journal of Pathology
Informatics, vol. 14, pp. 100218, 2023.
M. Aubreville, "Automatic Classification of Cancerous
Tissue in Laserendomicroscopy Images of the Oral
Cavity using Deep Learning," Scientific Reports, vol.
10, pp. 11754, 2020.
D. Tellez et al., "Whole-Slide Mitosis Detection in Breast
Cancer Histopathology Images using Deep Neural
Networks," in Proc. International Workshop on Breast
Imaging (IWBI), pp. 166-169, 2018.
B. Bertram et al., "A large-scale dataset for mitotic figure
assessment on whole slide images of canine cutaneous
mast cell tumor," Scientific Data, vol. 6, pp. 242, 2019.
A. Aksac, "Deep learning-based cell detection in
histopathological images of papillary thyroid
carcinoma," Computer Methods and Programs in
Biomedicine, vol. 179, pp. 104980, 2019.
W. Li et al., "Mitosis detection in breast cancer
histopathology images using deep cascaded networks,"
Medical Image Analysis, vol. 62, pp. 101700, 2020.
Chen, H., Dou, Q., Wang, X., Qin, J. and Heng, P., 2016,
February. Mitosis detection in breast cancer histology
images via deep cascaded networks. In Proceedings of
the AAAI conference on artificial intelligence (Vol. 30,
No. 1).
D. Malon et al., "Fast mitosis detection in breast cancer
histology images using parallel computation," in Proc.
IEEE International Symposium on Biomedical Imaging
(ISBI), pp. 776-779, 2013.
M. Veta et al., "Assessment of algorithms for mitosis
detection in breast cancer histology images," Medical
Image Analysis, vol. 20, no. 1, pp. 179-194, 2015.
S. Paul and D. Mukherjee, "Detection of mitotic cells in
breast cancer histopathology images using
morphological operators," in Proc. IEEE International
Conference on Intelligent Computing, Networking, and
Services (ICNS), pp. 36-40, 2013.
H. Irshad et al., "Methods for nuclei detection,
segmentation, and classification in digital
histopathology: A review—Current status and future
potential," IEEE Reviews in Biomedical Engineering,
vol. 7, pp. 97-114, 2014.
Early Diagnosis of Ovarian Cancer by the Integration of Whole Side Images and Deep Learning Models
469
