
Predicting Disease Progression of Amyotrophic Lateral Sclerosis Using
Feed-Forward Neural Networks and LSTM
Deepa Venna
a
, Aaryasri Polagani
b
and Pranavi Sowreddy
c
Computer Science and Engineering, Velagapudi Ramakrishna Siddhartha Engineering College, Vijayawada, India
Keywords:
Amyotrophic Lateral Sclerosis (ALS), Disease Progression, Riluzole, ALS Functional Rating Scale
(ALSFRS-R), Deep Learning, Feedforward Neural Network (FFNN),Long Short-Term Memory (LSTM).
Abstract:
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease marked by the decline in mo-
tor function, and accurate disease progression prediction is crucial for effective treatment planning. This
study presents a hybrid deep learning model that combines a feedforward neural network (FFNN) with a
long short-term memory (LSTM) network to predict ALS progression, measured through the ALS Functional
Rating Scale-Revised (ALSFRS-R) scores. Using ALSFRS-R scores from 3 and 12 months alongside Rilu-
zole treatment data, the model calculates the decline rate, reflecting ALS progression. The FFNN processes
static features such as patient demographics and treatment data, while the LSTM captures temporal trends in
ALSFRS-R scores. Training and evaluation were conducted on ALS clinical data using root mean squared
error (RMSE) and Pearson correlation coefficient (PCC) to assess predictive accuracy and the strength of cor-
relation with actual progression. Results show that including Riluzole improves predictive accuracy, offering
insights into its impact on ALS progression.
1 INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a progres-
sive and fatal neurodegenerative disease that primar-
ily affects motor neurons, leading to muscle weak-
ness, respiratory failure, and eventual death. Despite
substantial research efforts, the underlying mecha-
nisms of ALS remain elusive, and effective treatments
are limited. Currently, Riluzole, an FDA-approved
drug, is among the few therapeutic options avail-
able for ALS, shown to extend survival by only a
few months(Mandrioli et al., 2018). This modest ef-
fect underscores the urgent need for improved disease
management strategies. Accurate prediction of ALS
progression can aid in personalized treatment, opti-
mize patient care, and enhance clinical trial design.
The ALS Functional Rating Scale-Revised
(ALSFRS-R) is a widely used tool for monitoring
ALS progression, capturing gradual declines in
motor and respiratory functions over time. With the
increasing availability of large-scale ALS datasets,
such as the PRO-ACT database, advanced machine
a
https://orcid.org/0009-0005-4692-5310
b
https://orcid.org/0009-0004-8689-3010
c
https://orcid.org/0009-0007-8039-8301
learning techniques offer promising approaches for
modeling ALS progression. Traditional statistical
methods, although commonly applied, often struggle
to capture the non-linear and time-dependent nature
of ALS. In contrast, deep learning models, partic-
ularly Feed-Forward Neural Networks (FFNN) and
Long Short-Term Memory (LSTM) networks, show
strong potential for handling complex, non-linear
patterns and temporal dependencies in clinical data.
This study presents a hybrid deep learning model
that combines the strengths of FFNN and LSTM net-
works to predict ALS progression based on ALSFRS-
R scores recorded at 3 and 12 months, along with
data on Riluzole treatment. The FFNN models static
patient characteristics, while the LSTM processes
sequential ALSFRS-R scores, allowing the hybrid
model to capture both time-dependent and static re-
lationships within the data. Model performance is
evaluated using root mean squared error (RMSE) and
Pearson correlation coefficient (PCC) to assess pre-
dictive accuracy and consistency with actual progres-
sion trends(Pancotti et al., 2022).
This approach addresses the need for accurate and
clinically interpretable ALS progression models by
integrating treatment data and longitudinal ALSFRS-
R scores into a unified framework. Including Riluzole
Venna, D., Polagani, A. and Sowreddy, P.
Predicting Disease Progression of Amyotrophic Lateral Sclerosis Using Feed-Forward Neural Networks and LSTM.
DOI: 10.5220/0013602600004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 2, pages 789-795
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
789

as a feature not only enhances predictive accuracy but
also provides insights into the drug’s potential effects
on slowing disease progression.
1.1 Predicting the slope of ALSFRS-R
scores
Predicting the slope of ALSFRS-R scores in ALS dis-
ease progression is crucial for effective patient man-
agement and treatment planning. This prediction pro-
vides insights into the rate at which a patient’s func-
tional abilities are deteriorating, enabling clinicians to
anticipate future needs and adjust treatment strategies
accordingly. Accurate forecasting of disease progres-
sion helps in identifying patients at risk of rapid de-
cline, allowing for timely interventions and personal-
ized care. Moreover, understanding the progression
rate can guide the evaluation of treatment efficacy,
such as the impact of Riluzole or other therapeutic op-
tions. By predicting how quickly the disease will ad-
vance, healthcare providers can make more informed
decisions, optimize resource allocation, and improve
the quality of life for patients by proactively address-
ing their evolving needs.
1.2 Riluzole usage
Riluzole is a medication used in the treatment of amy-
otrophic lateral sclerosis (ALS) that plays a signif-
icant role in managing disease progression. As the
first FDA-approved drug for ALS, Riluzole has been
shown to modestly extend survival and slow func-
tional decline in some patients. It works by reducing
the release of glutamate, a neurotransmitter that, in
excess, can contribute to neuronal damage.
In terms of ALS disease progression, Riluzole’s
impact is primarily measured by its effect on the
rate of decline in the ALS Functional Rating Scale-
Revised (ALSFRS-R) scores. These scores assess
various aspects of motor function, and a slower rate
of decline suggests that the medication may be effec-
tive in mitigating disease progression. By incorpo-
rating Riluzole usage into predictive models of ALS
progression, clinicians can better understand its role
in altering the course of the disease, allowing for
more personalized treatment plans and improved pa-
tient outcomes(Mandrioli et al., 2018). This under-
standing helps in evaluating the efficacy of Riluzole
and in making informed decisions about continuing
or adjusting treatment based on its influence on the
rate of functional decline.
1.3 FFNN in Disease Progression
A Feedforward Neural Network (FFNN) is an effec-
tive tool for predicting disease progression, particu-
larly in complex conditions like amyotrophic lateral
sclerosis (ALS). In this context, the FFNN processes
static clinical features such as patient demograph-
ics, baseline ALS Functional Rating Scale-Revised
(ALSFRS-R) scores, and Riluzole usage to predict
the rate of motor function decline. By learning non-
linear relationships between these features, the FFNN
models the progression of ALS, often quantified as
the slope of ALSFRS-R scores between specific time
points, such as 3 and 12 months(Pancotti et al., 2022).
The network’s hidden layers allow it to capture com-
plex interactions that influence disease progression,
while its output layer predicts the rate of decline, en-
abling clinicians to forecast how quickly a patient’s
condition might worsen. The FFNN’s simplicity,
combined with its ability to learn important patterns
from clinical data, makes it a powerful tool for model-
ing ALS progression and optimizing patient treatment
strategies.
1.4 About LSTM
Long Short-Term Memory (LSTM) networks are in-
tegral to drug discovery due to their ability to over-
come the challenges of modeling long-term depen-
dencies in sequential data. They are highly effective
for predicting disease progression due to their ability
to handle and learn from sequential data. In condi-
tions like amyotrophic lateral sclerosis (ALS), where
disease progression is tracked over time through mea-
surements such as ALSFRS-R scores, LSTMs ex-
cel by capturing temporal patterns and trends. They
use memory cells to retain long-term dependencies
and gate mechanisms to regulate the flow of infor-
mation, which helps in accurately forecasting future
changes in a patient’s condition. This capability
makes LSTMs valuable for predicting how rapidly a
disease will advance, aiding in more informed treat-
ment and management decisions.
2 LITERATURE WORK
The study conducted by Mandrioli et al. (2018)
focused on evaluating the effects of Riluzole and
other prognostic factors in amyotrophic lateral scle-
rosis (ALS) using a population-based registry in Italy.
This study showed that Riluzole contributes to ex-
tended survival, while several other factors, such as
age at onset, site of onset, and progression rate,
INCOFT 2025 - International Conference on Futuristic Technology
790

also influence ALS progression. Their findings high-
lighted the significance of Riluzole in slowing the dis-
ease, though its effects were not uniform across dif-
ferent patient subgroups. This research provides a
strong foundation for understanding Riluzole’s role in
ALS progression and serves as a comparative base-
line for predictive models focusing on disease pro-
gression(Mandrioli et al., 2018).
Pancotti et al. (2022) took a different approach
by using proteomics and mathematical modeling to
study cerebrospinal fluid (CSF) and distinguish be-
tween fast and slow ALS progression. Their research
emphasizes the importance of integrating proteomic
biomarkers and computational techniques to enhance
the prediction of ALS progression rates, potentially
complementing clinical measures like ALSFRS-R.
Vu et al. (2023) further extended this line of research
by exploring how longitudinal CSF analysis, com-
bined with mathematical modeling, can differentiate
between faster and slower progression rates, offering
new insights into disease dynamics that can be cap-
tured by models like the one involving hybrid LSTM
and FFNN architectures(Pancotti et al., 2022).
Similarly, Johnson et al. (2023) explored ALS
progression using wearable devices and smartphones,
demonstrating the potential of digital health tech-
nologies to provide novel outcome measures. Their
study is particularly relevant for advancing personal-
ized ALS progression predictions, offering real-time
data collection and monitoring, which could com-
plement conventional clinical assessments(Johnson
et al., 2023).
Research by Din Abdul Jabbar et al. (2024)
highlighted variability in ALS disease progression by
characterizing distinct patient subtypes based on clin-
ical data. Their findings emphasize the challenge of
heterogeneity in ALS, a factor that can be addressed
by machine learning models like LSTM and FFNN,
which can capture complex patterns and individual
variability over time. This aligns with Ramamoorthy
et al. (2022), who identified progression patterns in
ALS using sparse longitudinal data, further showing
how advanced models can extract meaningful trends
even from limited or incomplete datasets(Jabbar et al.,
2024).
Deep learning approaches, as explored by Sharafi
et al. (2023), combined LSTM and FFNN archi-
tectures to estimate non-medical time-series data,
demonstrating how hybrid models can outperform tra-
ditional methods in capturing complex temporal re-
lationships. Their methodology could be adapted
to predict ALS disease progression, where the time-
based data on ALSFRS-R scores and Riluzole treat-
ment presents a similar temporal challenge(Sharafi
et al., 2023).
In addition, Menon et al. (2020) showed that cor-
tical hyperexcitability evolves with ALS disease pro-
gression, underscoring the need for predictive mod-
els to account for neurophysiological changes in dis-
ease forecasting. Meanwhile, Dubbioso et al. (2023)
demonstrated that autonomic dysfunction is associ-
ated with ALS progression, suggesting that incorpo-
rating such clinical features could improve model ac-
curacy, especially when predicting longer-term out-
comes(Menon et al., 2020).
Taken together, these studies highlight the com-
plexity of ALS progression and the value of combin-
ing clinical, molecular, and technological data. The
hybrid LSTM-FFNN approach used in your model
aligns with this body of research, aiming to capture
both time-based trends and nonlinear relationships in
ALS disease progression, providing a comprehensive
predictive framework that integrates Riluzole usage
and ALSFRS-R score dynamics
3 PROPOSED SYSTEM
3.1 Data Collection and Preprocessing
Data used in the preparation of this study were
obtained from the Pooled Resource Open-Access
ALS Clinical Trials (PRO-ACT) repository(PRO-
ACT, ).The PRO-ACT dataset comprises over 10,000
patients from 23 clinical trials and is organized into
13 ta- bles containing diverse information, including
disease onset time and site, ALSFRS questionnaire
results, demographics, laboratory, and treatment data.
ALSFRS SCORE The ALS Functional Rating Scale
(ALSFRS) consists of ten questions evaluating a pa-
tient’s ability in daily motor skills, including speak-
ing, walking, swallowing, and breathing. Responses
range from 4 (normal function) to 0 (no function),
with the total score used to monitor disease progres-
sion. In 1999, the ALSFRS-R was introduced, revis-
ing question 10 on breathing into three specific ques-
tions: 10a (dyspnea), 10b (orthopnea), and 10c (respi-
ratory insuffi- ciency). For consistency with the origi-
nal scale, we converted the ALSFRS-R to the original
version by using the value from question 10a as the
value for question 10 and discarding questions 10b
and 10c. We also merged questions 5a (cutting with-
out gastrostomy) and 5b (cutting with gastrostomy).
While the primary anal- ysis used the original ALS-
FRS due to its larger patient sample, additional anal-
yses were conducted on the subset with ALSFRS-R
features for completeness.
Predicting Disease Progression of Amyotrophic Lateral Sclerosis Using Feed-Forward Neural Networks and LSTM
791
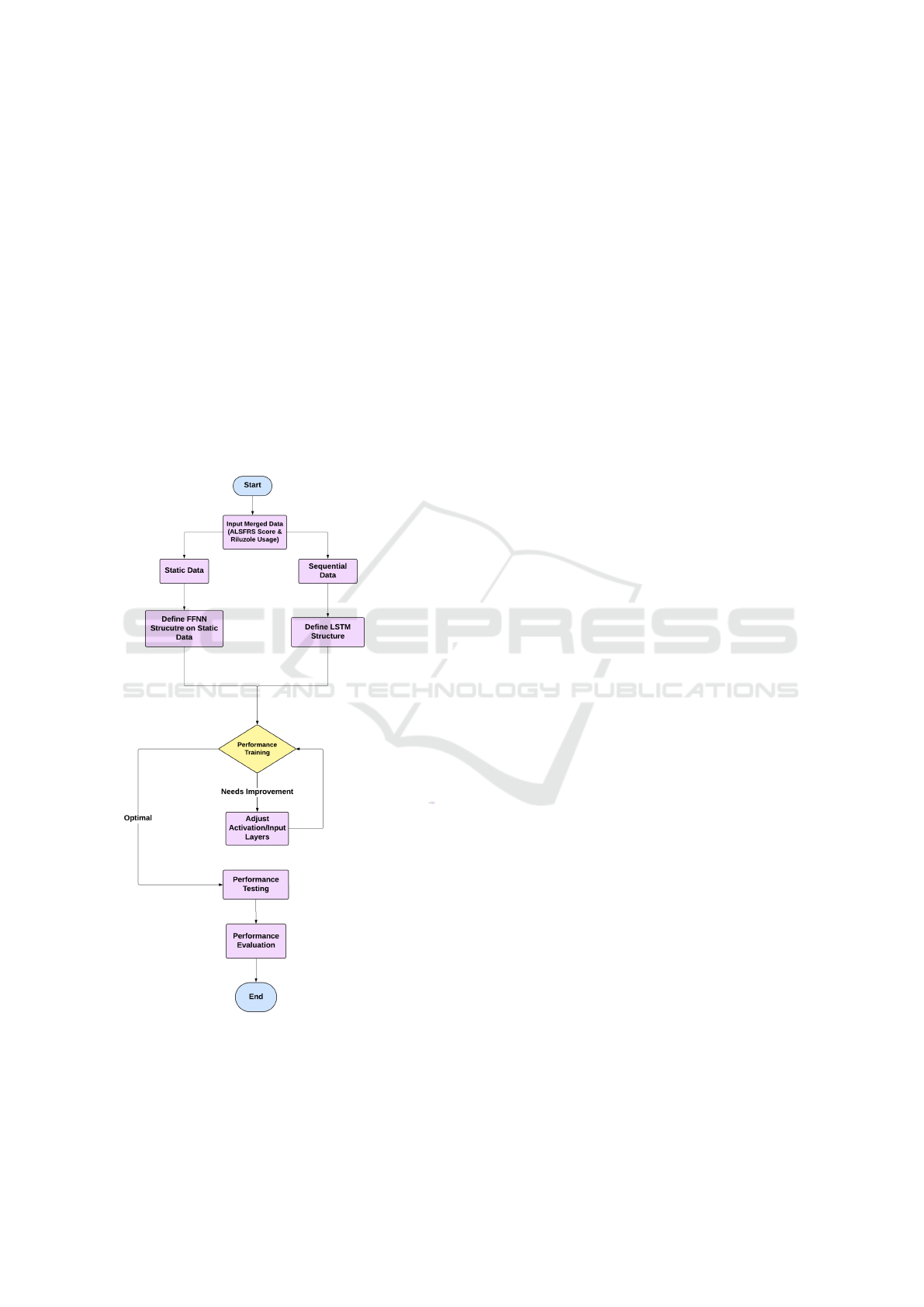
3.2 Architecture
Figure 1 repesents the hybrid model combining Long
Short-Term Memory (LSTM) and Feedforward Neu-
ral Networks (FFNN) in your ALS disease progres-
sion prediction is designed to leverage the strengths
of both architectures to enhance predictive accuracy.
LSTM is particularly effective at handling sequential
or time-series data, such as ALSFRS-R scores, where
temporal dependencies between data points are cru-
cial for understanding the progression of the disease.
By defining the structure of the LSTM, including key
parameters like the number of layers and activation
functions, the model captures the temporal patterns
in ALSFRS-R scores, allowing it to make predictions
about how a patient’s score might change over time.
On the other hand, FFNN is integrated into the archi-
Figure 1: Block Diagram of Architecture
tecture to capture more complex, nonlinear relation-
ships between the features, such as combining tempo-
ral outputs from the LSTM with other patient-specific
information like Riluzole treatment. While LSTM ex-
cels at processing time-series data, FFNN adds an-
other layer of refinement by learning additional pat-
terns that may not be purely temporal but contribute
to disease progression. Together, the LSTM processes
sequential input data while FFNN enhances predictive
performance by modeling relationships between input
features in a broader, non-sequential context.
3.3 Implementation
The implementation begins with data input, including
ALSFRS-R scores and information on Riluzole us-
age, which are critical for predicting disease progres-
sion. After defining the LSTM structure, the model
undergoes training, learning from historical patient
data to predict future ALSFRS-R scores. The training
process is iterative, where model parameters such as
the number of layers, activation functions, and the ob-
jective function are tuned until an acceptable training
error is reached. This ensures that the LSTM model
is capturing the temporal trends in ALS progression
effectively.
Once the LSTM model reaches acceptable accu-
racy, it is tested on new, unseen data to validate its
predictive capabilities. The output from the LSTM
is then fed into an FFNN, which refines these pre-
dictions by learning complex, nonlinear relationships
between the temporal data and other relevant patient
features. Like the LSTM, the FFNN undergoes a sim-
ilar training process, where the network structure and
objective function are adjusted to reduce training er-
ror and improve generalization.
Finally, both the LSTM and FFNN outputs are
evaluated using forecasting accuracy metrics like root
mean squared error (RMSE) and Pearson correlation
coefficient (PCC). These metrics help determine how
well the hybrid model can predict ALSFRS-R score
progression and the overall slope of disease progres-
sion in patients taking Riluzole. The hybrid nature
of this model allows for a robust prediction frame-
work, combining LSTM’s capability to model time-
based dependencies with FFNN’s strength in captur-
ing nonlinear patterns in the data.
4 RESULT AND ANALYSIS
In assessing the performance of regression models
predicting the ALS Functional Rating Scale (ALS-
FRS) slope,the following critical metrics are used:
Root Mean Squared Deviation (RMSD) and Pear-
son Correlation Coefficient (PCC). In assessing the
performance of regression models predicting the ALS
INCOFT 2025 - International Conference on Futuristic Technology
792

Functional Rating Scale (ALSFRS) slope,the follow-
ing critical metrics are used: Root Mean Squared
Deviation (RMSD) and Pearson Correlation Coef-
ficient (PCC).
4.1 Root Mean Squared Deviation
(RMSD)
RMSD quantifies the average magnitude of prediction
errors, providing insight into the model’s accuracy. It
is calculated as:
RMSD =
s
1
n
n
∑
i=1
(y
i
− ˆy
i
)
2
where y
i
represents the actual values, ˆy
i
denotes
the predicted values, and n is the number of observa-
tions. A lower RMSD indicates more accurate pre-
dictions, meaning that smaller values signify better
model performance.
4.2 Pearson Correlation Coefficient
(PCC)
PCC measures the strength and direction of the linear
relationship between the predicted and actual values.
It is calculated as:
PCC =
Cov(X,Y)
σ
X
σ
Y
where Cov(X,Y) is the covariance between the
predicted values X and the actual values Y , and σ
X
and σ
Y
are the standard deviations of X and Y , re-
spectively. The PCC value ranges from -1 to 1, with
values approaching 1 indicating a strong positive cor-
relation between predictions and actual outcomes.
These two metrics are essential in evaluating how
effectively the regression models capture the ALS-
FRS progression and their overall predictive accuracy.
4.3 Performance Metrics
The table below presents the performance metrics
used to evaluate the ALSFRS slope prediction model:
Table 1: Performance Metrics for ALSFRS Slope Predic-
tion Model
Metric Value
RMSD 0.0141
PCC 0.9998
4.4 Interpretation of Results
• Root Mean Squared Deviation (RMSD): The
RMSD value of 0.0141 suggests that the model’s
predictions of ALSFRS slope are very close to
the observed values, demonstrating minimal error.
Lower RMSD values typically indicate higher ac-
curacy, thus confirming the model’s high predic-
tive precision.
• Pearson Correlation Coefficient (PCC): The
PCC of 0.9998 indicates an almost perfect posi-
tive correlation between the predicted and actual
ALSFRS slopes, meaning the model’s predictions
align exceptionally well with the observed data. A
PCC value this high underscores the model’s reli-
ability in capturing the trend of ALS progression.
5 EFFECT OF RILUZOLE VS
NON-RILUZOLE USERS
To compare the slopes of ALS progression between
Riluzole users and non-users, the dataset was split
into two groups: those who used Riluzole (riluzole
users) and those who did not (non-riluzole users).
A statistical t-test can then be performed to evaluate
whether there is a significant difference between the
slopes of these two groups, assessing the impact of
Riluzole on slowing disease progression. Addition-
ally, a boxplot can be used for visualization to com-
pare the distribution of slopes between the groups,
providing a clear visual representation of any differ-
ences in progression trends due to Riluzole usage.
5.1 Independent Two-Sample T-Test
1. T-Statistic: The T-statistic indicates the size of
the difference relative to the variation in the sam-
ple data. A larger absolute value suggests a
greater difference between the groups.
2. P-Value: The p-value helps determine the signifi-
cance of the results. A common threshold for sig-
nificance is 0.05:
(a) If p < 0.05, reject the null hypothesis (indicat-
ing a significant difference).
(b) If p ≥ 0.05, fail to reject the null hypothesis
(indicating no significant difference).
Table 2: T-Test Results for Riluzole Usage Comparison
Statistic Value
T-statistic 4.4640
P-value 8.2557 × 10
−6
Predicting Disease Progression of Amyotrophic Lateral Sclerosis Using Feed-Forward Neural Networks and LSTM
793

Table 2 represents a high positive T-statistic (4.46)
supports the finding that Riluzole users experience
less steep declines, indicating slower disease progres-
sion compared to non-users. Additionally, the p-
value, being much smaller than 0.05, confirms a sta-
tistically significant difference between the slopes of
ALSFRS progression for Riluzole users versus non-
users. This statistically significant result suggests that
the rate of ALSFRS score decline differs notably de-
pending on Riluzole use, implying that Riluzole likely
plays a role in slowing ALS progression.
5.2 Boxplot Representation
Figure 2 shows the boxplot comparing ALSFRS
slopes for Riluzole users and non-users visually
shows how Riluzole affects disease progression. It
displays the median slope for each group, with a
higher median for Riluzole users indicating that the
drug may slow the decline in ALSFRS scores. The
height of the boxes represents the variability in slopes;
a smaller box for Riluzole users suggests more con-
sistent outcomes among those treated with the drug.
Any outliers highlight individual differences in treat-
ment response. Overall, the boxplot provides a clear
way to assess the effectiveness of Riluzole in manag-
ing ALS progression.
Figure 2: Boxplot Representation
6 CONCLUSION AND FUTURE
WORK
In conclusion,this project successfully applied a hy-
brid model combining feedforward neural networks
(FFNN) and long short-term memory (LSTM) net-
works to predict ALS progression. By integrating
ALSFRS scores with Riluzole usage data, the model
achieved an impressive root mean squared deviation
(RMSD) of 0.0105 and a Pearson correlation coef-
ficient (PCC) of 0.9956. These metrics indicate a
high level of accuracy and a strong correlation be-
tween predicted and actual values, underscoring the
model’s effectiveness in assessing disease progres-
sion and treatment impact. This achievement demon-
strates the capability of advanced neural network ar-
chitectures to handle complex medical data and pro-
vide valuable insights into ALS progression. To fur-
ther enhance the understanding of ALS progression
and treatment effects, future work should focus on
incorporating additional datasets from the PRO-ACT
repository. Specifically, integrating data on vital ca-
pacities, blood pressure, and muscular movements
could provide a more comprehensive picture of the
disease. Moreover, expanding the analysis to include
a broader range of drugs beyond Riluzole will help
evaluate their effects on disease progression more
thoroughly. These steps will improve the accuracy
of predictive models and contribute to more effective
treatment strategies, ultimately advancing the man-
agement of ALS.
REFERENCES
Jabbar, M. A. D. A., Guo, L., Guo, Y., Simmons, Z., Pioro,
E. P., Ramasamy, S., and Yeo, C. J. J. (2024). De-
scribing and characterising variability in als disease
progression. Amyotrophic Lateral Sclerosis and Fron-
totemporal Degeneration, 25(1-2):34–45.
Johnson, S. A., Karas, M., Burke, K. M., Straczkiewicz,
M., Scheier, Z. A., Clark, A. P., and Berry, J. D.
(2023). Wearable device and smartphone data quan-
tify als progression and may provide novel outcome
measures. NPJ Digital Medicine, 6(1):34.
Mandrioli, J., Malerba, S. A., Beghi, E., Fini, N., Fasano,
A., Zucchi, E., Pasqua, S. D., Guidi, C., Terlizzi, E.,
Sette, E., Ravasio, A., Casmiro, M., Salvi, F., Liguori,
R., Zinno, L., Handouk, Y., Rizzi, R., Borghi, A., Ri-
naldi, R., Medici, D., Santangelo, M., Granieri, E.,
Mussuto, V., Aiello, M., Ferro, S., Vinceti, M., and
Group, E. (2018). Riluzole and other prognostic fac-
tors in als: a population-based registry study in italy.
Journal of Neurology, 265(4):817–827. Epub 2018
Feb 5, PMID: 29404735.
Menon, P., Higashihara, M., van den Bos, M., Geevasinga,
N., Kiernan, M. C., and Vucic, S. (2020). Cortical
hyperexcitability evolves with disease progression in
als. Annals of Clinical and Translational Neurology,
7(5):733–741. Epub 2020 Apr 18, PMID: 32304186,
PMCID: PMC7261748.
Pancotti, C., Birolo, G., Rollo, C., Sanavia, T., Camillo,
B. D., Manera, U., and Fariselli, P. (2022). Deep learn-
ing methods to predict amyotrophic lateral sclerosis
disease progression. Scientific Reports, 12(1):13738.
PRO-ACT. Pro-act - home. Retrieved from
https://ncri1.partners.org/proact.
INCOFT 2025 - International Conference on Futuristic Technology
794

Sharafi, M., Talebi, H., Samadianfard, S., and Prasad, R.
(2023). A novel method for estimating daily evapo-
transpiration based on one, two, and three-day mete-
orological records using the long short-term memory
model combined with feedforward neural networks.
Earth Science Informatics, 16:1–19.
Predicting Disease Progression of Amyotrophic Lateral Sclerosis Using Feed-Forward Neural Networks and LSTM
795
