
Optimizing Brain Tumor Segmentation Using Attention U-NET and
ASPP U-NET
Mohana Saranya S, Sowmiya S, Vinieth S S, Savitha S, Mohanapriya S and Dinesh K
Department of CSE, Kongu Engineering College, Perundurai, Erode, Tamil Nadu, India
Keywords: BT Segmentation, 2D Images, Attention U-Net, ASPP U-Net.
Abstract: This study analyzes the performance evaluation of different deep learning models such as Attention U-Net,
and ASPP U-Net for segmentation of brain tumor (BT) in 2D MRI scans. It is an integral part of diagnosis
and treatment of tumors in the brain region. The traditional U-Net uses encoder-decoder paths for accurate
localization. In this paper, we have done comparison between Attention U-Net and ASPP U-Net. The
Attention U-Net enhances performance by an attention mechanism that highlights relevant tumor areas. The
ASPP U-Net also improves segmentation using Atrous Spatial Pyramid Pooling (ASPP) to capture multi-
scale features. The results of this work also indicate that the Attention U-Net is superior to ASPP U-Net on
accuracy and most importantly better improving BT segmentation.
1 INTRODUCTION
BT segmentation in medical images is one of the most
important tasks in the field of medical image studies.
Mainly to accurately the tumor regions from healthy
tissue in brain scans. To diagnose and plan
treatments, the automated segmentation models can
significantly help radiologists. This paper compares
various performances of deep neural networks
including Attention U-Net and ASPP U-Net designs
focused on BT segmentation. The U-Net model has
been popular in segmentation of medial images due
to its encoder-decoder structure that captures the
features for precise localization of objects. Attention
U-Net is another version of U-Net where the attention
function is applied to U-Net thereby allowing the
model to learn to ignore irrelevant portions and attend
more on pertinent tumor areas. ASPP U-Net improves
the quality of the segmentation process drastically by
using ASPP.
We desire to evaluate their segmentation
performance in terms of accuracy, dice coefficient,
and robustness to tumors of various sizes and shapes
by comparing the models. This comparison will show
which model achieves the optimal accuracy and
provides the most reliable technique for BT
segmentation.
2 BRATS DATASETS
BraTS dataset is mainly used in the medical fields
particularly for BT segmentation. The BraTS dataset
consists of a database of BT MRI brain scans
collected from multiple medical centres within the
region. Creating and evaluating BT segmentation and
diagnostic techniques, the BraTS dataset was
developed. It includes several medical imaging data
for the purpose of BT segmentation. The standard
modalities are T1-weighted (T1), T2-weighted (T2),
T1-weighted with contrast enhancement (T1c), and
Fluid-attenuated Inversion Recovery (FLAIR). The
T1 provides anatomical information and highlight the
contrast between normal brain tissues and tumor
tissue. The T1c images highlight regions of active
tumor growth and angiogenesis. The T2 emphasizes
the differences in water content between brain tissues
and useful for identifying edema surrounding the
tumor. The FLAIR suppresses cerebrospinal fluid
(CSF) signals, making it easier to detect and visualize
lesions, such as edema and tumor boundaries. The
BRATS 2020 dataset contained 660 images, with 369
images used for training, 125 images for validation
and 166 for testing.
Saranya S, M., S, S., S S, V., S, S., S, M. and K, D.
Optimizing Brain Tumor Segmentation Using Attention U-NET and ASPP U-NET.
DOI: 10.5220/0013589800004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 2, pages 223-228
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
223

3 RELATED WORKS
Brain tumors, especially gliomas, are known for their
aggressive nature and the difficulty in detecting them
due to their irregular shapes and indistinct
boundaries. Traditional methods for manually
segmenting tumors in MRI scans can be labor-
intensive and prone to errors, highlighting the need
for automated solutions. Numerous studies have
proposed deep learning-based models that
incorporate innovative architectures and optimization
techniques to enhance segmentation accuracy. For
example, the Bridged U-Net-ASPP-EVO model
(Yousef, Khan, et al. 2023)utilizes ASPP, squeeze-
and-excitation blocks, and evolving normalization
layers to improve multi-scale feature extraction and
downsampling, surpassing most advanced models on
the MICCAI BraTS datasets. Two versions of this
architecture showed better dice scores across various
tumor regions. The SPP-U-Net model (Vijay, Guhan,
et al. 2023), which employs spatial pyramid pooling
and attention mechanisms, achieves 7.84 as
Hausdorff distance on the BraTS 2021 dataset,
providing a competitive and effective solution for
brain tumor segmentation.
Numerous studies have explored the challenges
posed by the shapes, sizes, and class imbalances of
tumors that affect segmentation accuracy. The RD2A
U-Net model (Ahmad, Jin, et al. 2021) effectively
preserves contextual information for smaller tumors,
achieving average dice scores of 84.5 on the BraTS
2018 dataset, and 81.54 on BraTS 2019. A refined 3D
UNet that incorporates Transformer architecture
(Nguyen-Tat, Nguyen, et al. 2024), featuring a
Contextual Transformer (CoT) and double attention
blocks, improves long-range dependencies and
feature extraction, significantly surpassing existing
most advanced models. Multi-threshold attention U-
Net model (Awasthi, Pardasani, et al. 2020), that is
specially designed for the segmentation of several
regions of a tumor, achieved dice coefficients of 0.64
in the test dataset. The attention residual U-Net
(Zhang, Zhang, et al. 2020) combines attention units
and residual connections to boost segmentation
performance on small tumor regions, attaining high
scores on BraTS challenges from the years 2017 and
2018. Another model, the deep supervised U-
Attention Net (Xu, Teng, et al. 2021), merges U-Net
and attention networks to effectively capture both
low- and high-resolution features, achieving dice
coefficients of 0.81 on the training dataset.
Adversarial learning methods and ensemble
learning frameworks have shown significant potential
in improving segmentation performance and survival
prediction. A 3D segmentation network ( Peiris,
Chen, et al. 2021) that utilizes dual reciprocal
adversarial learning strategies reported dice scores of
85.84% on the BraTS 2021 dataset, along with better
Hausdorff Distances. In a similar vein, the VGG19-
UNet architecture (Nawaz, Akram, et al. 2021),
which features a pre-trained VGG19 encoder and an
ensemble classifier for survival outcomes, achieved
dice coefficients of 0.85 with a survival prediction
accuracy of 62.7%. The AML-Net (Aslam, Raza, et
al. 2024), which introduces an attention-based multi-
scale lightweight architecture, recorded an F1-score
0.909 and a sensitivity as 0.939, outperforming
established models like U-Net and CU-Net.
Additionally, the Hybrid UNet Transformer (HUT)
model (Soh, Yuen, et al. 2023), which combines both
UNet and Transformer pipelines, further improves
lesion segmentation accuracy with a 4.84% increase
in Dice score compared to the SPiN network on the
ATLAS dataset. These advancements have propelled
BT segmentation forward, enhancing diagnostic
workflows and patient outcomes while also reducing
computation time and improving multi-modality
analysis.
4 PROPOSED WORK
The steps involved in this approach are: dataset
loading, data preprocessing, extracting features and
segmentation of tumor regions.
Figure 1: Proposed work flowchart
Figure 1 illustrates the flowchart associated with
the implementation of the proposed techniques –
Attention U-Net and ASPP U-Net.
4.1.1 Attention U-Net
The Attention U-Net algorithm used in enhancing the
BT segmentation in this project, significantly
contributing to the accuracy of tumor detection.
Attention U-Net is advancement of U-Net
architecture for segmenting the medical images by
using attention mechanisms. By using attention
mechanism, it is helpful in focusing the most relevant
INCOFT 2025 - International Conference on Futuristic Technology
224
regions of the input BT image, so that the overall
performance for segmentation of tumor regions will
be improved.
Attention U-Net works by learning the weights
from different regions within the feature maps. By
incorporation of skip connections in the attention
gates, only the important features are passed from
encoder to decoder. Assigning higher weights to the
most relevant regions, allowing the model to focus on
essential feature by suppressing the irrelevant regions
from the image. Attention gates filter out the feature
from the encoder before passing them through skip
connections to the decoder.
Attention U-Net's ability to integrate spatial
information through skip connections further
strengthens its accuracy. These connections help in
retaining fine details from the input image, which is
crucial for identifying subtle differences in tissue
structures. The attention mechanism also enhances
the model’s interpretability by making the decision-
making process more transparent and reliable.
This, in turn, supports early diagnosis and enables
more tailored treatment strategies for patients. By
highlighting its potential in clinical environments,
Attention U-Net establishes itself as a valuable tool in
advancing BT analysis, ultimately contributing to
improved patient outcomes and advancements in
healthcare technology. Using this Attention U-Net
algorithm we achieve the accuracy as 95.1%.
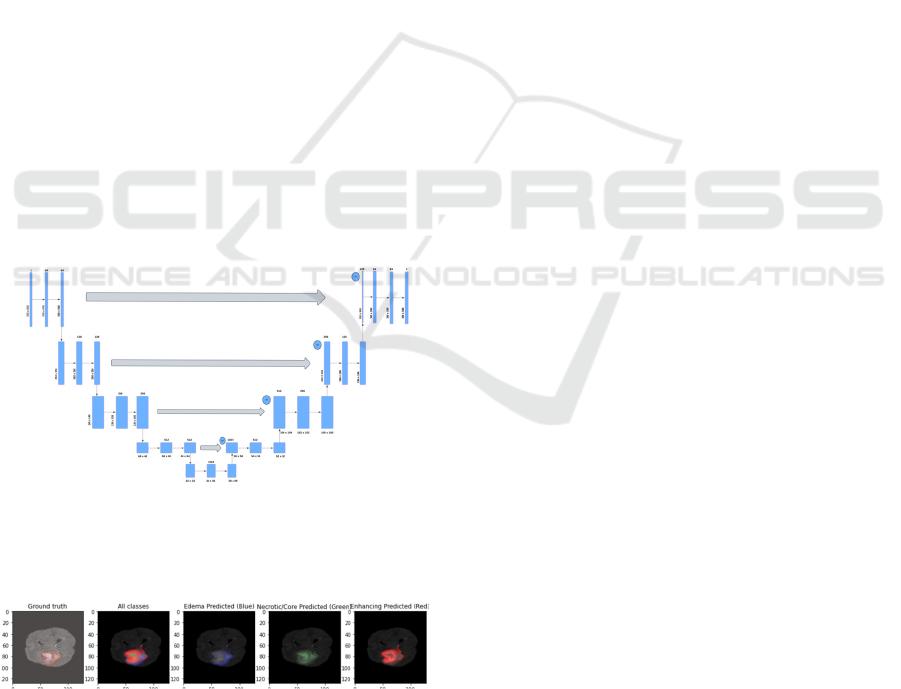
Fig.2 Architecture diagram of Attention U-Net
The figure 2 denotes architecture diagram of
Attention U-Net.
Figure 3 Attention U-Net
The figure 3 represents the segmented output of
Attention U-Net for edema, core and enhancing
tumor.
4.1.1 ASPP U-Net
The ASPP U-Net plays a vital role in enhancing
BT segmentation in this project, significantly
improving accuracy by capturing multi-scale
contextual information. The incorporation of ASPP
allows the model to efficiently gather features at
various scales, making it well-suited for detecting
tumors of different sizes and shapes in MRI images.
ASPP U-Net's architecture combines the strengths
of a traditional U-Net with advanced feature
extraction capabilities from ASPP. The encoder-
decoder structure effectively captures both local
details and global context, while ASPP layers ensure
that features are extracted from multiple receptive
fields. This enables the model to focus on fine details
in the tumor regions without losing the broader
context of the surrounding tissue.
By utilizing ASPP U-Net in this project, we
achieved enhanced segmentation accuracy and
demonstrated its ability to handle the complex nature
of brain tumor images. The proficiency of the model
in encompassing the information at various scales led
to better delineation of tumor boundaries, supporting
early diagnosis and enabling more personalized
treatment plans.
4.1.2 ASPP U-Net Variations
In BT segmentation, altering the dilation rates in
different variations of the ASPP U-Net can greatly
modify the ability of the model to learn features
across various spatio-temporal scales. By varying the
dilation rates in the ASPP block, the model can
capture both fine details of smaller tumors and the
broader context for larger and irregular tumor
regions.
Using dilation rates (ASPP M1) of 1, 2, and 4, the
network progressively captures larger features while
maintaining the spatial information. The 1 and 2 focus
on fine details and 4 captures broader regions. For
dilation rates (ASPP M2) of 2,4 and 8 allowing the
model to capture features from large areas. The
combination of dilation rates (ASPP M3) of 1,6 and
12 travels a wide range of receptive fields. The
dilation rate of 1 handle fine details and larger dilation
rates 6 and 12 cover more global features. This makes
the model adaptable to structures of different
scales.We use this ASPP U-Net M1, M2, M3 and
achieve the accuracies as 94.05,94.03, and 94.12.
Optimizing Brain Tumor Segmentation Using Attention U-NET and ASPP U-NET
225
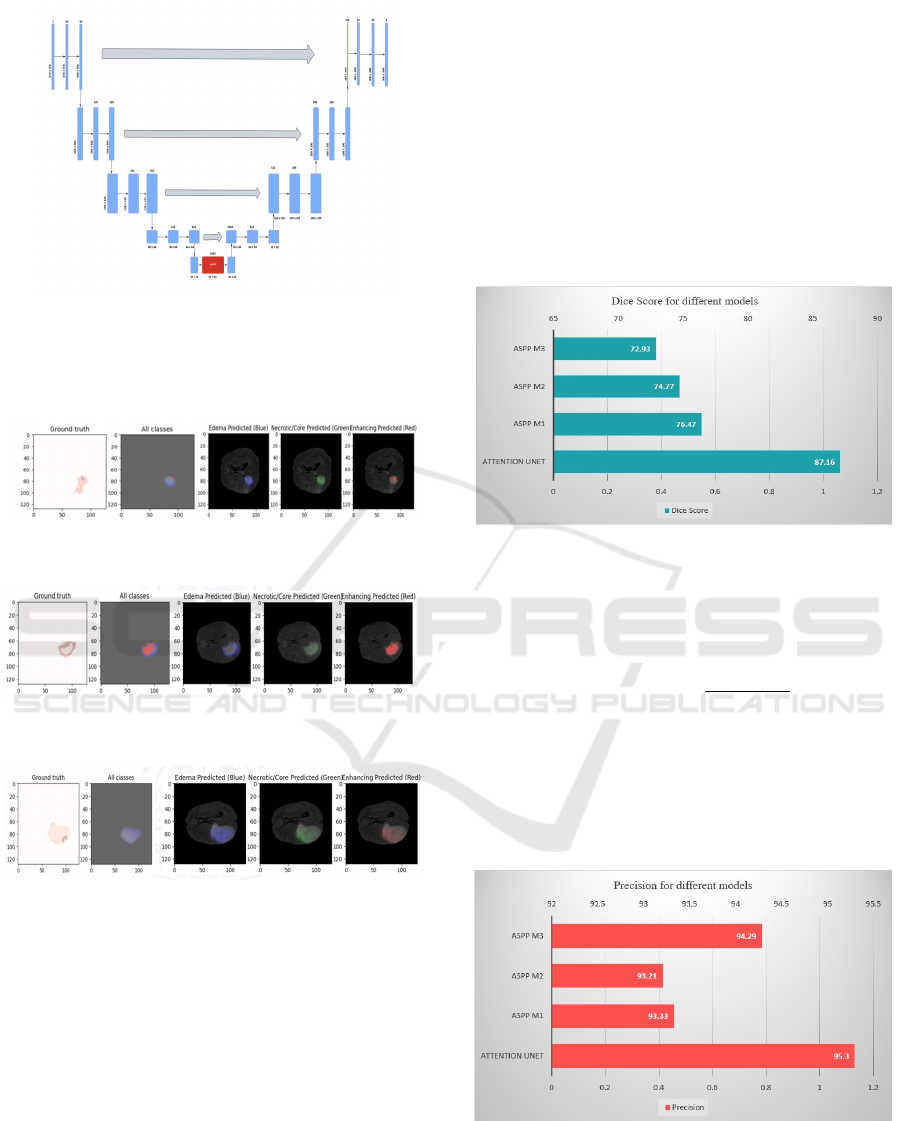
Figure 4 Architecture diagram of ASPP U-Net
Figure 4 represents the architecture diagram of ASPP U-
Net.
Figure 5: ASPP U-Net M1 (dilation rate -1, 2, 4)
Figure 6: ASPP U-Net M2 (dilation rate - 2, 4, 8)
Figure 7: ASPP U-Net M3 (dilationrate -1, 6, 12)
The figure 5 represents the segmented output of
ASPP U-Net M1 (dilation rate -1, 2, 4) for edema,
core and enhancing tumor.
The figure 6 represents the segmented output of
ASPP U-Net M2 (dilation rate - 2, 4, 8) for edema,
core and enhancing tumor.
The figure 7 represents the segmented output of
ASPP U-Net M3 (dilation rate -1, 6, 12) for edema,
core and enhancing tumor.
5 RESULT AND DISCUSSION
The model performance was evaluated using metrics,
which include accuracy, specificity, sensitivity,
precision, and dice score.
Here the TP, TN, FN and FP are described as
TP: True Positive
TN: True Negative
FN: False Negative
FP: False Positive
Figure 8: Dice Score
The Dice similarity Coefficient is an appropriate
statistical measure that is used to compute the
closeness of the predicted set with the actual set. It’s
described in following Equation (1).
𝐷𝑆𝐶
(1)
The Dice Score gets the highest value for the
Attention U-Net with 87.16 value. The outcome of
ASPP M1 is 76.47 whereas the outcome of ASPP M2
is 74.77. The ASPP M3 gets the lowest value of
72.93.
Figure 9: Precision
Calculating the proportion of true positive
outcomes against the total number of positive
INCOFT 2025 - International Conference on Futuristic Technology
226
outcomes is called Precision. It is described in the
following Equation (2).
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛
(2)
In precision, the Attention U-Net gets highest
value of 95.3 and ASPP M3 also get closely to this
with 94.29. The ASPP M1 and M2 gets the value of
93.33 and 93.21.
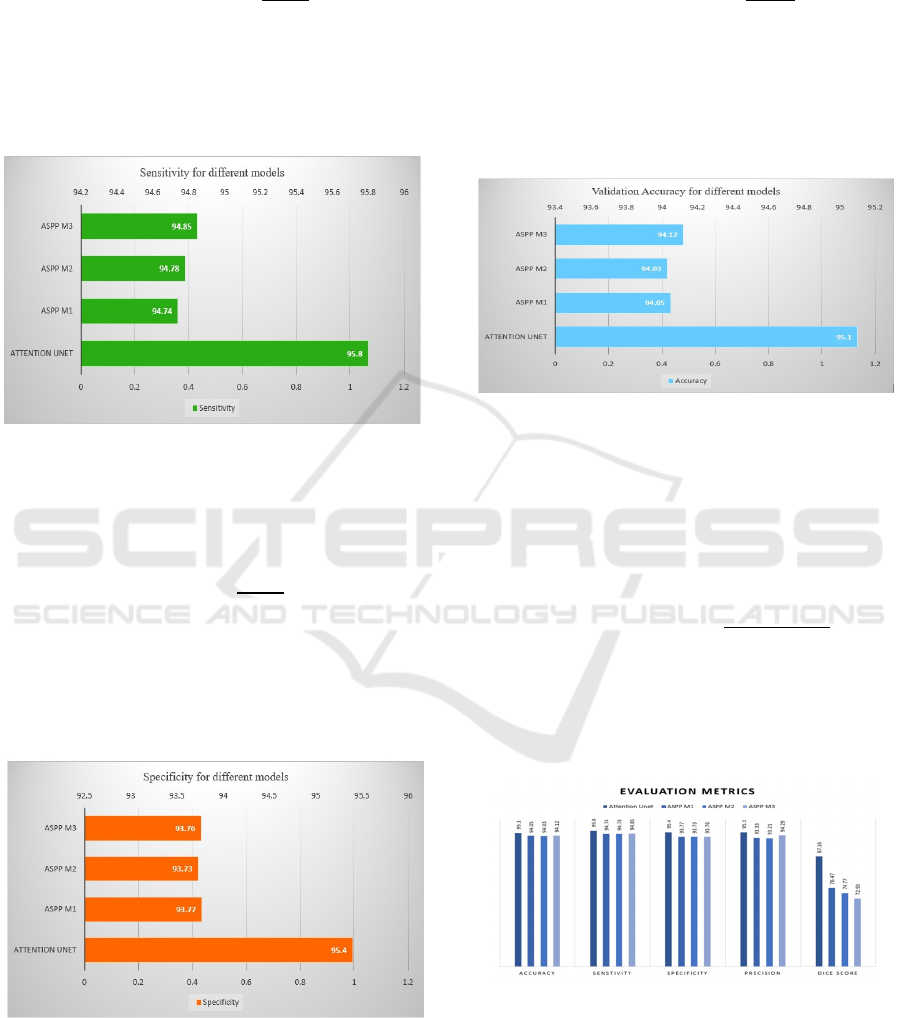
Figure 10: Sensitivity
Sensitivity assesses how many actual positive
cases a model has accurately captured. The definition
is described in the following Equation (3).
𝑆𝑒𝑛𝑠𝑖𝑡𝑖𝑣𝑖𝑡𝑦
(3)
The Sensitivity also highest for the Attention U-
Net with value of 95.8. The ASPP M3, ASPP M2 and
ASPP M1 also follows closely with 94.85, 94.78 and
94.74.
Figure 11: Specificity
The Specificity measures the percentage of
correct negative values divided by the number of
negative results. The definition of specificity is
described in the following Equation (4).
𝑆𝑝𝑒𝑐𝑖𝑓𝑖𝑐𝑖𝑡𝑦
(4)
Here also Attention U-Net reach first in
specificity with 95.4. Comparatively ASPP M1 reach
second with closest value of 93.77, ASPP M3 reach
third with 93.76 value and ASPP M2 reach last with
93.73.
Figure 12: Accuracy
Accuracy is expressed as a percentage of
predictions made by a model that are accurate. It is
calculated by comparing the total correct predictions
made to total predictions. It is described in the
following equation (5).
𝐴𝑐𝑐𝑢𝑟𝑎𝑐𝑦
(5)
In accuracy, the highest value is achieved by
Attention U-Net with 95.1%. The ASPP M3, ASPP
M1 and ASPP M2 also show good performance with
94.12%, 94.05% and 94.03%, which is closest to the
highest value.
Figure 13. Evaluation Metrics
Figure 8 depicts the performance of the model
with respect to various measures such as accuracy,
sensitivity, specificity, precision and dice score.
Optimizing Brain Tumor Segmentation Using Attention U-NET and ASPP U-NET
227
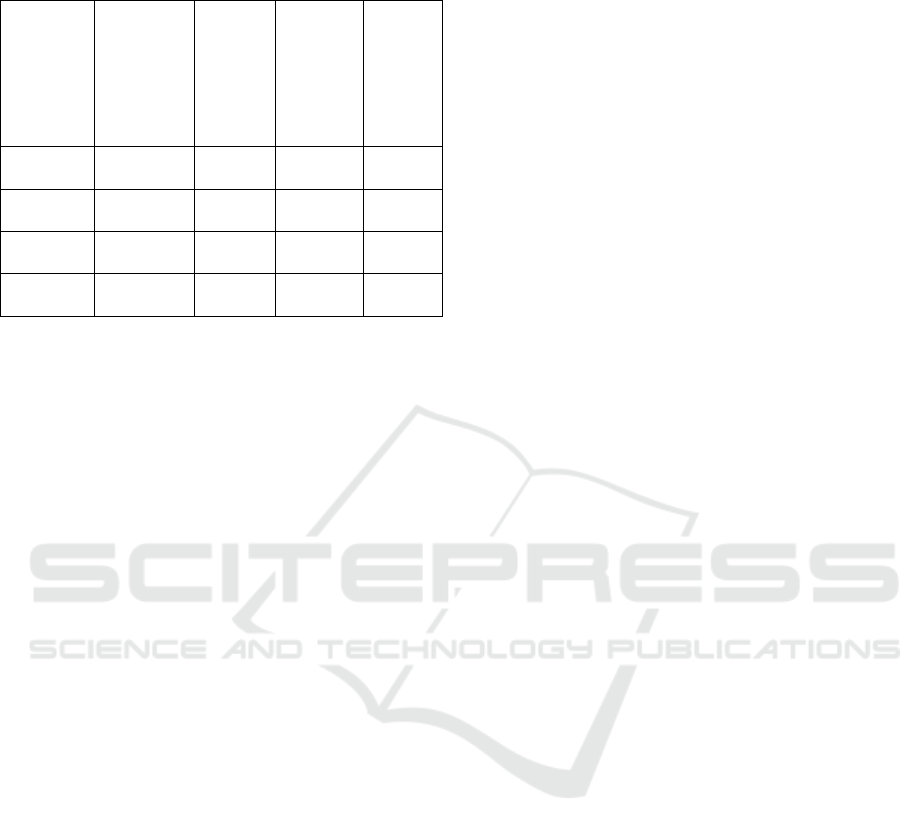
Table 1: Comparison between Attention U-Net and ASPP
U-Net variations
Algorith
m
Parameter
s
(in
Millions)
Trainin
g
Time
(approx
. in
hours)
Accurac
y
Dice
Score
co-
efficien
t
Attention
U-Net
28,634,12
0
6 95.1 87.16
ASPP U-
Net M1
21,221,09
2
4 94.05 76.47
ASPP U-
Net M2
21,221,09
2
4 94.03 74.77
ASPP U-
Net M3
21,221,09
2
4 94.12 72.93
The Attention U-Net is the best algorithm for the
segmentation task that achieves the highest accuracy
95.1, sensitivity 95.8, specificity 95.4, Dice Score
87.16 and precision 95.3. Comparatively the ASPP
M2 performed with the lowest accuracy of 94.03. In
overall performance, the Attention U-Net is the best
choice. The ASPP M3 and ASPP M1 are used as
alternatives.
6 CONCLUSION AND FUTURE
WORK
This paper shows that Attention U-Net performed
best for BT segmentation. The highest accuracy
(95.1%), sensitivity (95.8%), specificity (95.4%),
Dice Score (87.16%) and precision (95.3%) are
achieved by Attention U-Net, which performed better
than both ASPP M3 and ASPP M1, which also
performed well. The ASPP M2 also performed well
but comparatively it gets lowest values in metrices. In
enhancing segmentation accuracy for tumors of
varying sizes and shapes implemented by Attention
U-Net, these results highlight the importance by using
attention mechanism on the most relevant regions.
Further improvements in BT segmentation for
future work by integrating advanced attention
mechanism with U-Net architecture to enhance focus
on smaller, complex regions. Also, improve
segmentation performance by testing with different
attention strategies and more complex encoder-
decoder architectures. Testing these models on larger
and varied datasets provides insights into their
adaptability and further improves the models to
increase their accuracy in real-world medical
challenges.
REFERENCES
R. Yousef, S. Khan, G. Gupta, B. M. Albahlal, S. A.
Alajlan, and A. Ali, "Bridged-U-Net-ASPP-EVO and
deep learning optimization for brain tumor
segmentation," Diagnostics, vol. 13, p. 2633, 2023.
S. Vijay, T. Guhan, K. Srinivasan, P. D. R. Vincent, and C.-
Y. Chang, "MRI brain tumor segmentation using
residual Spatial Pyramid Pooling-powered 3D U-Net,"
Frontiers in public health, vol. 11, p. 1091850, 2023.
P. Ahmad, H. Jin, S. Qamar, R. Zheng, and A. Saeed, "RD
2 A: Densely connected residual networks using ASPP
for brain tumor segmentation," Multimedia Tools and
Applications, vol. 80, pp. 27069-27094, 2021.
T. B. Nguyen-Tat, T.-Q. T. Nguyen, H.-N. Nguyen, and V.
M. Ngo, "Enhancing brain tumor segmentation in MRI
images: A hybrid approach using UNet, attention
mechanisms, and transformers," Egyptian Informatics
Journal, vol. 27, p. 100528, 2024.
N. Awasthi, R. Pardasani, and S. Gupta, "Multi-threshold
attention u-net (mtau) based model for multimodal
brain tumor segmentation in mri scans," in Brainlesion:
Glioma, Multiple Sclerosis, Stroke and Traumatic
Brain Injuries: 6th International Workshop, BrainLes
2020, Held in Conjunction with MICCAI 2020, Lima,
Peru, October 4, 2020, Revised Selected Papers, Part
II 6, 2021, pp. 168-178.
J. Zhang, X. Lv, H. Zhang, and B. Liu, "AResU-Net:
Attention residual U-Net for brain tumor
segmentation," Symmetry, vol. 12, p. 721, 2020.
J. H. Xu, W. P. K. Teng, X. J. Wang, and A. Nürnberger,
"A deep supervised U-attention net for pixel-wise brain
tumor segmentation," in Brainlesion: Glioma, Multiple
Sclerosis, Stroke and Traumatic Brain Injuries: 6th
International Workshop, BrainLes 2020, Held in
Conjunction with MICCAI 2020, Lima, Peru, October
4, 2020, Revised Selected Papers, Part II 6, 2021, pp.
278-289.
H. Peiris, Z. Chen, G. Egan, and M. Harandi, "Reciprocal
adversarial learning for brain tumor segmentation: a
solution to BraTS challenge 2021 segmentation task,"
in International MICCAI Brainlesion Workshop, 2021,
pp. 171-181.
A. Nawaz, U. Akram, A. A. Salam, A. R. Ali, A. U.
Rehman, and J. Zeb, "VGG-UNET for brain tumor
segmentation and ensemble model for survival
prediction," in 2021 International Conference on
Robotics and Automation in Industry (ICRAI), 2021,
pp. 1-6.
M. Zeeshan Aslam, B. Raza, M. Faheem, and A. Raza,
"AML‐Net: Attention‐based multi‐scale lightweight
model for brain tumour segmentation in internet of
medical things," CAAI Transactions on Intelligence
Technology, 2024.
W. K. Soh, H. Y. Yuen, and J. C. Rajapakse, "HUT: Hybrid
UNet transformer for brain lesion and tumour
segmentation," Heliyon, vol. 9, 2023.
INCOFT 2025 - International Conference on Futuristic Technology
228
