
An Effective Convolutional Learning Model with Fine-Tuning for
Medicinal Plant Leaf Identification
Chaitrashree R
1
, Shashikala S V
1
and Sharathkumar Y H
2,3
1
BGSIT, ACU University, B G nagara, Karnataka India
2
Department of CSE, BGSIT, ACU University, B G nagara, Karnataka India
3
Department of ISE, MITM, VTU University, Karnataka India
Keywords: Convolutional Neural Network (CNN), Rectified Linear Unit (ReLU).
Abstract: In this work has successfully presented the custom CNN model and AYURNet approach for Classification.
We used a convolution neural network (CNN) to solve this problem of Multi-Class Classification of Plant
Leaves of some Andhra Ayurvedic Plants. The convolution layer part of CNN was used for feature extraction
and the fully connected dense layer part of CNN was used for Multiclass Classification. Using the known best
practices, a very simple and elegant CNN model was designed and built using Keras to solve the Plant leaf
Multi-Class Classification problem. The model was trained on the training dataset for 200 epochs. Using the
weight parameters obtained in each epoch, the model was tested against the validation dataset. Training
accuracy and Validation accuracy were compared at each epoch and the model with the best weight
parameters was chosen. The logic used to choose the AYUR-Best model was high accuracy above the chosen
threshold of 99% and the least possible difference between training accuracy and validation accuracy. This
was to ensure that the accuracy of the model was very high while ensuring that there was no overfitting. The
model chosen using this method performed very well on the test dataset too and it resulted in an accuracy of
99.88%. Similar high accuracies were achieved by leveraging popular pre-trained models like DenseNet169,
EfficientNetB6, InceptionResNetV2, ResNet152V2, VGG16 and Exception, but it is seen that the respective
models are heavy with a large number of parameters when compared to the custom CNN model described in
this work. Due to the small size of the Custom CNN model, it is suitable for the development of the mobile
application for Ayurvedic plant species identification based on the respective leaf images..
1 INTRODUCTION
The survival of human society is largely dependent
on the ecological resources on this planet. For
instance, the natural habitat and natural resources
around us are being utilized for various human needs.
In particular, human society has more dependencies
on plant habitat. Hence, there is a tremendous
magnitude of discoveries in the biological sciences,
which enables humans in understanding the
complexity in the biodiversity of plant life.
Taxonomists and botanists conduct their exploratory
expeditions to explore the natural habitat of plant life,
which leads to enormous amount of information
regarding the plant life in the natural habitat. This
leads to vast museums, libraries, biological gardens
(Bisby, 2000),(Edwards, Lane, et al. , 2000) and
herbariums. In recent years, we have witnessed the
revolution of digital world and virtualization,
particularly, the development of virtual herbariums.
Virtual herbariums contain all the information of
plant species in the form of text, images, videos and
other multimedia modalities. Classification and
retrieval of information from such virtual herbariums
is a challenging task. Image processing and pattern
recognition techniques can be explored to devise
methods for automatic classification and retrieval of
information from virtual herbariums. Ayurvedic
health care system uses medicinal plant resources like
leaves, flowers, barks, fruits and roots for the
preparation of medicines to prevent and cure common
diseases, so it is very much essential to possess
knowledge about the taxonomy and usage of
medicinal plants. Though there are some experts who
possess knowledge about the taxonomy of medicinal
plants, their knowledge is generally restricted to only
those plants, which are available in their respective
regions. The recent developments in ayurvedic
medicine have proved that the ayurvedic medicines
are very effective for certain diseases and do not have
212
R, C., S V, S. and Y H, S.
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification.
DOI: 10.5220/0013589700004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 2, pages 212-222
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

any side effects. Since medicinal plants can be grown
naturally and made available for a common man
easily, it is appropriate to provide knowledge about
medicinal plants and their usage. It is very difficult
for a common man to possess a complete knowledge
about all the medicinal plants and their usage as there
are millions of species in the nature, which have
medicinal value. Also, sometimes, even an expert in
ayurvedic medicine may fail to classify certain
species correctly due to intra-class dissimilarity and
inter-class similarity. Therefore, it is obvious that one
can think of an automated system to correctly classify
plant species in a large collection. In view of this, in
this work, we made an attempt to propose a medicinal
plants classification system based on leaves by
exploring some novel feature extraction techniques to
characterize plant leaves and efficient representation
technique to effectively classify medicinal plants.
2 RELATED WORK
A new technique was presented to find the ideal
structure of CNN for texture recognition. Dixit and
Ujjawal has suggested a strategy of texture
classification using CNN and optimized with the
whale optimization algorithm. This method applies
WOA's ideas to optimize convolutional layer filter
values and dense layer weight and bias values. For the
testing purpose, this method was applied on three
distinctive benchmark datasets viz. Kylberg v1.0,
Brodatz, and Outex_TC_00012. The outcome
demonstrated that the model performed fair enough
when compared with the existing methods and
accomplishes classification precision for the Brodatz
dataset. This method proves CNN is more powerful
and is a successful technique in texture recognition
(Dixit and Ujjawal, 2019). Mehdipour Ghaz et al. By
optimizing the transfer learning parameters, authors
presented a method of plant identification using a
deep neural network. Three amazing and famous deep
learning designs, namely Google Net, AlexNet, and
VGGNet, are utilized for the purpose. Transfer
learning is utilized to improve the pre-trained models.
To reduce over fitting, increase procedures are
applied on image transforms such as rotation,
translation, reflection, and scaling. Moreover, the
networks' parameters are balanced and various
classifiers are combined to improve overall
execution. The results of the plant identification
campaign LifeCLEF 2015 shows that the general
validation accuracy of the top system has improved
by 15% and the general inverse position value of the
test set has improved by 10%. The framework
recently got exceptionally a 2 nd place in the Plant
CLEF 2016 (Ghazi, Yanikoglu, et al. , 2017). The
recognition of plant species mainly depend upon the
leaf characteristics. CNN was introduced to improve
the recognition capacity of plant leaves in the
complex environment. Xiaolong Zhu et al. have
suggested a improved deep convolutional neural
network for identification of leaves. These techniques
has taken advantage of the Inception V2 with Batch
Normalization (BN) instead of convolutional neural
layers. In addition, the first image is cut to a
predefined size in numerical order, and the segmented
images are continuously loaded into the proposed
network successively. After the precise classification
via SoftMax and bounding field repressor, the divided
snap shots with unique labels are spliced collectively
as a final output snap shots. Research results show
that this method is more accurate than Faster RCNN
when it comes to recognizing leaf species in complex
backgrounds (Zhu, Zhu, et al. , 2018). The proposed
strategy used a deep belief network (DBN), and the
process of preparing the network included
unsupervised feature learning followed by fine-
tuning of the supervised network. Samadhi et al.
Presented a supervised deep learning-based method
for detecting changes in radar images with synthetic
apertures (SARs). From a general point of view, the
prepared DBN produces a change location map as the
output. Studies on DBNs exhibit that they don’t
create a perfect strategy without a proper dataset.
Then it provides a dataset containing the appropriate
amount and various information to train the DBN
using the input images and the images obtained by
applying morphological operators. The disadvantage
of deep learning algorithms are time-consuming. Test
results show that the method has acceptable
implementation time in addition to the desired
execution and precision (Samadi, Farnaam, et al. ,
2019) Ubbens et al. recommended using deep
learning to apply on rosette plants. Extended the
capabilities of Deep Convolutional Neural Networks
to perform leaf counting tasks. Deep learning systems
generally require a large and diverse data set to train
a generalized model without providing pre-designed
algorithms to perform the task. This paper has
introduced a new way to extend plant phenotypic
datasets using rendered images of manufactured
plants. It has been shown that the demonstration of
the leaf counting task can be modified by expanding
the dataset with high quality 3D synthetic plants. The
ability of the model to generate an arbitrary
distribution of phenotypes has been shown to mitigate
the problem of dataset changes when training and
testing on multiple datasets. After all, when training
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification
213

neural networks to do the job of counting leaves, real
and synthetic plants are importantly interchangeable
(Ubbens and Jordan, 2018). Preserving plants has
become a vital task. It is very important, as few plants
have incredible medicinal properties. Plants can be
recognized by leaves, bark, seeds, fruits, flowers, etc.
Lochan et al. proposed a method for detecting and
classifying plants using a high-speed region-based
convolutional neural network. Methodology that
takes into account is identification of plants by leaf
characteristics. The plants considered are medicinal
plants that can be introduced in individual locations
such as Himalayan and vegetable gardens. Author
used a regional convolutional neural network
(RCNN) to identify plants. The system uses a fast
RCNN model that uses a convolutional system to
extract features and classifies using support vector
machine (Lochan, Naga, et al. , 2020). Yang et al.
presented a method for recognizing semantic image
information via MultiFeature Fusion and SSAE-
based Deep Network. Effectively used the
Convolutional Neural Network in the field of visual
recognition and data augmentation techniques for
small datasets to get the right number of training
datasets. The author uses low-level features of the
image to help extract advanced features that are
naturally learned from deep networks in order to
obtain successful emotional features of the image. At
this point, the Stack Sparse auto-encoding system is
used to sense the emotions caused by the image.
Finally, a semantically enlightening high-level
phrase containing the emotions of the image is
delivered. Experiments are performed using
dimensional space IAPS and GAPED datasets and
discrete space art photo datasets (Yang and Xiaofeng,
2020). Taxon identification is an important step in
many plant biology studies. Pierre Barré et al.,
introduced semi-automatic system that can
significantly improve your productivity and
reproducibility. However, in most cases, it relies on a
hand-crafted algorithm to extract a previously
selected set of characteristics to distinguish between
the types of selected taxa. As a result, such
frameworks are limited to these taxa and also require
the involvement of experts to provide taxonomic
knowledge for the reproduction of such tailor-made
systems. The purpose of the study was to set up a deep
learning framework for learning to distinguish
features from leaf images, as well as a classifier for
identifying plant species. In contrast, the results with
Leaf Snap show that learning highlights via a
convolutional neural network improves the feature
representation of leaf images, as opposed to
handmade features. The analysis uses published Leaf
Snap, Flavia, and Foliage datasets (Barré, Stöver, et
al. , 2017). It is extremely challenging when the leaf
images are similar in size, shape and texture. J. Hu et
al recommended a method of multiscale fusion
convolutional neural network for plant leaf
recognition. First, the input image using the random
biprimary interpolation task is reduced to a low
resolution image. At this point, these input images of
different scales are stepped into the MSFCNN design
to learn identifiable points at different depths. In this
phase, the fusion of features between the two different
scales is confirmed by a join operation that connects
the maps captured with images of different scales
from the channel view. In addition to the depth of
MSFCNN, multiscale images are dynamically
processed and the corresponding highlights are
combined. Third, the final layer of MSFCNN sums
all the identification data to get the final predictor of
the plant species in the input image. Test results show
that the presented MSFCNN method is superior to
some state-of-the-art plant leaf detection methods in
the Malaya KewLeaf and LeafSnap datasets (Hu,
Chen, et al. , 2018). Authors have suggested DPCNN
and BOW methods for leaf recognition. The work
focuses predominantly on feature extraction,
particularly on textural feature extraction. Currently,
new methods of leaf recognition rely on the word of
bag (BOW) and entropy sequence (EnS). First, EnS
is attained by a dual-output pulse-coupled neural
system and later improved by BOW. A linear coding
strategy with locality constraints was used for sparse
coding and SVM used as classifier. Some
representative datasets and existing techniques were
assessed to understand the the impact of the methods
implemented. Finally, the results showed the
accuracy of the method is superior to that of existing
methods (Wang, Sun, et al. , 2017). Though Leaves
are helpful markers in identifying the plants, a
significant downside is that numerous biological and
environmental factors are likely to easily harm them.
A method of fragmented plant leaf recognition
presented by Chaki and Jyotismita uses fuzzy-colour,
Bag-of-features, and edge-texture histogram
descriptors with multilayer perception. Divided leaf
images cannot be perceived based on the feature of
shape. A unique methodology was brought in by
using the combination of edge-texture histogram and
fuzzy-colour to recognise divided leaf images.
Initially, by using bag-of-feature, the images that
were similar in appearance were recognized. To
produce the feature vector, the consolidated element
was utilized at that point. Since less information was
given by the divided leaves, the method also aimed at
achieving fragment size threshold. Using a multi-
INCOFT 2025 - International Conference on Futuristic Technology
214

layer-perceptron classifier, the effectiveness of the
proposed approach was considered. Since divided
image public database is not available, a technique
was developed to produce fragmented leaf image
from the whole one (Chaki and Jyotismita, 2019). In
general, Agriculturists and farmers have been facing
repeated challenges in agriculture, one of it is being
the diseases faced in rice. The extreme case of rice
sickness is being no harvest. So, we need a more cost
effective, Simpler and Automatic technique to
identify sickness in rice. Using deep transfer learning
Chen and Junde presents a technique to detect rice
plant diseases. An outstanding result in classification
and image processing is demonstrated by deep
learning approach for settling the task. Taking
advantage of both, the inception and Dense Net
pretrained on the image net modules were chosen to
be used in the network. Using this technique results
in best performance in comparison with other state of
the art methods. It achieves an average result of at
least a public dataset. This average accuracy becomes
high for the prediction of class for rice disease images
even when different diseases were considered. The
validity of suggested approach was shown by the test
outcome. It is applied for the effective rice disease
detection (Chen and Junde, 2020). Computer
Researchers helped botanists by creating the plant
identification systems to identify and perceive strange
plant species more quickly. For the use of plant
predictive modelling, different studies so far focused
on algorithms or strategies that amplify the use of leaf
databases. A technique of learning leaf features were
presented by Lee and Sue Han. Leaf characteristics
legitimately form the basic input data representations
using CNN, and gain a knowledge of selected features
on a Deconvolutional Network (DN) approach. The
most typical feature undertaken in this research was
veins of leaf. A multi-level representation of leaf data
shows a hierarchical transition from a lower level trait
abstraction to a higher level in relation to species.
These results shown are consistent with the various
botanical implications of leaf features. The results
provides insight into the technology of hybrid feature
extraction models that further enhance the
discriminating ability of plant classification systems
(Lee and Han, 2017). To resolve the issue of plant
identification from patterns of the leaf veins, Grinblat
et al., has proposed a deep convolutional neural
network (CNN). In specific, three diverse legume
species are considered, namely soybean, red bean and
white bean. The presentation of a CNN evades the
utilization of feature extractors which are handcrafted
as it is standard in best in class pipeline. Moreover,
this approach of deep learning altogether improves
the precision of the pipeline. We additionally show
that the announced accurateness is reached by
extending the depth of the model. At last, by studying
the models with a basic visualization method, we can
disclose pertinent vein designs (Grinblat, Guillermo,
2016). Ferentinos et al has utilized leaf images of
diseased and healthy plants. convolutional neural
network models were created to detect disease in
plants through methods of deep learning. A large
dataset images were trained using 25 to 58 classes,
including disease-free plants. The research
undertaken has proved the model an remarkably
helpful tool with good accuracy rate. The work
carried can further be extended to work on real time
plant disease (Ferentinos and Konstantinos, 2018).
Based on venation fractal dimension and outline
fractal dimension a new method is proposed
portraying the features of plant leaf Du, Ji-xiang et al.
Initially to separate leaf vein and edge, and get
multiple veins, multiple threshold edge detection
technique was used. Later, 2 dimensional fractal
dimension of the multiple vein picture and leaf edge
image will be calculated, and also adopted a fresh ring
projection fractal image for shape of the leaf. Then,
to classification and recognition of plant leaves, two
kinds of fractal dimension characteristics are utilized.
The test results showed the adequacy of the fractal
dimension characteristic technique(Du, Xiang, et al. ,
2016). Atabay et al., has presented a method of
convolutional neural network with a new architecture
applied to leaf classification. A new CNN design was
introduced and applied on leaf classification. The
utilization of the recently introduced Exponential
Linear Unit (ELU) instead of Rectified Linear Unit
(ReLU) as the non-linearity function of CNN was
investigated. The structure has been analysed on
Flavia and Swedish leaf datasets and the
classification outcomes show that the presented CNN
is effective for leaf classification (Atabay, Agh.,
2016). For an effective weed management, data on
the weed species in farming land is very important.
By using a convolutional neural network, a method is
proposed to recognise the plant species images in
colour Dyrmann et al., 10413 images of weed and
crop species at initial stages of growth is tested and
trained having 22 species to build the network from
scratch. These pictures used from six distinctive
datasets, which have varieties like resolution,
lighting, goal, and soil type. Also incorporates
pictures taken under controlled conditions concerning
camera stabilization and illumination, and pictures
shot with cell phones which were hand-held in fields
with distinctive soil types and changing lighting
conditions. For these 22 species, A classification
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification
215

accuracy of 86.2% is achieved by the network
(Dyrmann, Mads, et al. , 2016). Authors have
reviewed with the upsides and downsides of each
technique with respect to input information (Crop
type) Iniyan et al. In developing countries like India,
economy is depending majorly on agriculture.
Expansion in agro-items influences the GDP of the
country to a decent degree. To expand the
profitability in farming, early identification of
sicknesses is required. In the developmental work, we
have to limit our discussion and detect the crop issues
using technologies like machine learning. with the
help of artificial neural network and support vector
machine (Iniyan, et al. , 2020).
3 PROPOSED SYSTEM
In this chapter, a custom CNN architecture for
APLC and an innovative AYUR-Best model
approach to achieve the highest classification
accuracy have been proposed. For experimentation, a
comprehensive results analysis takes place using the
custom leaf dataset. Classification of plant species is
important to be able to take full advantage of the
benefits provided by the respective species. Given the
huge number of plant species, the classification of
plant species requires knowledge and expertise. An
expert botanist has the skill to classify plant species
based on morphological characteristics. Manual
techniques to classify plants are time-consuming and
demand expert knowledge. Classification of plant
species based on leaf images has become an active
area of research. Due to advances in image processing
and artificial intelligence techniques, it is possible to
solve the complex problem of APLC. CNN's have
gained popularity for the last 10 years with the
availability of supporting hardware and software
platforms. The problem chosen in this work is
medicinal plant species identification by the
classification of respective leaf images. It is not
always possible for humans to come up with explicit
patterns and features that can be fed as input to
computer programs for image classification of non-
geometric figures like leaves. CNN's are a good fit for
solving this problem since they can come up with
models that can identify some patterns and features
which may not be understandable and interpretable
by humans but can do a good job of image
classification. A large data set of images of leaves of
various plants to be classified was first collected. A
CNN architecture was designed which resulted in a
model that was able to classify the chosen leaves with
targeted accuracy.
3.1 Pre-processing
The only preprocessing step required in this work
was to rescale the leaf images from 4000x3000x3
pixel size to 150x150x3 pixel size. This image
rescaling could have been done as a part of the
main Keras code itself. The only reason for
implementing it separately in the local system
using python was to reduce the size of images
before uploading them to google drive. This is to
increase the speed of upload from the local system
to google drive. This is also keeping in view of the
upper limit of 15GB free storage availability in
google drive.
3.2 Building the CNN Model
The ML techniques of leaf image classification rely
on data from hand-crafted features. The leaf images
are run through several pre-processing steps. Hand-
crafted features are a set of features extracted and
derived from leaf images by researchers, to help the
machine differentiate one leaf class from another.
This feature data is collected from several leaves
belonging to various plants. The leaf feature data
and the corresponding leaf labels are fed to relevant
ML classification algorithms and training is done. So,
the usual steps involved are pre-processing, feature
extraction, and classification. Some of the
classification algorithms used are the Multiclass
support vector machine (MSVM) and the Random
Forest classifier. Upon training, the classification
algorithms can predict the leaf label based on the
input feature data of the new leaves which were
previously not part of the training dataset. In feature
engineering, hand-crafted features are the set of
features that the researchers assume will help
humans implicitly differentiate one image class
from another. We are still not at a stage where we can
fully decipher how the human brain recognizes and
identifies objects. So the better option thought by
researchers for classification was to let the machine
itself figure out what features would be best suitable
for classification when it is given a set of training
images of various classes. Deep CNN is better
suitable for this task of image classification and has
been used in the leaf image classification work
described in this work. The basic concept behind
CNN is to create a model that best maps the input
training images to the corresponding known image
labels. This model created based on the training
image set and labels, is then used for image class
prediction. The model consists of convolutional
layers and max-pooling layers followed by a multi-
INCOFT 2025 - International Conference on Futuristic Technology
216

layer DNN. Convolutional layers are filters that act as
feature identifiers or feature extractors. After the
Convolutional layer, we usually have the Max-
Pooling layer which helps in down-sampling of the
feature representation received from the preceding
Convolutional layer. The multi-layer neural network
consists of fully connected layers also known as
Dense layers. Classification of images is a very
complex task. The image data involved is not linearly
separable. A simple mathematical function is not
enough for achieving this image classification task.
The extremely complex relationship between the
input image data and output labels is needed for this
purpose. These cannot be expressed as direct
mathematical formulas. The solution lies in coming
up with a combination of basic building blocks that
can model the required extremely complex
relationship. The building blocks are the artificial
neurons. Artificial neurons are mathematical
functions that are the summation of products of
inputs and weights. That means they are weighted
aggregations of the inputs. A non-linear activation
function like sigmoid, ReLU, or Tanh is then applied
to this sum to introduce non-linearity. A DNN
consisting of layers of several such artificial neurons
is built. As per the Universal Approximation
Theorem, such a kind of deep artificial neural
network can be tailored to approximately represent
any complex relationship between a set of inputs and
outputs. The final layer of this network is the output
layer which is used for prediction. In this case of
image classification, it is the image class or label
which is predicted. This process of modeling the
neural network is not a simple exercise. It is a
complex engineering exercise that involves trying out
several CNN architectures for the problem at hand.
Study and usage of known best practices are done for
faster convergence to the solution. To start with, the
weight parameters associated with the various
neurons in the network are initialized with some
values. The inputs to this network are numerical pixel
data corresponding to the images. The input data is
processed through the network and the prediction is
done at the output layer. A comparison of the
predicted output classes is done with the known true
classes of the training data set of images. It is highly
improbable that the prediction would be perfect in
just one forward pass through the network. The
difference between the predicted values and the true
values is calculated using loss functions. If there is a
loss or difference, then it means that the neural
network model does not correctly define the
relationship between the input and output. The task at
hand is thus to minimize the loss and improve the
model so that predicted values are as near to true
values as possible. The way to improve the model is
to adjust the weight parameters. This is done using the
learning process. The updates to weights should not
be done randomly. The weight adjustment should be
done in a principled way based on the loss function.
It is mathematically proven that the weight
adjustment should be based on the derivative of the
loss function for the weight. The derivative of loss for
weight is known as the gradient. By moving in a
direction opposite to the gradient, we can reduce the
loss. This is known as Gradient Descent. In DNNs,
the derivative of loss function needs to be calculated
for all the weights corresponding to all the artificial
neurons starting from the first layer till the output
layer. The paths over which the derivatives of various
weights for loss needs to be calculated using the chain
rule may be complicated. These computations of
derivatives of all the weights in the network can be
done efficiently using the backpropagation method.
In the backpropagation method, the computation is
done backward from the outward layer for reuse and
efficiency. This processing of inputs through the
network and the backpropagation of weights is done
until the required accuracy is achieved. Since the
amount of input data required to train these networks
is large, we do not wait until all the input data is
processed. The weight updates are done after a certain
batch size of data processing is complete. When the
processing of the full input training data set is
complete, it is known as the completion of one epoch.
Several such epochs are required before the required
prediction accuracy is achieved. If the chosen model
architecture does not achieve the required accuracy
even after training for a large number of epochs, then
another suitable architecture needs to be designed and
the training process needs to be repeated. This
exercise needs to be carried out until the desired
accuracy is achieved. After the training of the model
is completed and the desired training accuracy is
achieved, the evaluation of the model against
previously unseen test data needs to be done. Only
when the test accuracy similar to training accuracy is
achieved, the model is considered as successful.
Sometimes, much less accuracy may be achieved
during testing when compared to training. This is due
to overfitting. Overfitting means that the model has
memorized the training data instead of coming up
with a proper relationship that will achieve similar
accuracy on any test data. In such cases, changes to
the model will again be required. CNN modeling
aims to come up with the simplest possible network
design which will give the highest possible accuracy
for the chosen image classification task. For the
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification
217
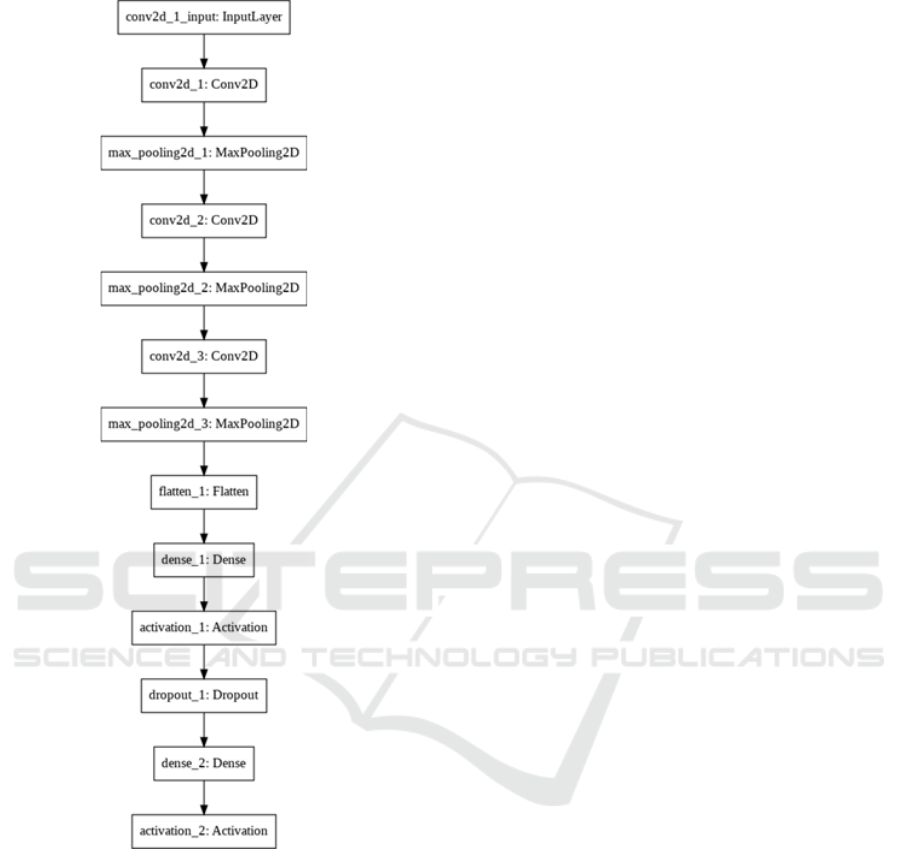
APLC task, we designed the CNN model as shown in
figure 1.
Figure 1:Graphical Description Of The CNN For
Ayurvedic Leaf Image Classification
For the sake of simplicity, this CNN is designed
as a sequential model with a linear stack of layers.
Several hyperparameters are used in the design of the
CNN model. These hyperparameters can be tuned to
improve the accuracy of the CNN model. One of the
hyperparameters is the activation function. These
activation functions in neural networks are nonlinear.
Without the non-linear activation functions, the
output of the neural network would just be a linear
transformation of the input even if the network is
deep with many layers. A linear transformation
cannot solve complex problems involving non-
linearly separable data. The representation power of
the Universal Approximation Theorem and thus the
DNNs is owing to the non-linear activation functions.
The usual activation function that comes to mind
in the case of a neural network is the sigmoid
function. One problem with the sigmoid function is
that it saturates when it reaches its peak value or
bottommost value during training. This means that
the derivative of the function becomes zero. When the
neuron gets saturated, the weight update stops
happening and learning gets affected. This is what is
known as the vanishing gradient problem. One of the
other problems with the sigmoid function is that it is
computationally expensive since it contains the
exponential function.
The recommended default activation function for
CNN is ReLU. ReLU is the short form for the
rectified linear unit. It is represented in Equation 1.
When the input value is positive, then the ReLU
function will output the input value itself. If the input
value is negative, then the ReLU function will output
zero. ReLU is thus better than sigmoid function in the
sense that it does not saturate for positive values and
it is easy to compute. In the proposed CNN model,
ReLU is used as the activation function for both the
convolutional layers as well as the fully connected
hidden layers.
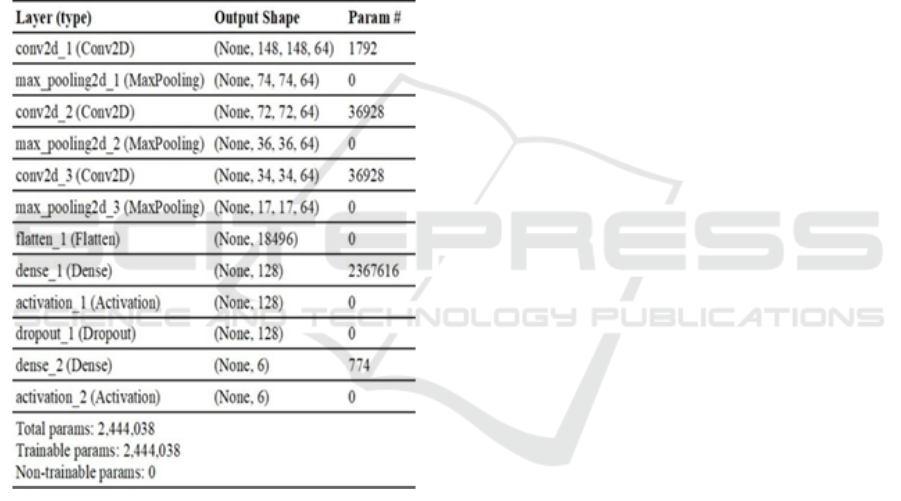
This model was designed based on the known best
practices and several trials with various CNN
architectures. Figure 2 shows the model summary
of the proposed network with the details of the
number of parameters involved.
f(x)=max(0,x) (1)
Softmax is used as the activation function in the
output layer.
3.3 Choice of Loss Function
The whole idea behind CNN modeling is to find
the approximate relationship which is as near as
possible to the true complex relationship between
input and output. In our case of training the CNN
model for leaf classification of multiple classes, the
inputs are images of leaves of different known
classes and the outputs are the leaf class labels of
the respective leaf images. During the training
process, we get the predicted output. For the training
dataset, we know the true output. To assess the
correctness of the model, we find the summation of
INCOFT 2025 - International Conference on Futuristic Technology
218
the difference between the true output and the
predicted output. This difference is calculated using a
loss function. During the training, the loss is
calculated using the loss function for all the data in
the training dataset and it is summed up. It is this loss
value that helps us choose the model which better
approximates the relationship between the input data
and output. It is obvious that the lower is the loss
value, the better is the model. Some of the loss
functions used in neural networks are squared error
loss, cross-entropy loss, and KL divergence. For this
case of multi-class classification problem of leaves
where we are using softmax function as the
activation function in the fully connected output
layer, the loss function which we chose is the
categorical cross-entropy.
Figure 2:Summarry of the Cnn fort Ayurvedic Plant Leaf
Image Classification
3.4 Learning
Artificial neurons in the neural network consist of
inputs, weights, bais, and activation functions. Once
we chose a neural network model to tackle a
problem, we need to find the weights and bais which
are the parameters that define the model. The values
of these parameters are not known at the time of
choosing the model. The best possible parameter
values need to be found through the process of
learning. To start with, the parameters for the chosen
model will be initialized with some values. The inputs
are then processed by the model and the outputs are
predicted. During the training process, the inputs are
the training dataset for which the outputs are known.
The predicted output would not match with the
known output after the first pass through the
network. The loss function is used to find the
difference between the actual known output and the
predicted output. The aim of the learning process to
minimize the loss such that we arrive at the parameter
values which results in predicted output to be as near
to the known output as possible. This will result in the
required model which will best describe the
relationship between the input and the output. For the
complex relationship between the inputs and the
outputs, it is just not possible to determine the values
of a large number of parameters by guesswork or trial
and error method or brute force search method. The
purpose of the learning algorithm is to arrive at those
values of the parameters which minimize the loss.
This is hence an optimization problem that involves
computing the parameters as efficiently as possible.
The algorithms used for solving this problem are
mostly based on calculus and linear algebra. Some of
the popular learning algorithms are Gradient Descent,
Adagrad, RMSProp, and Adam. The optimization
algorithm used in this work for compiling the model
is Adam. Adam is the short form for adaptive moment
estimation. This algorithm was proposed by Diederik
P. Kingma from OpenAI and Jimmy Lei Ba from the
University of Toronto. Adam combines the
advantages of two other optimization algorithms
named AdaGrad and RMSProp. Adagrad is a gradient
based optimization algorithm and a short form for the
adaptive gradient. Adagrad performs small updates to
parameters associated with dense features and it does
larger updates to sparse features. Thus, it allows the
learning rate to adapt based on the parameters. It,
however, has the problem of shrinking of learning
rate as the training progresses. This problem is
overcome in another optimization algorithm named
RMSProp by dividing the learning rate by
exponentially decaying average of squared
gradients. RMSProp chooses a different learning rate
for each parameter. RMSProp is the short form for
Root Mean Square Propagation and was proposed by
Geoffrey Hinton. Adam's optimization algorithm
used in the work described in this work combines the
advantage of AdaGrad‟s ability to deal with sparse
gradients and RMSProp‟s ability to deal with non-
stationary objectives.
3.5 Evaluation
In the leaf classification problem described in this
work, the dataset is perfectly balanced. Since the
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification
219
dataset was created exclusively for this work, it was
made sure that the count of images in the training,
validation, and test dataset of different classes of
leaves is exactly equal. Since the count of images of
different classes are exactly equal, the dataset is
perfectly balanced. The advantage of a balanced
dataset is that accuracy can be used as the metric to
evaluate this model. The advantage of accuracy as a
metric is that it is very easy and intuitive to
understand. Both the overall accuracy as well as per
class accuracy have been calculated for this model.
The formula for calculation of accuracy is given in
equation 1.
Accuracy =
Number of correct predictions (1)
Total number of prediction
3.6 Methodology
In this work, the self-created data set of leaf images
of six different species of locally available ayurvedic
plants are used. For each of the six plant species,
1000 leaf images are taken. The total dataset size is
hence 6000 leaf images. For each of the 1000 plant
species images, the data set is split into training,
validation, and test dataset in the ratio 70:15:15. The
training and validation data set are used by our keras
cnn program while building and training the
model. The leaf images are rescaled to 150x150x3
pixel size. The rescaled leaf images are uploaded to
google drive in the standard folder format as expected
by the designed keras cnn program. The keras cnn
program is coded in the google colab environment.
The google drive is mounted locally to the google
colab environment so that the leaf image dataset on
the google drive is accessible to the keras cnn
program. Keras cnn program is trained using the
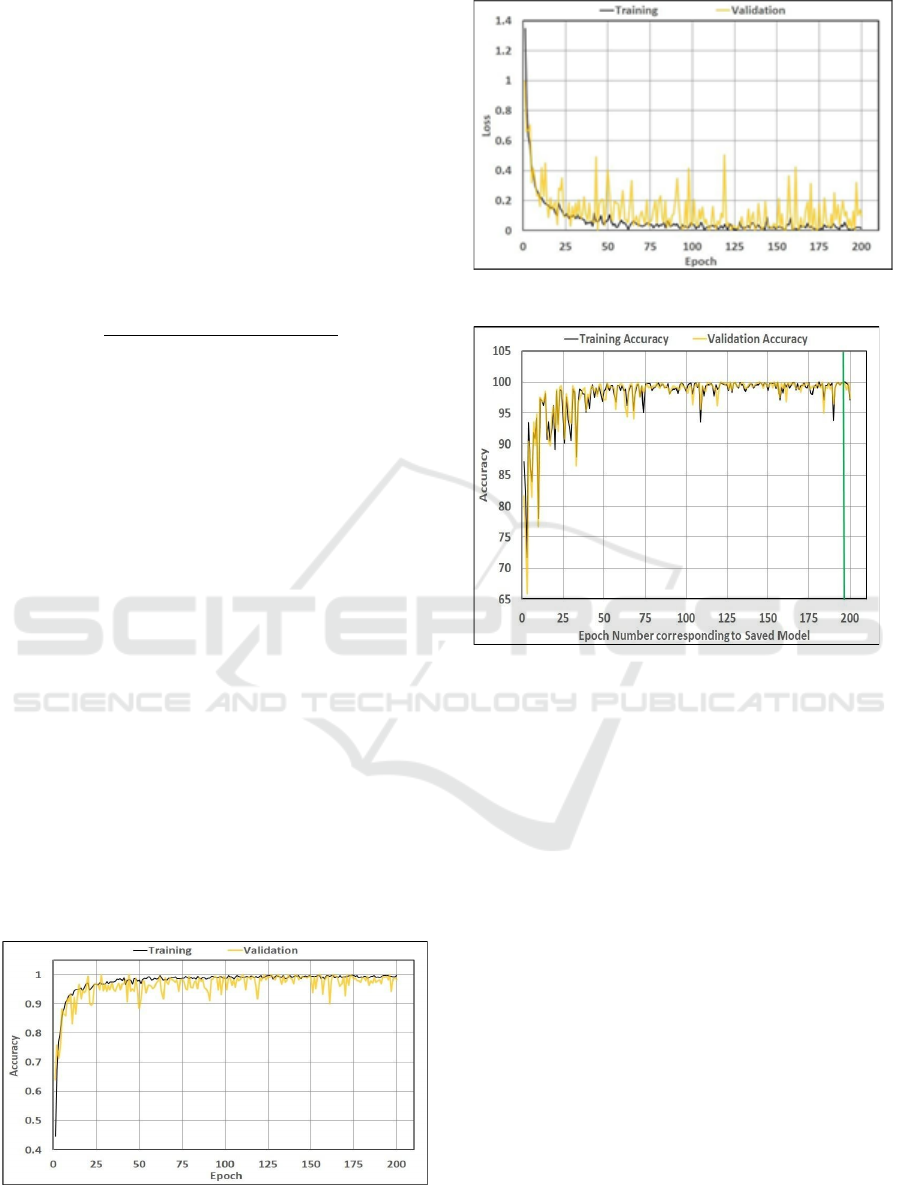
training dataset for 200 epochs. For every epoch, the
training accuracy of the model is calculated. With the
parameter weights calculated for each epoch, the
Figure 3:Model Accuracy
Figure 4:Model Loss
Figure 5:Training Validation accuracy-Checkpoint Saved
Models
model is evaluated against the validation dataset and the
validation accuracy is calculated. Using the checkpointing
capability provided by keras, the model with parameter
weights is saved at the end of each epoch. While the training
is in progress, the training loss, training accuracy,
validation loss and validation accuracy for each epoch is
monitored.
4 RESULTS AND DISCUSSIONS
To start the process of plant species identification,
the first step involved collecting the dataset. A
qualitative study was conducted to identify suitable
medicinal leaf dataset sources and determine the
dataset format. The selection of a dataset is influenced
by several factors, including the nature of the
problem, the availability of data, the diversity of the
data, and the relevance to the application. In order to
generalize our approach, we used the benchmark
dataset of medicinal plant leaf classification, i.e.,
Mendeley Medicinal Leaf. The dataset, representing
images from 30 different medicinal plants, was
INCOFT 2025 - International Conference on Futuristic Technology
220

selected for this study. The selected dataset contained
a total of 1835 leaf images. A system’s performance
is affected by factors like the dataset size, class
distribution, and data quality. To address this
concern, data preprocessing was employed to clean
the dataset. Figure 6 depicts some of the leaf samples
of the Mendeley Medicinal Leaf Dataset.
Figure 6: Showcases a sample of the medicinal leaf dataset
images, illustrating the diverse range of plant images
incorporated for classification purposes. (a) Alpinia
Galanga (Rasna), (b) Amaranthus Viridis (Arive-Dantu),
(c) Artocarpus Heterophyllus (jackfruit), (d) Azadirachta
Indica (neem), (e) Basella Alba (Basale), (f) Brassica
Juncea (Indian mustard), (g) Carissa Carandas (Karanda),
(h) Citrus Limon (lemon), (i) Ficus Auriculata (Roxburgh
fig), (j) Ficus Religiosa (peepal tree), (k) Jasminum
(jasmine), (l) Mangifera Indica (mango).
The custom CNN model described in this work
for APLC was freshly designed, trained and tested
using the AYUR-Best model approach. It is also
possible to leverage pre-trained CNN models for
Ayurvedic leaf image classification. This method of
reuse of pre-trained CNN models is known as
Transfer learning. There are many popular pre-
trained CNN models available in TensorFlow. As a
part of the work presented, a comparison of the
described CNN model is done with repurposed CNN
models based on pre-trained models like
DenseNet169, EfficientNetB6, Inception ResNetV2,
ResNet152V2, VGG16 and Xception. The results
are presented in Table 2. The observation is that
classification accuracy achieved on the test dataset is
similar for the custom CNN model (developed using
the AYUR-Best model approach) and the repurposed
CNN models. The custom CNN model (developed
using the AYUR-Best model approach) is better in
terms of the number of parameters and the size of the
respective saved model. When compared to the
repurposed CNN models, the number of parameters
in the custom CNN model is the lowest and the size
of the respective saved model is the smallest.
5 CONCLUSION AND FUTURE
ENHANCEMENT
This work has successfully presented the custom
CNN model and ayurnet approach for aplc. The
motivation behind the work was to come up with an
automatic computer vision based system to identify
locally available ayurvedic plants. In this work, the
part of the plant chosen for plant identification is the
leaf. The leaves of various plants are distinguishable
from each other due to morphological differences.
Leaves are the most easily available part of the plant
and are available throughout the year. Several
previous works done about plant leaf classification
were reviewed. We used a convolution neural
network (cnn) to solve this problem of multi-class
classification of plant leaves of some andhra
ayurvedic plants. The convolution layer part of cnn
was used for feature extraction and the fully
connected dense layer part of cnn was used for
multiclass classification. Using the known best
practices, a very simple and elegant cnn model was
designed and built using keras to solve the plant leaf
multi-class classification problem. The model was
trained on the training dataset for 200 epochs. Using
the weight parameters obtained in each epoch, the
model was tested against the validation dataset.
Training accuracy and validation accuracy were
compared at each epoch and the model with the best
weight parameters was chosen. The logic used to
choose the ayur-best model was high accuracy above
the chosen threshold of 99% and the least possible
difference between training accuracy and validation
accuracy. This was to ensure that the accuracy of the
model was very high while ensuring that there was no
overfitting. The model chosen using this method
performed very well on the test dataset too and it
resulted in an accuracy of 99.88%. Similar high
accuracies were achieved by leveraging popular pre-
trained models like densenet169, efficientnetb6,
inceptionresnetv2, resnet152v2, vgg16 and xception,
but it is seen that the respective models are heavy with
a large number of parameters when compared to the
custom cnn model described in this work. Due to the
small size of the custom cnn model, it is suitable for
the development of the mobile application for
ayurvedic plant species identification based on the
respective leaf images.
An Effective Convolutional Learning Model with Fine-Tuning for Medicinal Plant Leaf Identification
221

REFERENCES
Bisby, F. A. (2000). The quiet revolution: biodiversity
informatics and the internet. Science, 289(5488):2309–
2312.
Edwards, J. L., Lane, M. A., and Nielsen, E. S. (2000).
Interoperability of biodiversity databases: biodiversity
information on every desktop. Science,
289(5488):2312–2314.
Dixit and Ujjawal, "Texture classification using
convolutional neural network optimized with whale
optimization algorithm." SN Applied Sciences 1.6
(2019): 655
M. Mehdipour Ghazi, B. Yanikoglu and E. Aptoula, "Plant
identification using deep neural networks via
optimization of transfer learning parameters",
Neurocomputing, vol. 235, pp. 228-235, 2017
X. Zhu, M. Zhu and H. Ren, "Method of plant leaf
recognition based on improved deep convolutional
neural network", Cognitive Systems Research, vol. 52,
pp. 223-233, 2018.
Samadi, Farnaam, Gholamreza Akbarizadeh, and Hooman
Kaabi. "Change detection in SAR images using deep
belief network: a new training approach based on
morphological images." IET Image Processing 13.12
(2019): 2255-2264.
Ubbens and Jordan, "The use of plant models in deep
learning: an application to leaf counting in rosette
plants." Plant methods 14.1 (2018).
Lochan, Raja Naga, Anoop Singh Tomar, and R.
Srinivasan. "Plant Detection and Classification Using
Fast Region-Based Convolution Neural Networks."
Artificial Intelligence and Evolutionary Computations
in Engineering Systems. Springer, Singapore, 2020.
623-634
Yang and Xiaofeng, "Recognizing Image Semantic
Information through Multi-Feature
Fusion and SSAE-Based Deep Network." Journal of
Medical Systems 44.2 (2020).
P. Barré, B. Stöver, K. Müller and V. Steinhage, "LeafNet:
A computer vision system for automatic plant species
identification", Ecological Informatics, vol. 40, pp. 50-
56, 2017
J. Hu, Z. Chen, M. Yang, R. Zhang and Y. Cui, "A
Multiscale Fusion Convolutional Neural Network for
Plant Leaf Recognition", IEEE Signal Processing
Letters, vol. 25, no. 6, pp. 853-857, 2018.
Z. Wang, X. Sun, Z. Yang, Y. Zhang, Y. Zhu and Y. Ma,
"Leaf Recognition Based on DPCNN and BOW",
Neural Processing Letters, vol. 47, no. 1, pp. 99-115,
2017.
Chaki and Jyotismita, "Fragmented plant leaf recognition:
Bag-of-features, fuzzy-color and edge-texture
histogram descriptors with multi-layer perceptron."
Optik 181 (2019): 639-650.
Chen and Junde, "Detection of rice plant diseases based on
deep transfer learning." Journal of the Science of Food
and Agriculture (2020).
Lee and Sue Han, "How deep learning extracts and learns
leaf features for plant classification." Pattern
Recognition 71 (2017): 1-13.
Grinblat, Guillermo L., "Deep learning for plant
identification using vein morphological patterns."
Computers and Electronics in Agriculture 127 (2016):
418- 424.
Ferentinos, Konstantinos P. "Deep learning models for
plant disease detection and diagnosis." Computers and
Electronics in Agriculture 145 (2018): 311-318.
Du, Ji-xiang, Chuan-Min Zhai, and Qing-Ping Wang.
"Recognition of plant leaf image based on fractal
dimension features." Neurocomputing 116 (2016): 150-
156.
Atabay, Habibollah Agh. "A convolutional neural network
with a new architecture applied on leaf classification."
IIOAB J 7.5 (2016): 226-331.
Dyrmann, Mads, Henrik Karstoft, and Henrik Skov
Midtiby. "Plant species classification using deep
convolutional neural network." Biosystems
Engineering 151 (2016): 72-80.
Iniyan, S., et al. "Plant Disease Identification and Detection
Using Support Vector Machines and Artificial Neural
Networks." Artificial Intelligence and Evolutionary
Computations in Engineering Systems. Springer,
Singapore, 2020. 15-27.
Ho, T. K. (1995, August). Random decision forests. In
Proceedings of 3rd international conference on
document analysis and recognition (Vol. 1, pp. 278-
282). IEEE
INCOFT 2025 - International Conference on Futuristic Technology
222
