
Unsupervised Approach for Named Entity Recognition in Biomedical
Documents Using LDA-BERT Models
Veena G
1 a
, Deepa Gupta
2 b
and V D Vivek
2
1
Department of Computer Science and Applications, Amrita Vishwa Vidyapeetham,
Amritapuri, India
2
Department of Computer Science and Engineering, Amrita Vishwa Vidyapeetham,
Bengaluru, India
Keywords:
Unsupervised Approach, Named Entity Recognition, SciBERT, Biomedical, Topic Modeling, LDA.
Abstract:
Named Entity Recognition (NER) is a crucial task in biomedical text mining, enabling the identification and
extraction of entities such as genes, proteins, diseases, and drugs. Existing NER approaches often rely on
supervised learning with labeled data, which may be limited and expensive to obtain. This research proposes
a novel semantic model featuring weighted distributions focused on unsupervised NER within the biomedical
domain. Our methodology leverages an enhanced SciBERT model incorporating LDA topic modeling (SciB-
ERT+LDA) for NER. The model specifically targets identifying three significant entities: Disease, Chemical,
and Protein. The assessment of the proposed method demonstrates its effectiveness in recognizing named
entities within the biomedical domain. Our proposed approach demonstrates promising results, attaining a
macro-average F-measure of 86%. Moreover, the proposed approach can readily be expanded to encompass
the recognition of more domain-specific entities.
1 INTRODUCTION
Biomedical Named Entity Recognition (BioNER) is
a specialized task within Natural Language Process-
ing (NLP) and computational biology that involves
identifying and classifying specific entities or terms in
biomedical texts. It recognizes the major named enti-
ties present in medical records, research articles, and
other healthcare documents. Identifying these entities
is vital in downstream NLP tasks such as knowledge
graph creation, clinical decision support systems, and
drug discovery and development. Significant research
efforts have been focused on developing open-domain
NER systems that utilize cutting-edge machine learn-
ing models. Identifying named entities within spe-
cific domains poses a notably more challenging task
((Gridach, 2017), (Wei et al., 2019), (Zhang et al.,
2021)) due to the semantic complexity of the entities.
Current open NER models failed to identify domain-
specific entities due to their training on distinct cor-
pora. Furthermore, the challenges of transferability
and generalizability persist as significant obstacles. In
this work, we formulate BioNER as a classification
a
https://orcid.org/0000-0003-3513-9304
b
https://orcid.org/0000-0002-1041-5125
problem instead of sequence labeling problem, which
is used in State-of-the-art NER models. Our method-
ology uses the extended SciBERT coupled with LDA
(SciBERT+LDA) model to generate a Global Vector
for each entity. We first use topic modeling to identify
the latent topics within the corpus. Specifically, we
adopt the widely recognized LDA approach for topic
modeling (Blei et al., 2003a). Subsequently, these
identified topics undergo vectorization using SciB-
ERT, a contextualized word embedding model tai-
lored for the biomedical and scientific domains. SciB-
ERT captures the context and semantics of scientific
language, including the specific vocabulary and struc-
tures commonly found in academic papers, research
articles, and other scientific documents. By combin-
ing LDA and SciBERT, we capitalize on the distinc-
tive capabilities inherent in each model. Furthermore,
we extend SciBERT’s tokenizer to recognize domain-
specific keywords. Additionally, in our process, we
use the weighted scores obtained from the LDA mod-
elm to calculate local input word vectors. In the final
step of our methodology, we compare these locally
generated input vectors and the Global Vectors. This
comparison uses the cosine similarity metric to ascer-
tain the ultimate entity tag for the given input phrase.
The primary contributions of our work include:
G, V., Gupta, D. and V D, V.
Unsupervised Approach for Named Entity Recognition in Biomedical Documents Using LDA-BERT Models.
DOI: 10.5220/0013588500004664
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 3rd International Conference on Futuristic Technology (INCOFT 2025) - Volume 2, pages 155-162
ISBN: 978-989-758-763-4
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
155

• We introduce an unsupervised approach for
named entity labeling within the Biomedical do-
main. The SciBERT+LDA model incorporates el-
ements from both SciBERT and LDA to provide
a robust solution for identifying and categorizing
named entities in biomedical texts.
• Our approach produces domain-specific word
vectors through an enhanced SciBERT, suitable
for application in tasks related to relation extrac-
tion.
The remainder of the document is organized as
follows: Section 2 provides a review of related works.
The proposed method is detailed in Section 3. Experi-
mental results and analysis are discussed in Section 4.
In conclusion, Section 5 summarizes the paper, pro-
viding concluding remarks and suggesting potential
directions for future research.
2 RELATED WORKS
In the present era, state-of-the-art performance
in NER systems is accomplished by utilising deep
learning models, which demand minimal feature en-
gineering (Brundha et al., 2023), (Srivastava et al.,
2022). We conducted an exhaustive literature review
to develop a NER system tailored to the biomedical
domain, concentrating on unsupervised approaches
for identifying named entities. These models au-
tonomously uncover latent features within raw text
and have found applications across diverse domains
(Shahina et al., 2019). Their versatility has enabled
effective application in various fields, ranging from
biomedical research and healthcare to finance and le-
gal domains. A deep neural network based NER
model in the Biomedical Domain (BioNER) is pre-
sented in (Yao et al., 2015). Bio-NER reported a
71.01% F-measure on the GENIA corpus. Wei et
al.(Wei et al., 2016) present a Conditional Random
Field (CRF)-based neural system designed for identi-
fying disease entities in PubMed abstracts. Gopalakr-
ishnan et al. (Gopalakrishnan et al., 2019) pro-
posed an RNN, LSTM, and GRU approach on GE-
NIA version 3.02 corpus and achieved an F score
of 90%. Chiu and Eric Nichols (Chiu and Nichols,
2016) proposed an approach utilizing bidirectional
LSTM-CNN for named entity identification, achiev-
ing a 91.62% F-measure on the CoNLL-2003 dataset.
Li et al. (Li et al., 2020) propose a detailed survey of
deep learning based NER systems.
Several recent research works have explored Pre-
trained language models for downstream NLP ap-
plications, yielding substantial success (Veena et al.,
2023b), (G et al., 2023), (Veena et al., 2023a). Bidi-
rectional Encoder Representations from Transform-
ers for Biomedical Text Mining (BioBERT) is a
domain-specific language representation model pre-
trained on large-scale biomedical corpora (Lee et al.,
2020). BioBERT is fine-tuned on specific biomed-
ical NLP tasks, NER, relation extraction, and clas-
sification (Zhu et al., 2020), (KafiKang and Hen-
dawi, 2023), etc. ABioNER, a BERT-based model
to identify biomedical named entities in the Arabic
text, is proposed in (Boudjellal et al., 2021). A de-
tailed survey of transformer based pretrained models
for biomedical NER systems can be found in (Kalyan
et al., 2022).
Conventional supervised approaches necessitate
the availability of manually annotated datasets for
training, a process that can be both resource-intensive
and time-consuming. In contrast, unsupervised mod-
els offer a more flexible and adaptable alternative
by alleviating the dependency on annotated data, en-
abling them to be applied across diverse domains and
languages. Unsupervised methodologies are specif-
ically crafted to unveil latent patterns or structures
inherent within datasets lacking explicit labeling. A
notable illustration of such an approach is exem-
plified by Zhang and Elhadad (Zhang and Elhadad,
2013), who introduce an unsupervised technique for
biomedical named-entity recognition. The efficacy
of their method is assessed on the i2b2 and GENIA
corpora, yielding reported accuracies of 69.5% and
53.8%, respectively. Another unsupervised strategy,
CycleNER (Iovine et al., 2022), is centered around
learning the mapping between sentences and entities.
This approach undergoes evaluation on the CoNLL
and BC2GM datasets, with resulting average F mea-
sures of 68.6% and 38%, respectively. After the lit-
erature survey, we identified the following research
gaps:
• Traditional supervised approaches require manu-
ally annotated datasets for training, which can be
resource-intensive and time-consuming to create.
• Models trained on specifically labeled datasets
may have difficulty generalizing to new, unseen
data that differs significantly from the training set.
• Supervised models can be influenced by biases
present in annotated data.
Considering these gaps, we propose an unsuper-
vised approach for SciBert with LDA. After applying
LDA, each document can be represented as a distri-
bution over topics, and each topic is represented as a
distribution over words. These distributions can serve
as features for the documents. Detailed explanations
of the proposed architecture and the analysis of results
INCOFT 2025 - International Conference on Futuristic Technology
156
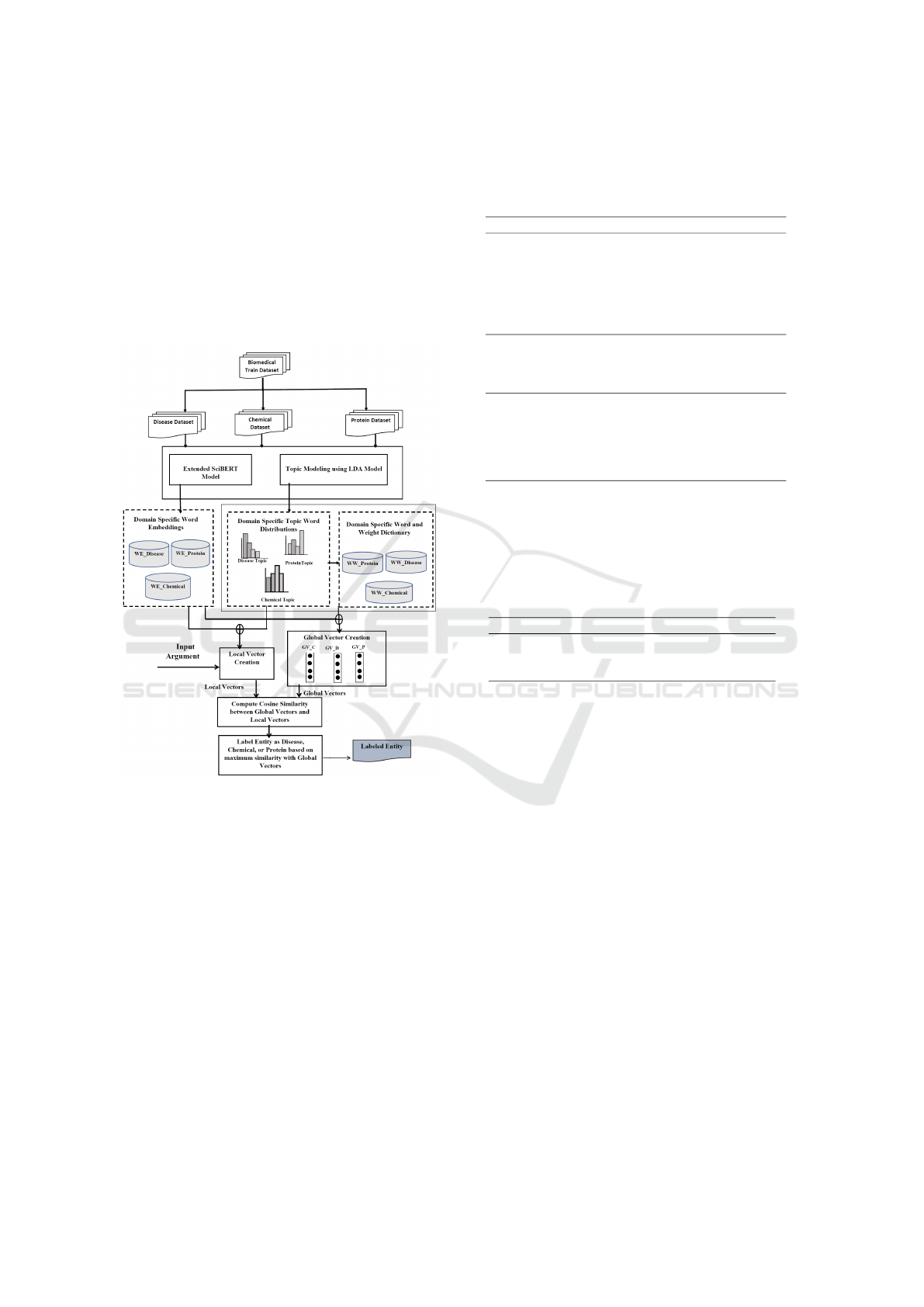
are provided in the following sections.
3 METHODOLOGY
The subsequent steps involve the labeling of enti-
ties within the sentence by the proposed model, cat-
egorizing them into three distinct classes, viz., Dis-
ease, Chemical, and Protein.
Figure 1: The Pipeline model of the Proposed Unsupervised
Named Entity Recognition in Biomedical text.
3.1 Dataset Creation Module
We created distinct datasets for each Biomedical sub-
domain: Disease, Chemical, and Protein. To ex-
tract data, we used key phrases associated with Dis-
ease, Chemical, and Protein subdomains, focusing
on BC4CHEMD ((Krallinger et al., 2015)), NCBI-
Disease((Do
ˇ
gan et al., 2014)), and BC2GM ((Smith
et al., 2008)) datasets. The list of keyphrases con-
tains 550 disease names, 250 names of genes, and 100
chemical names. Examples of key phrases include
{‘arthralgia’, ‘atherogenesis’, ‘Duchenne muscular
dystrophy’, ‘pancreatic cysts and tumors’, ‘breast
and prostate cancer’, ‘Erythropoietin receptor’, ‘lep-
tin receptor’, ’phalloidin’, ’cyclin-dependent kinase
1’,..}. Examples of retrieved sentences related to each
subdomain are displayed in Table 3.
Table 1: Sample Sentences extracted using Keywords
Domain Phrases
Disease Duchenne muscular dystrophy (DMD) is a
severe type of muscular dystrophy affecting
boys. Retinoblastoma (Rb) is a rare cancer
developing in retinal cells. Soft-tissue sar-
coma (STS) is a malignant tumor in soft tis-
sue, often growing painlessly over months or
years.
Protein The long-chain fatty acyl-CoA ligase
enzyme activates fatty acid oxidation.
ACSL5 gene encodes the long-chain-fatty-
acid—CoA ligase 5 enzyme.
Chemical The glycine transporter-1 inhibitor
SSR103800 exhibits an antipsychotic-
like profile in mice. Schizophrenia is linked
to dopamine dysfunction. High doses of
catecholamines, sedatives, and relaxants via
a central venous catheter were ineffective.
In the process of constructing the training set, 80%
of the key phrases are used. Detailed information re-
garding the data statistics for the combined training
and test datasets within each subdomain is available
in Table 3.
Table 2: Statistics of the Dataset
Sub-domain Sentences Avg. Length Unique Words
Disease 2906 12.40 7836
Chemical 2837 11.88 16060
Protein 3503 11.90 18113
Following data acquisition, we undertake funda-
mental pre-processing procedures. In the subsequent
subsection, a detailed explanation of the Entity Iden-
tification process is provided.
3.2 Entity Identification Module
The test sentences undergo an Entity Identification
module, which recognizes meaningful noun phrases
representing entities within the input sentence. For
entity extraction, we employed Open Information Ex-
traction systems (Del Corro and Gemulla, 2013). Af-
ter obtaining the entities, the subsequent step involves
labeling each entity into one of the three categories or
assigning it to the ’OTHER’ category. This process is
further detailed in the subsequent subsection.
3.3 Biomedical Entity Recognization
using the extended SciBERT with
LDA model
The three major entities in the biomedical docu-
ments, namely, Disease, Chemical, and Protein en-
Unsupervised Approach for Named Entity Recognition in Biomedical Documents Using LDA-BERT Models
157
tities, are identified and labeled using the SciB-
ERT+LDA model. The overall architecture of the
SciBERT+LDA model is depicted in Figure 1. The
entire process is divided into four submodules and ex-
plained in the following subsections.
3.3.1 Subdomain Word Vectorization
The training datasets from the three biomedical sub-
domains are fed into the extended SciBERT and LDA
models. The extended SciBERT model produces
domain-specific word vectors that capture semantic
information. At the same time, the LDA model cre-
ates domain-specific topic distributions and word dis-
tributions, offering insights into the thematic content
of the respective subdomains.
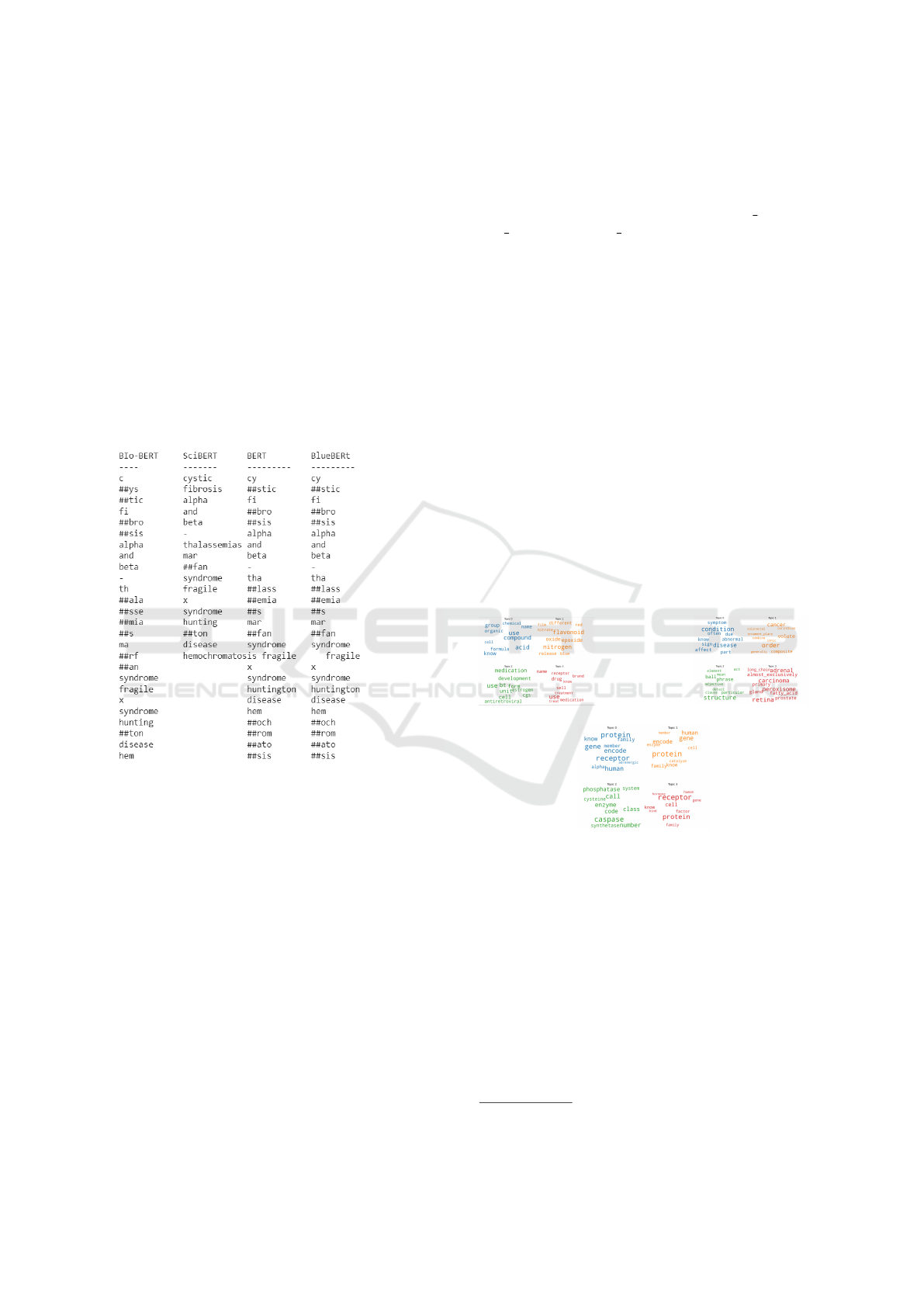
Figure 2: Comparison of different versions of BERT Mod-
els
SciBERT (Beltagy et al., 2019)is an extension of
the BERT model tailored for scientific and biomedi-
cal text. Its architecture retains the transformer-based
design of BERT but incorporates domain-specific pre-
training and an expanded vocabulary to better han-
dle scientific language characteristics. As depicted in
Figure 2, the number of tokens generated by various
BERT models for the following list of words cystic fi-
brosis alpha and beta-thalassemias Marfan syndrome
fragile X syndrome Huntington disease hemochro-
matosis are higher compared to SciBERT. This is
because of generating more subwords by the BERT
models. Consequently, for our experiments, we opted
for the SciBERT model. However, the SciBERT
model presents challenges, particularly concerning
the inclusion of domain-specific terms. To address
this issue, in our approach we augment the SciBERT
tokenizer’s vocabulary with domain-specific terms
((Tai et al., 2020)). Once the training dataset is vec-
torized, the resulting subdomain word vectors are
stored in separate dictionaries named WE Disease,
WE Protein, and WE Chemical. These dictionaries
are then used to generate both Global Vectors and Lo-
cal Vectors, as detailed in the following sections.
3.3.2 Topic Modeling using LDA Model
As shown in Figure 1, to identify the topics and as-
sociated word distributions, we employ topic model-
ing on the training datasets within each subdomain.
The main objective of topic modeling is to identify
the thematic structures within an extensive corpus of
text data. Common techniques for topic modeling
include Latent Dirichlet Allocation (LDA) and Non-
negative Matrix Factorization (NMF), Latent Seman-
tic Analysis (LSA) and Probabilistic Latent Seman-
tic Analysis (pLSA). We have used the most popular
approach, LDA (Blei et al., 2003b), for topic model-
ing. Subsequently, these topics undergo vectorization
using SciBERT, a semantic model leveraging the dis-
tributional properties of words. We used the two cru-
(a) Chemical (b) Disease
(c) Protein
Figure 3: Topic Distribution of Three LDA Models
cial hyperparameters in our experimental setup: α and
η. The parameter α governs document-topic density,
while η controls the topic-word density. For these hy-
perparameters, we opted for the default values pro-
vided by the Gensim library (
1
). Additionally, We
also focused on the determination of two critical pa-
rameters: the number of topics required (T ) and the
number of words required in a topic (K). In the con-
text of LDA, the optimal number of topics (T ) is se-
lected based on coherence scores. The topics identi-
fied for each subdomain are presented in Figures 3.
1
https://pypi.org/project/gensim/
INCOFT 2025 - International Conference on Futuristic Technology
158

3.3.3 Weighted Local Vector Generation
The Entity Identification module identifies the in-
put arguments to be labeled. Subsequently, these
input arguments undergo vectorization through the
weighted Local Vector creation module. We employ
a weighted scoring mechanism to adjust the signifi-
cance of words relative to each subdomain. Specif-
ically, if Arg represents an input argument with m
words (w
1
, w
2
, w
3
, . . . w
m
), we calculate the Local
Vector (LV) as the weighted average of all the m
words in Arg using the equation 1.
LV (Arg) =
∑
m
i=1
, W
wi
∗Vec(w
i
)
m
(1)
=
LV C; i f W
wi
∈ WW C & Vec(w
i
) ∈ W E C
LV D; i f W
wi
∈ WW D & Vec(w
i
) ∈ W E D
LV P; i f W
wi
∈ WW G & Vec(w
i
) ∈ W E P
GV (subdomain) =
∑
T
j=1
(
∑
K
i=1
W
w
ij
∗Vec(w
ij
))
T
(2)
=
GV C; i f W
w
ij
∈ TV C & Vec(w
i j
) ∈ W E C
GV D; i f W
w
ij
∈ TV D & Vec(w
i j
) ∈ W E D
GV P; i f W
w
ij
∈ TV P & Vec(w
i j
) ∈ W E P
In Equation (1), w
i
represents the ith word, and W
w
i
denotes the weight associated with the word w
i
. The
vector corresponding to each of the m words is de-
rived from the domain-specific word vectors which is
stored in the dictioneries WE C, WE D, and WE G.
These weights are obtained from the word-weight dic-
tionaries WW C (Chemical), WW D (Disease), and
WW P (Protein).
3.3.4 Global Vector(GV) Generation
The Global Vector creation commences with the ap-
plication of Topic modeling to the subdomain dataset,
where major topics are identified based on coherence
scores. The topics undergo vectorization, and the
Global Vector is created utilizing Equation (2). To
break down the steps further, the Global Vector is
derived by first calculating the average of all subdo-
main Topic Vectors, denoted as TV C, TV D, and
TV P. This involves computing the vector repre-
sentation for each word (w
i j
) within a topic, where
T signifies the total number of topics, K represents
the number of words in a topic, and W
wi j
denotes
the weight assigned to that word as defined by the
LDA in a subdomain. The function Vec(w
i j
) extracts
the embedding vector associated with the word w
i j
from the subdomain dictionaries. This ensures that
the contextual embeddings of the words, as captured
in the subdomain-specific dictionaries, contribute to
the overall composition of the Global Vector. This
entire process is repeated for each subdomain, lead-
ing to the generation of three distinct Global Vectors,
viz., GV C, GV D, and GV P.
3.3.5 Bio Named Entity Labeling- Chemical,
Disease, and Protein Entities
In this section, the labeling of the input argument is
determined based on its resemblance to subdomains.
As depicted in Figure 1, the input argument under-
goes a vectorization procedure, generating Local Vec-
tors tailored to each subdomain. Subsequently, these
Local Vectors are compared with the corresponding
set of four Global Vectors, namely GV C, GV D,
and GV P. The rationale behind this comparison
is to evaluate the relevance of the input argument
within the distinct contexts represented by the global
characteristics of each subdomain. In Equation (3),
Cos(LV, GV ) calculates the cosine similarity between
the Local and Global Vectors. Following the compu-
tation of the cosine similarity scores, the next step in-
volves identifying the Global Vector that exhibits the
maximum similarity with the Local Vector. The maxi-
mum similarity score is computed using Equation (4),
which shows the resemblance between the input argu-
ment and the identified subdomain’s Global Vector.
The final named entity label is determined by assign-
ing the tag (label) associated with the Global Vector,
as specified in Equation (5).
Label = argmax
i
{Cos(LV i, GV i) : i ∈ {C, D, G}}
(3)
Score = max
i
{Cos(LV i, GV i) : i ∈ {C, D, G}} (4)
ArgLabel=
C; i f Label = C and Score ≥ θ
chemical
D; i f Label = D and Score ≥ θ
disease
P; i f Label = P and Score ≥ θ
protein
OT HER or Place; Otherwise
(5)
Individual threshold values are assigned to establish
precise criteria for each subdomain. The threshold pa-
rameter, θ, is the average similarity score between the
training seed phrases and their corresponding Global
Vectors. To illustrate, consider the disease domain,
where the threshold value θ
disease
= 0.65 is derived.
This value is computed by determining the average
cosine similarity score across one thousand training
phrases compared to the Global Vector GV D repre-
sentative of the disease domain. Similar procedures
are employed to find threshold values for other do-
mains. In the chemical domain, the threshold is set
at θ
chemical
= 0.61, representing the average similarity
score between training phrases and the correspond-
ing Global Vector GV C for chemicals. Likewise,
in the protein domain, the threshold is determined as
Unsupervised Approach for Named Entity Recognition in Biomedical Documents Using LDA-BERT Models
159
θ
protein
= 0.60, reflecting the average cosine similarity
score obtained from the training phrases in relation to
the Global Vector GV P specific to proteins.
4 RESULTS ANALYSIS
This section provides a detailed examination of
the outcomes and assessment of the introduced un-
supervised BioNER model. A test corpus comprising
600 sentences from each subdomain is utilized to con-
duct the evaluation. The entities are identified using
the OpenIE system. In Table 3 the similarity score is
presented.
Table 3: Similarity Score of Global and Local Vectors
No. Phrase Cos Sim. GV D Cos Sim. GV C Cos Sim. GV P Pred. Actual
1 Abnormal
color vision
0.74 0.54 0.10 DISEASE DISEASE
2 Colorectal can-
cers
0.67 0.53 0.00 DISEASE DISEASE
3 Glycine 0.00 0.62 0.30 Chemical Chemical
4 Palinurin 0.00 0.00 0.00 Chemical Other
5 5-
hydroxytryptamin
0.53 0.63 0.43 Protein Protein
6 5-kinase 0.53 0.63 0.43 Chemical Protein
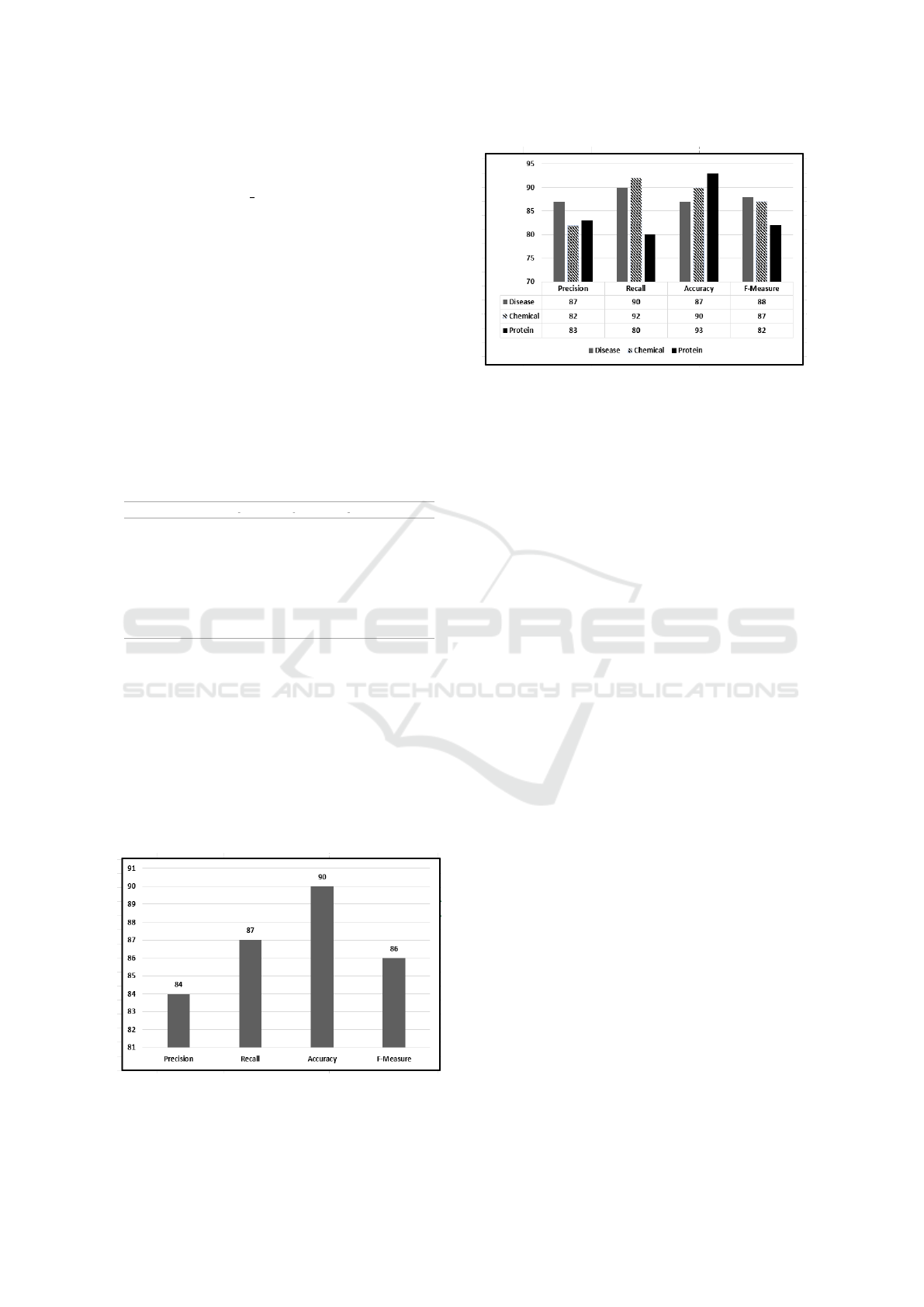
The model’s performance is rigorously assessed
using key metrics, including Precision, Recall, Accu-
racy, and F Score. The comprehensive performance
metrics of the proposed model are depicted in Figure
4. For a more in-depth understanding, we delve into
an entity-level analysis, the findings of which are pre-
sented in Figure 5. This analysis allows us to explore
the model’s performance at a granular level, offering
insights into how well it performs for each specific
entity.
Figure 4: Average performance metrics including Accuracy,
Precision, Recall, and F-Measure of the Proposed Unsuper-
vised BioNER Model.
Figure 5: Performance Metrics of the Proposed Unsuper-
vised BioNER Model for Identifying Disease, Chemical,
and Protein Entities
5 CONCLUSION
The presented methodology introduces an inno-
vative unsupervised approach for NER within the
Biomedical domain, known as the unsupervised
BioNER Model. This model specifically targets and
labels biomedical entities, including Disease, Chemi-
cal, and Protein. The core of the proposed approach
lies in the SciBERT+LDA model, which plays a piv-
otal role in the NER process. This model is designed
to create Global Vectors, essentially representative
vectors, for each distinct biomedical entity. These
Global Vectors encapsulate the overall characteristics
and significance of the respective entities within the
biomedical context. To enhance the model’s adapt-
ability to various domain-specific entities, a crucial
suggestion is made to extend the SciBERT tokenizer.
For the recognition of new inputs, the model gener-
ates Local Vectors using a sophisticated mechanism
involving a weighted score derived from the LDA
model and uses the cosine similarity of Local vec-
tors with Global vectors. Moreover, the proposed
BioNER Model offers seamless integration with any
Open Information Extraction (OIE) System. This
means that the model can be effortlessly incorporated
into existing OIE frameworks to carry out the label-
ing of arguments. The presented approach represents
a comprehensive and effective strategy for unsuper-
vised biomedical entity recognition. The suggested
methodology yielded an average F Measure of 86%, a
noteworthy achievement, particularly within the con-
text of an unsupervised setting. The attainment of an
86% F Measure implies a commendable level of ac-
curacy and effectiveness in the proposed approach for
the given task.
In our future endeavours, we aim to identify
entities, properties and relations that extend beyond
INCOFT 2025 - International Conference on Futuristic Technology
160

the confines of two sentences. The outcomes hold
significant promise for downstream applications,
particularly in creating knowledge bases. By in-
corporating the extracted information into existing
healthcare systems, professionals can access a wealth
of structured and interrelated data. This, in turn, can
contribute decision making regarding patient care
and treatment plans.
REFERENCES
Beltagy, I., Lo, K., and Cohan, A. (2019). SCIBERT: A pre-
trained language model for scientific text. In EMNLP-
IJCNLP 2019 - 2019 Conference on Empirical Meth-
ods in Natural Language Processing and 9th Inter-
national Joint Conference on Natural Language Pro-
cessing, Proceedings of the Conference.
Blei, D. M., Ng, A. Y., and Jordan, M. I. (2003a). Latent
Dirichlet allocation. Journal of Machine Learning Re-
search, 3(4-5).
Blei, D. M., Ng, A. Y., and Jordan, M. I. (2003b). Latent
Dirichlet allocation. Journal of Machine Learning Re-
search, 3(4-5).
Boudjellal, N., Zhang, H., Khan, A., Ahmad, A., Naseem,
R., Shang, J., and Dai, L. (2021). ABioNER: A BERT-
Based Model for Arabic Biomedical Named-Entity
Recognition. Complexity, 2021.
Brundha, J., Nair, P. C., Gupta, D., and Agarwal, J. (2023).
Name entity recognition for Air Traffic Control tran-
scripts using deep learning based approach. In 2023
IEEE 20th India Council International Conference,
INDICON 2023.
Chiu, J. P. and Nichols, E. (2016). Named Entity Recogni-
tion with Bidirectional LSTM-CNNs. Transactions of
the Association for Computational Linguistics, 4.
Del Corro, L. and Gemulla, R. (2013). ClausIE: Clause-
based open information extraction. In WWW 2013 -
Proceedings of the 22nd International Conference on
World Wide Web.
Do
ˇ
gan, R. I., Leaman, R., and Lu, Z. (2014). NCBI disease
corpus: A resource for disease name recognition and
concept normalization. Journal of Biomedical Infor-
matics, 47.
G, V., Gupta, D., and Kanjirangat, V. (2023). Semi Super-
vised Approach for Relation Extraction in Agriculture
Documents.
Gopalakrishnan, A., Soman, K. P., and Premjith, B. (2019).
A Deep Learning-Based Named Entity Recognition in
Biomedical Domain. In Lecture Notes in Electrical
Engineering, volume 545.
Gridach, M. (2017). Character-level neural network for
biomedical named entity recognition. Journal of
Biomedical Informatics, 70.
Iovine, A., Fang, A., Fetahu, B., Rokhlenko, O., and Mal-
masi, S. (2022). CycleNER: An Unsupervised Train-
ing Approach for Named Entity Recognition. In
WWW 2022 - Proceedings of the ACM Web Confer-
ence 2022.
KafiKang, M. and Hendawi, A. (2023). Drug-Drug Inter-
action Extraction from Biomedical Text Using Rela-
tion BioBERT with BLSTM. Machine Learning and
Knowledge Extraction, 5(2).
Kalyan, K. S., Rajasekharan, A., and Sangeetha, S. (2022).
AMMU: A survey of transformer-based biomedical
pretrained language models.
Krallinger, M., Rabal, O., Leitner, F., Vazquez, M., Salgado,
D., Lu, Z., Leaman, R., Lu, Y., Ji, D., Lowe, D. M.,
Sayle, R. A., Batista-Navarro, R. T., Rak, R., Huber,
T., Rockt
¨
aschel, T., Matos, S., Campos, D., Tang, B.,
Xu, H., Munkhdalai, T., Ryu, K. H., Ramanan, S. V.,
Nathan, S.,
ˇ
Zitnik, S., Bajec, M., Weber, L., Irmer, M.,
Akhondi, S. A., Kors, J. A., Xu, S., An, X., Sikdar,
U. K., Ekbal, A., Yoshioka, M., Dieb, T. M., Choi,
M., Verspoor, K., Khabsa, M., Giles, C. L., Liu, H.,
Ravikumar, K. E., Lamurias, A., Couto, F. M., Dai,
H. J., Tsai, R. T. H., Ata, C., Can, T., Usi
´
e, A., Alves,
R., Segura-Bedmar, I., Mart
´
ınez, P., Oyarzabal, J.,
and Valencia, A. (2015). The CHEMDNER corpus
of chemicals and drugs and its annotation principles.
Journal of Cheminformatics, 7.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., and
Kang, J. (2020). BioBERT: A pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4).
Li, J., Sun, A., Han, J., and Li, C. (2020). A Survey on
Deep Learning for Named Entity Recognition. IEEE
Transactions on Knowledge and Data Engineering.
Shahina, K. K., Jyothsna, P. V., Prabha, G., Premjith, B.,
and Soman, K. P. (2019). A Sequential Labelling Ap-
proach for the Named Entity Recognition in Arabic
Language Using Deep Learning Algorithms. In 2019
International Conference on Data Science and Com-
munication, IconDSC 2019.
Smith, L., Tanabe, L. K., Ando, R., Kuo, C. J., Chung, I. F.,
Hsu, C. N., Lin, Y. S., Klinger, R., Friedrich, C. M.,
Ganchev, K., Torii, M., Liu, H., Haddow, B., Stru-
ble, C. A., Povinelli, R. J., Vlachos, A., Baumgartner,
W. A., Hunter, L., Carpenter, B., Tsai, R. T. H., Dai,
H. J., Liu, F., Chen, Y., Sun, C., Katrenko, S., Adri-
aans, P., Blaschke, C., Torres, R., Neves, M., Nakov,
P., Divoli, A., Ma
˜
na-L
´
opez, M., Mata, J., and Wilbur,
W. J. (2008). Overview of BioCreative II gene men-
tion recognition.
Srivastava, S., Paul, B., and Gupta, D. (2022). Study
of Word Embeddings for Enhanced Cyber Security
Named Entity Recognition. In Procedia Computer
Science, volume 218.
Tai, W., Kung, H. T., Dong, X., Comiter, M., and Kuo, C. F.
(2020). exBERT: Extending pre-trained models with
domain-specific vocabulary under constrained train-
ing resources. In Findings of the Association for
Computational Linguistics Findings of ACL: EMNLP
2020.
Veena, G., Gupta, D., and Kanjirangat, V. (2023a). Semi-
Supervised Bootstrapped Syntax-Semantics-Based
Approach for Agriculture Relation Extraction for
Knowledge Graph Creation and Reasoning. IEEE Ac-
cess, 11.
Unsupervised Approach for Named Entity Recognition in Biomedical Documents Using LDA-BERT Models
161

Veena, G., Kanjirangat, V., and Gupta, D. (2023b).
AGRONER: An unsupervised agriculture named en-
tity recognition using weighted distributional seman-
tic model. Expert Systems with Applications, 229.
Wei, H., Gao, M., Zhou, A., Chen, F., Qu, W., Wang, C.,
and Lu, M. (2019). Named Entity Recognition from
Biomedical Texts Using a Fusion Attention-Based
BiLSTM-CRF. IEEE Access, 7.
Wei, Q., Chen, T., Xu, R., He, Y., and Gui, L. (2016). Dis-
ease named entity recognition by combining condi-
tional random fields and bidirectional recurrent neural
networks. Database, 2016.
Yao, L., Liu, H., Liu, Y., Li, X., and Anwar, M. W. (2015).
Biomedical Named Entity Recognition based on Deep
Neutral Network. International Journal of Hybrid In-
formation Technology, 8(8).
Zhang, Q., Sun, Y., Zhang, L., Jiao, Y., and Tian, Y. (2021).
Named entity recognition method in health preserving
field based on BERT. In Procedia Computer Science,
volume 183.
Zhang, S. and Elhadad, N. (2013). Unsupervised biomedi-
cal named entity recognition: Experiments with clini-
cal and biological texts. Journal of Biomedical Infor-
matics, 46(6).
Zhu, Y., Li, L., Lu, H., Zhou, A., and Qin, X. (2020).
Extracting drug-drug interactions from texts with
BioBERT and multiple entity-aware attentions. Jour-
nal of Biomedical Informatics, 106.
INCOFT 2025 - International Conference on Futuristic Technology
162
