
Brain Tumor Classification with Hybrid Deep Learning Models from
MRI Images
Dhouha Boubdellaha
1
, Raouia Mokni
2 a
and Boudour Ammar
1,3 b
1
Research Groups in Intelligent Machines (REGIM), National Engineering School of Sfax, University of Sfax, Tunisia
2
Advanced Technologies for Environment and Smart Cities(ATES Unit), University of Sfax, Tunisia
3
Department of Computer Management at the Higher Business School of Sfax (ESC Sfax), University of Sfax, Tunisia
Keywords:
Brain Tumor, Ensemble Learning, Deep Learning, MRI, Hybrid Models, Vision Transformers.
Abstract:
Brain tumor classification using MRI images plays a crucial role in medical diagnostics, enabling early detec-
tion and improving treatment planning. Traditional diagnostic methods are often subjective and time-intensive,
emphasizing the need for automated and precise solutions. This study explores hybrid deep learning mod-
els alongside fine-tuned pre-trained architectures for classifying brain tumors into four categories: glioma,
meningioma, pituitary tumor, and healthy brain tissue. The proposed approach incorporates both hybrid en-
semble models—Ens-VGG16-FT-InceptionV3, Ens-ViT-FT-InceptionV3, and Ens-CNN-ViT—and fine-tuned
architectures—FT-VGG16, FT-VGG19, and FT-InceptionV3. To enhance model robustness and generaliza-
tion, data augmentation techniques such as rotation and scaling were applied. Among these models, the hybrid
ensemble Ens-VGG16-FT-InceptionV3 achieved the highest accuracy and F1-score of 99%, outperforming
both standalone models and other hybrid configurations. These findings demonstrate the effectiveness of inte-
grating complementary architectures for improved brain tumor classification. Ultimately, this study highlights
the potential of hybrid ensemble learning to advance brain tumor diagnostics, providing more accurate, reli-
able, and scalable medical imaging solutions.
1 INTRODUCTION
Brain tumors are among the most serious and life-
threatening medical conditions, contributing signifi-
cantly to global morbidity and mortality. Recognized
by the World Health Organization (WHO) as a signif-
icant public health concern (Arabahmadi et al., 2022),
the increasing prevalence of brain tumors underscores
the urgent need for improved diagnostic techniques.
Early and precise diagnosis is essential for effective
treatment planning and better patient outcomes. Mag-
netic Resonance Imaging (MRI) is widely used for
detecting and analyzing brain abnormalities, offering
detailed insights into tumor size, location, and type
(Lee et al., 2020). However, manual interpretation
of MRI scans by radiologists is time-consuming and
subject to variability, particularly in distinguishing
subtle tumor variations (Rivkin and Kanoff, 2013).
Recent advancements in artificial intelligence (AI)
and deep learning have demonstrated significant po-
a
https://orcid.org/0000-0002-6652-5251
b
https://orcid.org/0000-0002-4934-1573
tential in automating this process (Bharadiya, 2023),
leading to faster and more accurate brain tumor clas-
sification (Pham et al., 2023).
Despite progress in medical imaging, brain tu-
mor classification remains a challenging task. Tra-
ditional diagnostic methods depend on manual eval-
uations, which are inherently subjective and require
extensive expertise (Sanvito et al., 2021). While ma-
chine learning has shown promise in automating clas-
sification, existing studies often face limitations such
as small datasets, simplistic architectures, or inade-
quate feature extraction (Iqbal et al., 2023), (Mokni
and Haoues, 2022). Additionally, many models strug-
gle to generalize across diverse patient populations,
emphasizing the need for robust, scalable solutions
capable of accurately classifying tumors into glioma,
meningioma, pituitary tumors, and healthy brain tis-
sue (Farmanfarma et al., 2019).
This study aims to address the challenges of brain
tumor classification from MRI scans by leveraging
advanced deep learning techniques. The primary goal
is to explore and compare the performance of various
hybrid deep learning models in detecting and catego-
Boubdellaha, D., Mokni, R., Ammar and B.
Brain Tumor Classification with Hybrid Deep Learning Models from MRI Images.
DOI: 10.5220/0013523700003967
In Proceedings of the 14th International Conference on Data Science, Technology and Applications (DATA 2025), pages 455-464
ISBN: 978-989-758-758-0; ISSN: 2184-285X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
455

rizing brain tumors into four classes: glioma, menin-
gioma, pituitary tumor, and no tumor. To achieve this,
the research focuses on:
• Applying Advanced Deep Learning Models: Uti-
lizing Convolutional Neural Networks (CNNs),
Vision Transformers (ViTs), and transfer learning
with fine-tuned pre-trained models (InceptionV3,
VGG16, and VGG19) to improve classification
accuracy and performance in brain tumor detec-
tion. The fine-tuned models are referred to as FT-
InceptionV3, FT-VGG16, and FT-VGG19.
• Exploring Hybrid Architectures and Ensemble
Learning: Developing and evaluating innovative
hybrid architectures by combining multiple mod-
els, such as CNN and ViT (Ens-CNN-VIT), ViT
and FT-InceptionV3 (Ens-VIT-FT-InceptionV3),
and FT-VGG16 and FT-InceptionV3 (Ens-FT-
VGG16-FT-InceptionV3). Ensemble learning en-
hances diagnostic precision by leveraging the
complementary strengths of different models,
mitigating their weaknesses, and improving over-
all robustness and generalization.
Through these objectives, the study aims to con-
tribute to the growing field of AI-driven medical di-
agnostics, ultimately enabling early, accurate detec-
tion and improved treatment of brain tumors, while
reducing the global burden of these life-threatening
conditions.
The remainder of this paper is organized as fol-
lows: Section 2 reviews related work on brain tumor
classification and the application of deep learning in
medical imaging. Section 3 outlines the proposed hy-
brid models. Section 4 presents the experimental re-
sults, followed by a comparative evaluation with pre-
vious studies in Section 5. Finally, Section 6 con-
cludes the paper and outlines future research direc-
tions.
2 RELATED WORK
In this section, we review the literature and findings
from research studies that focused on brain tumor de-
tection.
G
´
omez-Guzm
´
an et al.,(G
´
omez-Guzm
´
an et al.,
2023) evaluated seven deep CNN models for brain
tumor classification using MRI scans. Their study
compared a generic CNN with six pre-trained mod-
els, including InceptionV3, ResNet50, and Xception.
Among these, InceptionV3 achieved the highest ac-
curacy of 97.12%, demonstrating its ability to ex-
tract robust and distinctive features for classification.
Their findings emphasized the effectiveness of CNNs
in identifying brain tumors by capturing hierarchical
patterns in MRI images. This research contributed to
the advancement of automated brain tumor classifica-
tion, reinforcing the suitability of CNN-based archi-
tectures for medical imaging applications.
Khaliki and Basarslan(Khaliki and Bas¸arslan,
2024) advanced this research by incorporating trans-
fer learning techniques, leveraging pre-trained mod-
els like VGG16, VGG19, EfficientNetB4, and In-
ceptionV3. These architectures, initially trained on
large-scale datasets, were fine-tuned for brain tu-
mor classification tasks, achieving impressive accu-
racies of 98% and 97% with VGG16 and Efficient-
NetB4, respectively. The use of transfer learning sig-
nificantly reduced training time while enhancing the
models generalization capabilities on smaller medical
datasets. Their hybrid classification system demon-
strated that leveraging pre-trained networks could
bridge the gap between limited medical data availabil-
ity and high-precision diagnostics.
Balamurugan and Gnanamanoharan (Balamuru-
gan and Gnanamanoharan, 2023) and Fki et al., (Fki
et al., 2024)introduced a hybrid approach that com-
bined a Deep Convolutional Neural Network (DCNN)
with the enhanced LuNet algorithm for both segmen-
tation and classification of brain tumors. Their model
achieved unprecedented accuracies of 99.4% in seg-
mentation and 99.5% in classification, emphasizing
the efficacy of integrating advanced feature extraction
techniques with deep learning models.
Dhakshnamurthy et al., (Dhakshnamurthy et al.,
2024) proposed a hybrid model combining VGG16
and ResNet-50 architectures, achieving a ground-
breaking accuracy of 99.98%. This model outper-
formed standalone architectures such as AlexNet,
VGG16, and ResNet-50, emphasizing the effective-
ness of combining complementary strengths to en-
hance diagnostic precision. The integration of ResNet
skip connections with VGG16 deep feature extraction
layers facilitated improved gradient flow during train-
ing, addressing challenges associated with vanishing
gradients in deep networks.
Mahmud and Muntasir et al.,(Mahmud et al.,
2023) further contributed to this field by designing a
CNN-based approach for early detection of brain tu-
mors, achieving an accuracy of 93.3%. Their model
incorporated extensive preprocessing and data aug-
mentation techniques, including rotation, flipping,
and zooming, to enhance model robustness. By com-
paring their CNN model with architectures such as
ResNet-50 and InceptionV3, they demonstrated that
carefully designed CNN architectures could outper-
form more complex models in tasks requiring preci-
sion and computational efficiency.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
456
Asiri et al., (Asiri et al., 2023) explored the ca-
pabilities of Vision Transformers (ViTs) in medical
imaging by developing a fine-tuned Vision Trans-
former (FT-ViT) for brain tumor classification. The
FT-ViT achieved a classification accuracy of 98.13%
on the BraTS dataset, showcasing exceptional pre-
cision and recall across various tumor classes. This
approach marked a paradigm shift from convolution-
based methods to transformer-based architectures,
which rely on self-attention mechanisms to capture
global and local relationships in visual data. The find-
ings demonstrated the ViTs ability to process MRI
images as sequences of image patches, enabling a
more nuanced understanding of tumor morphology.
Table 1: Summary of previous studies on brain tumor clas-
sification.
Proposal Dataset Method Result
G
´
omez-
Guzm
´
an et
al.,(G
´
omez-
Guzm
´
an et al.,
2023)
Brain
Tumor
Classi-
fication
(MRI)
InceptionV3
ResNet50
InceptionResNetV2
Xception
MobileNetV2
EfficientNetB0
Generic CNN
97.12%
96.97%
96.78%
95.67%
95.45%
90.88%
81.08%
Khaliki and
Basarslan
(Khaliki and
Bas¸arslan,
2024)
Brain
Tumor
Classi-
fication
(MRI)
EfficientNetB4
InceptionV3
VGG19
VGG16
CNN
97%
95%
96%
98%
91%
Balamurugan
and Gnana-
manoha-
ran(Balamurugan
and Gnana-
manoharan,
2023)
BRATS DCNN-LuNet 99.50%
Fki et al., (Fki
et al., 2024)
BRATS DCNN-LuNet 99.50%
Dhakshnamurthy
et
al.,(Balamurugan
and Gnana-
manoharan,
2023)
Brain
Tumor
Classi-
fication
(MRI)
AlexNet
VGG16
ResNet50
VGG16–ResNet50
95.60%
97.66%
96.90%
98.80%
Mahmud et
al.,(Mahmud
et al., 2023)
Brain
Tumor
Classi-
fication
(MRI)
VGG16
ResNet-50
CNN
Inception V3
71.60%
81.10%
93.30%
80.00%
Asiri (Asiri
et al., 2023)
Brain
Tumor
MRI
Dataset
FT-ViT 98.13%
3 PROPOSED METHODOLOGY
This section presents a hybrid approach for brain
tumor classification using ensemble learning to im-
prove diagnostic accuracy. It combines the out-
puts of complementary deep learning models, in-
cluding (CNN and ViT), (ViT and FT-InceptionV3),
and (FT-VGG16 and FT-InceptionV3), to leverage
their strengths in pattern detection, feature extraction,
and representation learning. This ensemble learning
method reduces overfitting, mitigates biases, and en-
hances the robustness and generalization of the classi-
fication system. The integrated features are processed
through a dense layer to finalize tumor classifica-
tion. In fact, we developed three hybrid architectures:
Ens-CNN-ViT, Ens-FT-VGG16-FT-InceptionV3, and
Ens-ViT-FT-InceptionV3.These models demonstrated
remarkable accuracy in tumor classification, offering
a promising new approach for early and accurate brain
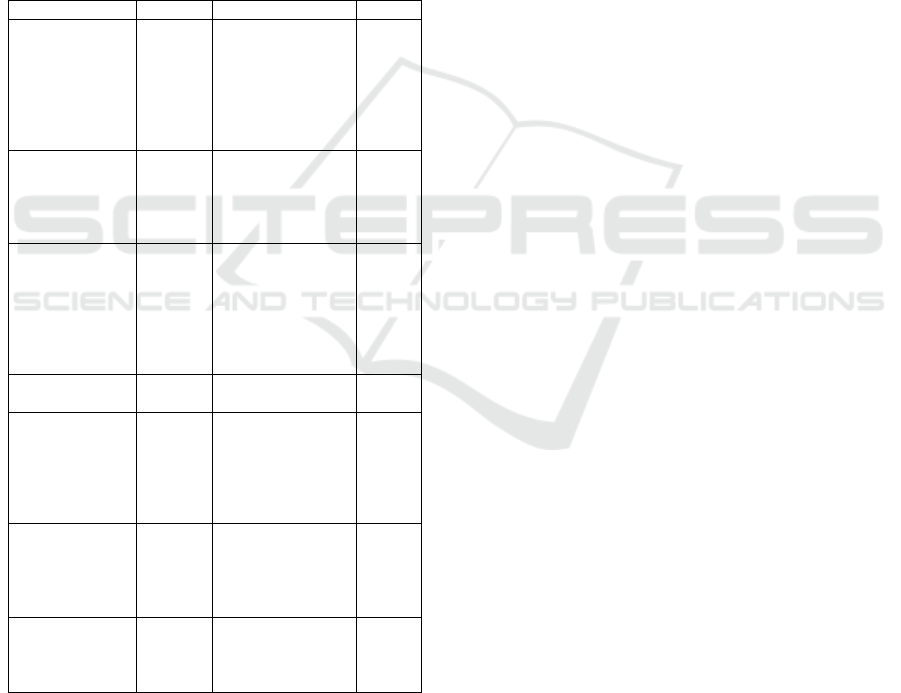
tumor diagnosis. Figure 1 represents an overview
of our proposed models for brain tumor classifica-
tion. This approach consists of six key steps: (1)
Dataset collection, (2) Data preprocessing, (3) Data
augmentation, (4) Deep learning-based model devel-
opment, (5) Ensemble learning-based Hybrid model,
and (6) Classification to classify brain tumors into
four stages.
3.1 Dataset Description
We used a publicly available ”Brain Tumor Classifi-
cation MRI” dataset from Kaggle (Bhuvaji, 2024) .
This dataset contains approximately 3,264 MRI im-
ages, classified into four categories: glioma, menin-
gioma, pituitary tumor, and no tumor, as illustrated in
Figure 2.
3.2 Dataset Preprocessing
This section describes the basic preprocessing steps
taken to prepare the data for model training. The data
preparation process involves loading brain tumor im-
ages from the specified folders (training and testing),
resizing them to 150 * 150 pixels, and then normaliz-
ing the image pixel values to the range [0, 1] by divid-
ing them by 255.0, which contributes to faster conver-
gence during training. Finally, we used the function
’Train test split’ which allows us to split the data into
training and testing sets, with 80% used for training
and 20% reserved for testing, allowing for a proper
evaluation of the model’s performance on unknown
data.
3.3 Dataset Augmentation
In our study, we applied data augmentation strate-
gies to enhance model performance by expanding the
dataset. The objective is to improve accuracy and op-
Brain Tumor Classification with Hybrid Deep Learning Models from MRI Images
457
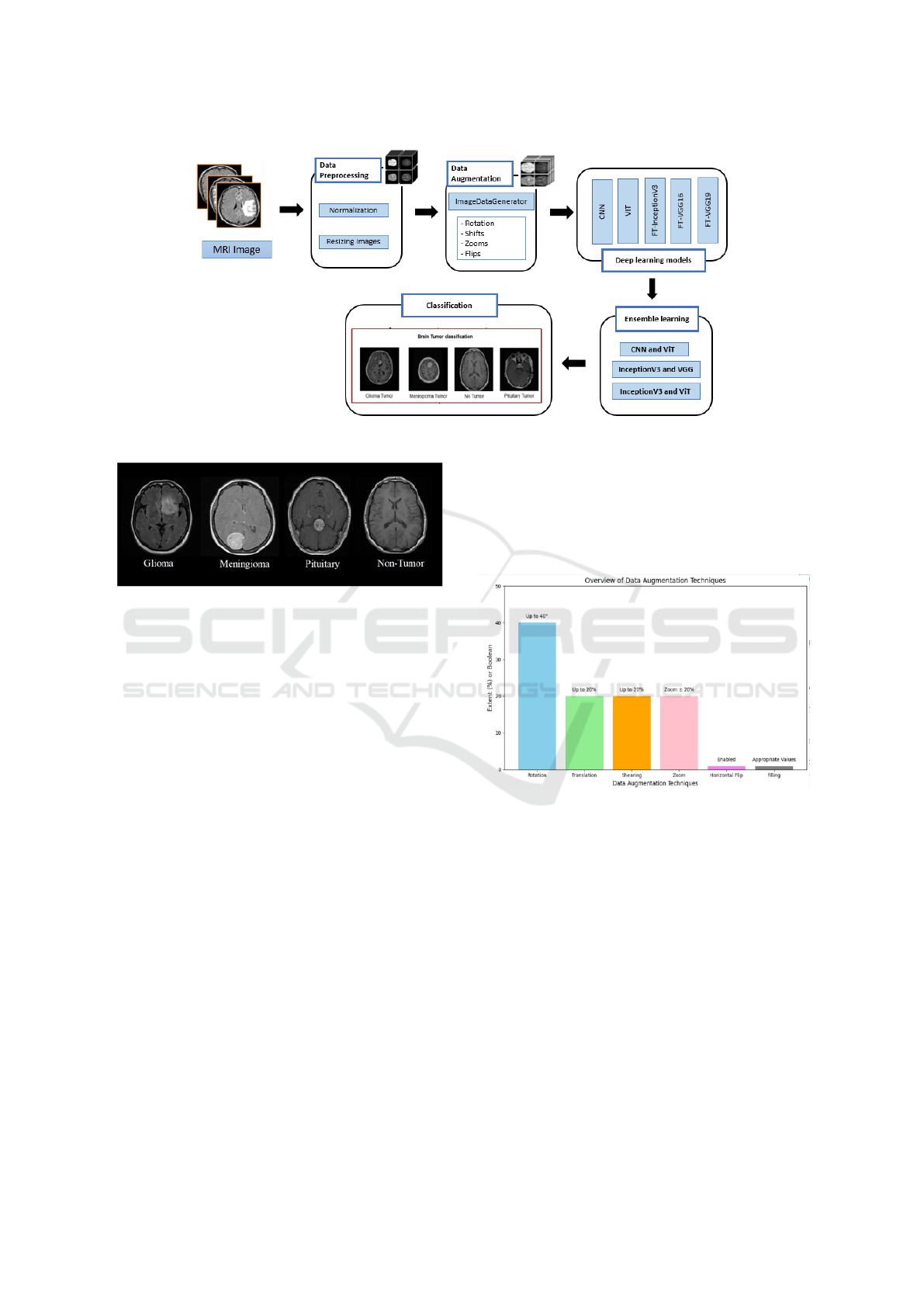
Figure 1: The proposed approach process for brain tumor classification.
Figure 2: Sample MRI images.
timize the model’s ability to classify brain tumor im-
ages. The key techniques used include:
1. The ’ImageDataGenerator’ class of Keras: ap-
ply alternative transformations to images during train-
ing including:
• Rotation: Images can be rotated up to 40 degrees.
• Translation: Horizontally and vertically deform
images (up to 20% of the large size and height).
• Shearing: Apply a cisaillement transformation
up to 20%.
• Zoom: Zoom in before or after 20%.
• Horizontal Flip: Images can be returned horizon-
tally.
• Filling: Use appropriate values to represent the
new pixels created during the transformations
Figure 3 presents the different Data augmentation
techniques.
2. Normalization: images are normalized to avoid
pixel values between 0 and 1, which helps the model
to approximate faster through training.
3. SMOTE (Synthetic Minority Oversampling
Technique): To generate synthetic examples from the
minority class to equalize the data game. This helps
to mitigate model bias in case of class imbalance.
3.4 Model Architecture Development
The methodology involves the application of both in-
dividual deep learning models and ensemble learning-
based hybrid architectures for the classification of
brain tumors.
Figure 3: Dataset Augmentation.
3.4.1 Convolutional Neural Networks (CNNs)
The proposed CNN for brain tumor classification
follows a structured architecture designed for effec-
tive feature extraction and classification. It begins
with multiple Conv2D layers, applying various fil-
ters to capture hierarchical features from input im-
ages. These layers are followed by BatchNormaliza-
tion to stabilize learning and MaxPooling2D to reduce
spatial dimensions while preserving key information.
The model then flattens the 2D feature map, convert-
ing it into a vector that passes through a Dense layer
with neurons, followed by a Dropout layer to prevent
overfitting. The final classification is handled by a
Dense output layer with 4 units and a Softmax activa-
tion function, categorizing brain tumors into glioma,
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
458

meningioma, non-tumor, and pituitary tumor. Figure
4 presents this structured approach.
Figure 4: Architecture of the Proposed CNN Model.
3.4.2 Vision Transformer (ViT)
This study utilizes ViT to classify brain tumors from
medical images.
Instead of processing entire images, ViT divides them
into patches, which are flattened and treated as to-
ken sequences, enabling the model to capture spa-
tial relationships and contextual information effec-
tively. The architecture incorporates multi-head self-
attention mechanisms, residual connections, and layer
normalization to enhance feature extraction and sta-
bilize training. Position embeddings preserve spatial
awareness, while feed-forward networks with ReLU
activations, dense layers, and dropout regularization
ensure generalization and prevent overfitting. The fi-
nal classification is performed through a dense layer.
ViT’s scalability and ability to learn complex patterns
make it a strong complement to CNNs, significantly
improving diagnostic accuracy in brain tumor classi-
fication. Figure 5 provides a detailed overview of the
proposed ViT architecture.
3.4.3 Transfer Learning with Pre-Trained
Models
In this paper, we explore the use of transfer learn-
ing and fine-tuning on pre-trained models (VGG16,
VGG19, and InceptionV3) to develop three en-
hanced architectures: FT-VGG16, FT-VGG19, and
FT-InceptionV3. Initially, these models, pre-trained
on the ImageNet dataset, leverage their learned fea-
ture extraction capabilities, enabling faster conver-
gence and improved performance. Their pre-trained
weights remain frozen during the initial training phase
to preserve the integrity of the learned features. Sub-
sequently, we apply a transfer learning approach by
modifying the architecture: first, the output is flat-
tened before passing through a single dense layer.
This structure is then refined by replacing the dense
layer with a GlobalAveragePooling2D layer, which
reduces the feature map size while retaining impor-
tant information. Additionally, two fully connected
dense layers with ReLU activation and Dropout reg-
ularization (0.5) are incorporated to enhance gener-
alization and mitigate overfitting. Finally, a soft-
max layer classifies images into four tumor cate-
gories: glioma, meningioma, pituitary tumor, and
no tumor. FT-InceptionV3 integrates these improve-
ments within the InceptionV3 architecture, while FT-
VGG16 and FT-VGG19 apply similar modifications
to their respective VGG architectures, ensuring opti-
mized accuracy and robustness. Figures 6, 7 , and
8 present the architectures of FT-InceptionV3, FT-
VGG16, and FT-VGG19, models, respectively.
3.4.4 Hybrid Ensemble Learning
We aim to enhance brain tumor classification perfor-
mance through a hybrid ensemble learning approach,
leveraging the strengths of multiple deep learning
models, such as CNNs, ViT, and pre-trained mod-
els like FT-VGG16 and FT-InceptionV3 for improved
accuracy and robustness. By combining these mod-
els, each excelling in different aspects of image pro-
cessing, our approach aims to capture both local and
global features of brain tumor images. CNNs spe-
cialize in detecting fine-grained spatial patterns, while
ViT excels at modeling long-range dependencies. Ad-
ditionally, FT-VGG16 and FT-InceptionV3 contribute
their expertise in multi-scale feature extraction and
advanced representation power.
This hybrid ensemble strategy mitigates the limi-
tations of individual models, improving classification
accuracy, reducing overfitting, and ultimately provid-
ing a more robust system for accurately classifying
brain tumors such as gliomas, meningiomas, pitu-
itary tumors, and non-tumor cases. Figure 9 presents
the architectures of the proposed hybrid models Ens-
CNN-VIT, Ens-FT-VGG16-FT-InceptionV3 and Ens-
vit-FT-InceptionV3 based on the application of the
ensemble learning.
– Ens-FT-VGG16-FT-InceptionV3 Model
The Ens-FT-VGG16-FT-InceptionV3 model is a hy-
brid ensemble designed to integrate the strengths of
FT-VGG16 and FT-InceptionV3. FT-VGG16 excels
at extracting fine-grained spatial features, while FT-
InceptionV3 captures multiscale patterns through its
inception modules. MRI images are processed sep-
arately through both models, and their extracted fea-
ture vectors are concatenated into a unified represen-
tation. This combined vector is passed through fully
connected layers and classified into four tumor cate-
gories using a softmax layer.
– Ens-VIT-FT-InceptionV3 Model
The Ens-ViT-FT-InceptionV3 model is a hybrid en-
semble architecture that combines the ViT and FT-
InceptionV3 models to enhance brain tumor classifi-
cation accuracy. Specifically, the ViT model captures
Brain Tumor Classification with Hybrid Deep Learning Models from MRI Images
459
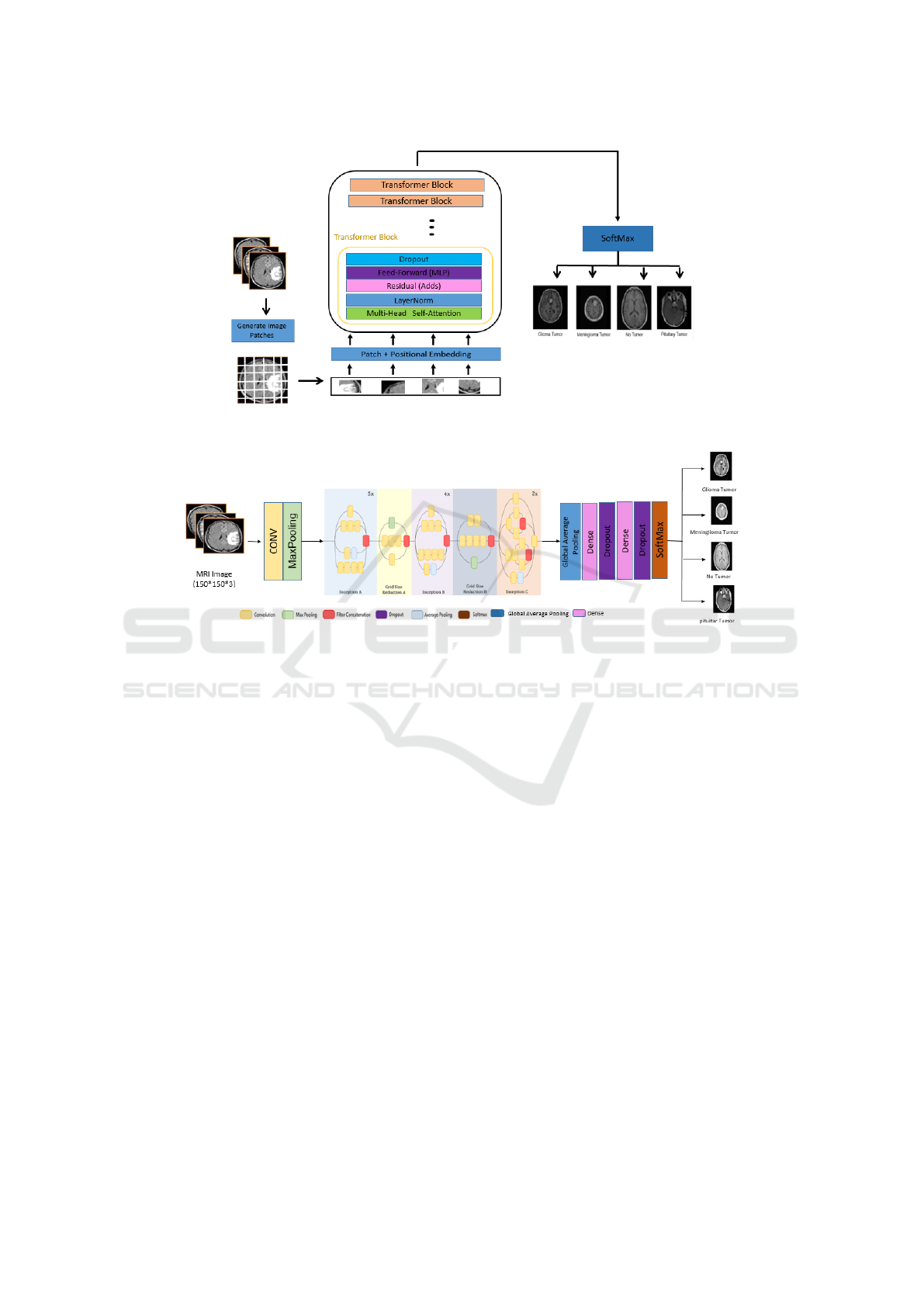
Figure 5: Architecture of the proposed Vit Model.
Figure 6: Architecture of the Proposed Fine-Tuned InceptionV3 Model.
spatial attention features, while InceptionV3 excels at
extracting high-level features. For each input image,
the outputs of both models are obtained—ViT gener-
ates class probabilities, and InceptionV3 provides fea-
ture maps. These outputs are then flattened and con-
catenated into a single feature vector, which is passed
to a dense layer for the final classification. The Adam
optimizer is used for training the ensemble model,
and the training process is carried out over 30 epochs.
This combination of two different models allows the
ensemble to take advantage of the complementary in-
formation provided by both networks, potentially im-
proving classification accuracy.
– Ens-CNN-VIT Model
The Ens-CNN-ViT model combines CNNs and ViTs
to enhance brain tumor classification. CNNs extract
local spatial features like edges and textures, and spa-
tial details through their convolutional and pooling
layers while ViTs excel at capturing global relation-
ships within the data using their self-attention mecha-
nism and process MRI images as patches, capturing
global context and long-range dependencies. MRI
images are processed through both models, generat-
ing feature maps that are then concatenated into a uni-
fied feature vector. This fused representation is passed
through fully connected layers, with a softmax layer
classifying images into four tumor categories. By
combining these two architectures, the model benefits
from both local and global feature extraction capabil-
ities, improving classification accuracy and reducing
the risk of overfitting.
4 EXPERIMENTAL RESULTS
In this section, we present and analyze the results ob-
tained from the hybrid models developed for brain tu-
mor classification using MRI images.
Performance evaluation was carried out using several
standard metrics, including accuracy, recall, F1-score,
and confusion matrices. These results demonstrate
the effectiveness and robustness of the proposed mod-
els.
Table 2 presents the evaluation results for brain
tumor classification using the proposed models, in-
cluding the Proposed-CNN, ViT model, Fine-Tuned
VGGNet16, Fine-Tuned VGGNet19, Fine-Tuned In-
ceptionV3, and the new three ensemble learning hy-
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
460
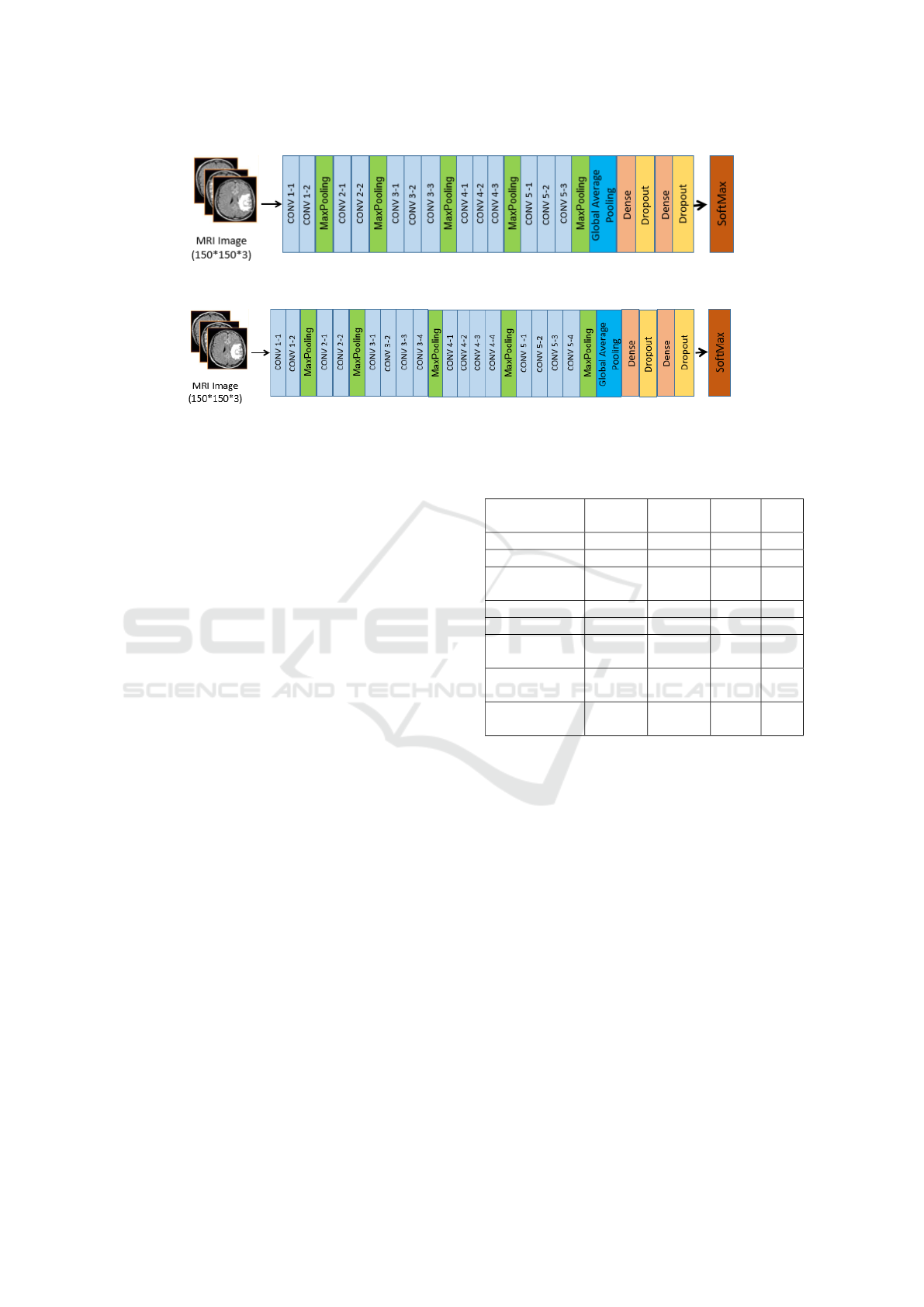
Figure 7: Architecture of the Proposed Fine-Tuned Vgg16 Model.
Figure 8: Architecture of the Proposed Fine-Tuned Vgg19 Model.
brid models, which are Ens-CNN-ViT, Ens-VGG16-
InceptionV3, and Ens-ViT-InceptionV3 models. This
table displays the performance metrics—accuracy,
precision, recall, and F1-score—for each model.
As indicated in the table, the Ens-VGG16-
InceptionV3 hybrid model achieved the highest over-
all performance with an accuracy of 99%, followed by
FT-InceptionV3, CNN, Ens-ViT-InceptionV3, Ens-
CNN-ViT, Vit model, FT-VGG16, and FT-VGG19
with 93%, 92%, 90%, 88%, 83%, 81% and 75%
respectively. The Ens-ViT-InceptionV3 model also
demonstrated a strong performance with an accuracy
of 90%, while the Ens-CNN-ViT model attained an
accuracy of 88%. These results highlight the su-
perior performance of the Ens-VGG16-InceptionV3
model, with a notable improvement of 11 % over
the Ens-CNN-ViT model and a 9% improvement over
the Ens-ViT-InceptionV3 Model. In comparison, the
CNN model achieved an accuracy of 92%, while
the ViT model reached 83%. Among the fine-tuned
pre-trained models, FT-VGG16 and FT-VGG19 per-
formed the least, with accuracies of 81% and 75%,
respectively.
Figure 10 illustrates the confusion matrices for
the three hybrid ensemble learning models: (A)
Ens-ViT-FT-InceptionV3, (B) Ens-FT-VGG16-FT-
InceptionV3, and (C) Ens-CNN-ViT. As an exam-
ple, Figure 10 (B) presents the matrix includes four
classes: suspected glioma, suspected meningioma,
not suspected tumor, and pituitary. The diagonal el-
ements represent the correct model predictions: 826
correct predictions for suspected glioma, 815 correct
predictions for suspected meningioma, 395 correct
predictions for suspected tumor, and 826 correct pre-
dictions for suspected pituitary. The off-diagonal ele-
ments indicate that there are some misclassifications,
but these misclassifications are rare.
Among them, 7 suspected meningiomas were mis-
Table 2: Evaluation of brain tumor classification using the
proposed CNN, ViT model, fine-tuned pre-trained models,
and three ensemble learning hybrid models.
Classifier Accuracy Precision Recall F1-
score
CNN 92% 92% 92% 92%
ViT 83% 85% 83% 84%
FT-
InceptionV3
93% 93% 93% 93%
FT-VGG16 81% 81% 81% 81%
FT-VGG19 75% 77% 75% 76%
Ens-CNN-
ViT
88% 89% 88% 88%
Ens-ViT-
InceptionV3
90% 91% 90% 90%
Ens-VGG16-
InceptionV3
99% 99% 99% 99%
classified as suspected gliomas, and 1 suspected pi-
tuitary tumor was misclassified as suspected glioma.
Overall, the model showed excellent accuracy and
correctly classified most cases with few misclassifi-
cations. This strong performance shows that the inte-
grated model (FT-VGG16-FT-InceptionV3) can dis-
tinguish these four categories very effectively.
Figure 11, shows the training and validation accu-
racy and loss curves for the hybrid ensemble model,
highlighting the superior performance of Ens-FT-
VGG16-FT-InceptionV3, which achieves the highest
validation accuracy and lowest loss, indicating its ro-
bustness.
5 COMPARATIVE EVALUATION
The primary objective of this study is to propose hy-
brid deep learning models to enhance the classifica-
tion of brain tumors from MRI images. These tu-
Brain Tumor Classification with Hybrid Deep Learning Models from MRI Images
461
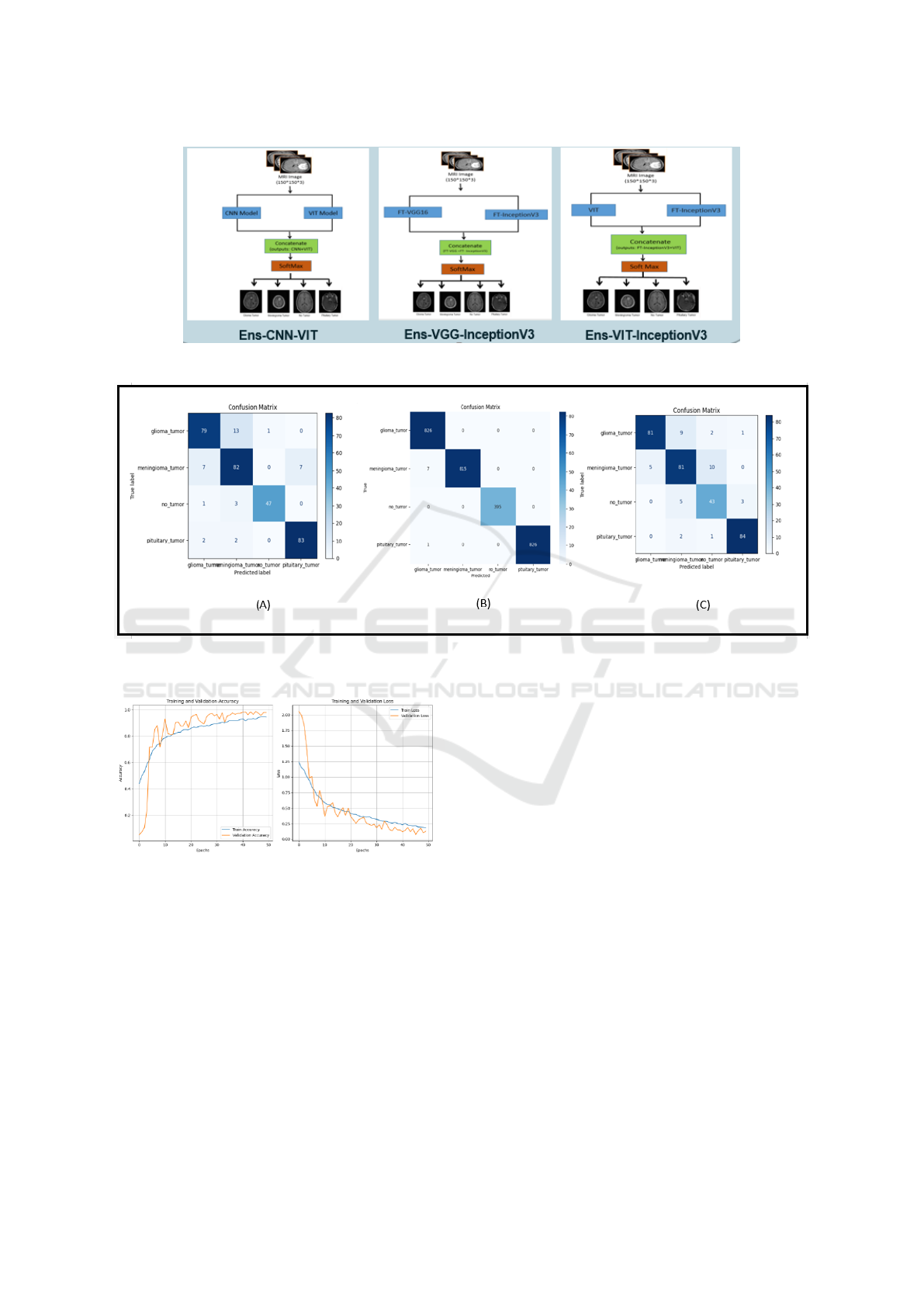
Figure 9: Architectures of the proposed hybrid models based on Ensemble learning.
Figure 10: Confusion Matrices obtained by (A) Ens-ViT-FT-InceptionV3 (B) Ens-FT-VGG16-FT-InceptionV3 (C) Ens-CNN-
ViT.
Figure 11: Training accuracy and loss curves for the Ens-
FT-VGG16-FT-InceptionV3 ensemble models.
mors—including glioma, meningioma, pituitary tu-
mor, and healthy brains—pose significant diagnos-
tic challenges due to their often similar characteris-
tics. Our results, summarized in Table 3, demon-
strate that the proposed ensemble models, particu-
larly Ens-VGG16-InceptionV3, achieved remarkable
performance, attaining 99% accuracy and a 99% F1-
score. These findings underscore the effectiveness of
leveraging multiple architectures to improve classifi-
cation performance and robustness.
Table 3 provides a comparative analysis of the
performance of our proposed models against previ-
ous works using the same Brain Tumor Classifica-
tion (MRI) dataset. Earlier studies, such as G
´
omez-
Guzm
´
an et al., which employed a combination of
generic CNN and InceptionV3 model, achieved an ac-
curacy of 97.12% and an F1-score of 96.59%. Sim-
ilarly, Zafer Khaliki et al., and Dhakshnamurthy et
al., reported comparable results, with both achieving
98% accuracy using VGG16 and VGG16-ResNet50
combination models, respectively. Mahmud et al., re-
ported a lower accuracy of 93.30% and an F1-score
of 91% using a standalone CNN. In contrast, our en-
semble models significantly outperform these earlier
methods, demonstrating the superiority of hybrid ap-
proaches in leveraging complementary features for
more precise brain tumor classification.
The superior performance of ensemble learning
hybrid lies in their ability to combine the feature ex-
traction strengths of different architectures. For ex-
ample, CNNs excel at detecting local features but may
struggle with global relationships, which are effec-
tively captured by Vision Transformers. By integrat-
ing these architectures, models like Ens-FT-VGG16-
FT-InceptionV3 and Ens-ViT-FT-InceptionV3 capi-
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
462
Table 3: Comparative analysis of proposed work with previous works.
Authors Models Best Model Accuracy (%) F1-
score
Proposed approaches Ens-FT-VGG16-FT-
InceptionV3
Ens-ViT-FT-InceptionV3
Ens-CNN-ViT
Ens-FT-VGG16-FT-
InceptionV3
99% 99%
G
´
omez-Guzm
´
an et
al.,(G
´
omez-Guzm
´
an
et al., 2023)
Generic CNN and six TL mod-
els
InceptionV3 97.12% 96.59%
Khaliki and
Basarslan(Khaliki
and Bas¸arslan, 2024)
EfficientNetB4
InceptionV3
VGG19
VGG16
CNN
VGG16 98% 97%
Dhakshnamurthy et
al.,(Dhakshnamurthy
et al., 2024)
AlexNet
VGG16
ResNet50
VGG16–ResNet50
VGG16– ResNet50 98% 98%
Mahmud et
al.,(Mahmud et al.,
2023)
VGG16
ResNet-50
CNN
nceptionV3
CNN 93.30% 91%
talize on both local and global feature extraction,
achieving balanced and superior performance.
6 CONCLUSION AND FUTURE
WORK
This study proposed a hybrid deep learning approach
for brain tumor classification from MRI images, inte-
grating CNNs, ViTs, and fine-tuned pre-trained mod-
els (FT-InceptionV3, FT-VGG16, and FT-VGG19).
By leveraging ensemble learning, we combined the
strengths of different architectures, leading to sig-
nificant improvements in classification accuracy, ro-
bustness, and generalization. The proposed hybrid
models—Ens-FT-VGG16-FT-InceptionV3, Ens-ViT-
FT-InceptionV3, and Ens-CNN-ViT—demonstrated
superior performance compared to standalone mod-
els and previous studies. Notably, Ens-FT-VGG16-
FT-InceptionV3 achieved the highest accuracy and
F1-score of 99%, underscoring the effectiveness of
hybrid architectures in medical imaging applications.
These findings highlight the potential of AI-driven so-
lutions to enhance early detection and classification of
brain tumors, minimizing diagnostic delays and im-
proving patient outcomes. Future research will focus
on enhancing model interpretability through Explain-
able AI (XAI), expanding datasets to improve gener-
alization across diverse populations, and optimizing
computational efficiency for real-time clinical deploy-
ment. This study reinforces the transformative impact
of deep learning in medical diagnostics, paving the
way for more precise, scalable, and reliable brain tu-
mor classification systems.
ACKNOWLEDGMENT
The research leading to these results has received
funding from the Ministry of Higher Education and
Scientific Research of Tunisia under grant agreement
number LR11ES48.
REFERENCES
Arabahmadi, M., Farahbakhsh, R., and Rezazadeh, J.
(2022). Deep learning for smart healthcare—a survey
on brain tumor detection from medical imaging.
Asiri, A. A., Shaf, A., Ali, T., Shakeel, U., Irfan, M.,
Mehdar, K. M., Halawani, H. T., Alghamdi, A. H.,
Alshamrani, A. F. A., and Alqhtani, S. M. (2023). Ex-
ploring the power of deep learning: fine-tuned vision
transformer for accurate and efficient brain tumor de-
tection in mri scans.
Balamurugan, T. and Gnanamanoharan, E. (2023). Brain tu-
mor segmentation and classification using hybrid deep
cnn with lunetclassifier.
Bharadiya, J. (2023). A comprehensive survey of deep
learning techniques natural language processing.
Bhuvaji, S. (2024). Brain tumor classification mri dataset.
https://www.kaggle.com/datasets/sartajbhuvaji/
brain-tumor-classification-mri. Accessed: 2024-05-
22.
Dhakshnamurthy, V. K., Govindan, M., Sreerangan, K., Na-
garajan, M. D., and Thomas, A. (2024). Brain tu-
Brain Tumor Classification with Hybrid Deep Learning Models from MRI Images
463

mor detection and classification using transfer learn-
ing models.
Farmanfarma, K. K., Mohammadian, M., Shahabinia, Z.,
Hassanipour, S., and Salehiniya, H. (2019). Brain can-
cer in the world: an epidemiological review.
Fki, Z., Ammar, B., and Ayed, M. B. (2024). Towards au-
tomated optimization of residual convolutional neural
networks for electrocardiogram classification. Cogni-
tive Computation, 16(3):1334–1344.
G
´
omez-Guzm
´
an, M. A., Jim
´
enez-Berista
´
ın, L., Garc
´
ıa-
Guerrero, E. E., L
´
opez-Bonilla, O. R., Tamayo-Perez,
U. J., Esqueda-Elizondo, J. J., Palomino-Vizcaino, K.,
and Inzunza-Gonz
´
alez, E. (2023). Classifying brain
tumors on magnetic resonance imaging by using con-
volutional neural networks.
Iqbal, S., N. Qureshi, A., Li, J., and Mahmood, T. (2023).
On the analyses of medical images using traditional
machine learning techniques and convolutional neural
networks.
Khaliki, M. Z. and Bas¸arslan, M. S. (2024). Brain tumor
detection from images and comparison with transfer
learning methods and 3-layer cnn.
Lee, S. H., Shin, H. J., and Moon, W. K. (2020). Diffusion-
weighted magnetic resonance imaging of the breast:
standardization of image acquisition and interpreta-
tion.
Mahmud, M. I., Mamun, M., and Abdelgawad, A. (2023).
A deep analysis of brain tumor detection from mr im-
ages using deep learning networks.
Mokni, R. and Haoues, M. (2022). Cadnet157 model:
fine-tuned resnet152 model for breast cancer diagno-
sis from mammography images. Neural Computing
and Applications, 34(24):22023–22046.
Pham, P., Nguyen, L. T., Nguyen, N. T., Kozma, R., and
Vo, B. (2023). A hierarchical fused fuzzy deep neural
network with heterogeneous network embedding for
recommendation.
Rivkin, M. and Kanoff, R. B. (2013). Metastatic brain tu-
mors: current therapeutic options and historical per-
spective.
Sanvito, F., Castellano, A., and Falini, A. (2021). Ad-
vancements in neuroimaging to unravel biological and
molecular features of brain tumors.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
464
