
Arbitrary Shaped Clustering Validation on the Test Bench
Georg Stefan Schlake
a
and Christian Beecks
b
FernUniversit
¨
at in Hagen, Chair of Data Science, 58084 Hagen, Germany
Keywords:
Machine Learning, Unsupervised Learning, Clustering, Clustering Validation, Arbitrary Shaped Clusters.
Abstract:
Clustering is a highly important as well as highly subjective task in the field of data analytics. Selecting a
suitable clustering method and a good clustering result is all but trivial and needs insight into not only the
field of clustering, but also the application scenario, in which the clustering is utilized. Evaluating a single
clustering is hard, especially as there exists a wide variety of indices to evaluate the quality of a clustering,
both for simple convex and for arbitrary shaped clusterings. In this paper, we investigate the ability of 11
state-of-the-art Clustering Validation Indices (CVI) to evaluate arbitrary shaped clusterings. To this end, we
provide a survey of the intuitive workings of these CVI and an extensive benchmark on newly generated
datasets. Furthermore, we evaluate both the Euclidean distance and the density-based DC-distance to quantify
the quality of arbitrary shaped clusters. We use the generation of novel datasets to evaluate the influence of a
number of metafeatures on the CVI.
1 INTRODUCTION
Clustering is a well known field in machine learning
with many application areas. The objective of cluster-
ing is to partition a collection of objects into multiple
groups of somehow similar objects. However, as the
notion of similarity differs across application settings,
there exists a wide variety of algorithms complying
with these different notions of similarity.
One way to view clusters is by defining a cen-
troid of each cluster and assigning each object to the
most similar centroid, which leads to convex clus-
ters and is a very popular choice with algorithms like
kMeans (McQueen, 1967) or kCenter (Lim et al.,
2005). Another view of clusters is to view them as
an area with a high density of objects, where this area
can have a varying shape. This so-called density-
connectivity view considers objects which are con-
nected over dense areas as part of the same cluster.
This view lead to algorithms like DBSCAN (Ester
et al., 1996), Optics (Ankerst et al., 1999) and HDB-
SCAN (Campello et al., 2015). Multiple other views
of clusterings exist, including hierarchical (Ward Jr,
1963) or fuzzy (Ruspini et al., 2019) clusterings,
which will not be part of this paper. As these dif-
ferent notions are hard to compare, it is challenging
(i) to find a singular “best” clustering (von Luxburg
a
https://orcid.org/0009-0008-5714-1804
b
https://orcid.org/0009-0000-9028-629X
et al., 2012) and (ii) to evaluate the “goodness” of a
clustering, even under clear assumptions.
A plethora of Clustering Validation Indices (CVIs)
has been engineered to value the goodness of a
density-connectivity based clustering. However,
many of these works lack in a comparison with other
CVIs following the same notion (Bay
´
a and Granitto,
2013; Liu et al., 2013; Moulavi et al., 2014; Hu and
Zhong, 2019; Rojas Thomas and Santos Pe
˜
nas, 2021)
or with a very limited selection (Xie et al., 2020;
Guan and Loew, 2022; S¸enol, 2022). Even in re-
cent works, which aim at a comparative evaluation of
CVIs (Schlake and Beecks, 2024b), the lack of con-
trolled high-dimensional datasets makes it difficult to
find meaningful qualitative findings on the existing
CVIs. These findings might facilitate to identify weak
spots in state-of-the-art CVIs, enabling the design and
engineering of customized CVIs and also the selec-
tion of a suitable CVI for a given dataset, yielding to
potentially better clustering results. By making use of
a novel dataset generator like the recently proposed
Densired (Jahn et al., 2024), it is possible to specifi-
cally design datasets with different metafeatures and
to evaluate the impact of these datasets on the differ-
ent CVIs. In this paper, we will thus investigate 11
different CVIs, 8 of which are specifically designed
for density based clusterings, and elucidate how these
CVIs react to a change in different metafeatures like
the number of clusters, dimensions, the ratio of out-
Schlake, G. S., Beecks and C.
Arbitrary Shaped Clustering Validation on the Test Bench.
DOI: 10.5220/0013495500003967
In Proceedings of the 14th International Conference on Data Science, Technology and Applications (DATA 2025), pages 363-373
ISBN: 978-989-758-758-0; ISSN: 2184-285X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
363

Figure 1: Three different datasets, which can be clearly split
to two clusters, which are however not findable using con-
vex algorithms.
liers or overlaps between clusters. To this end, we fo-
cus on simple interpretability and comparability of the
individual CVIs and abstract from many implementa-
tion details so as to provide a guide for data scientists
and practitioners alike.
2 RELATED WORK
Clustering is a very prominent field of unsupervised
learning. A dataset of multiple objects is split into
different groups, which contain similar objects. Both
the notion of similarity and the idea, how to split the
dataset are highly subjective and depend on the de-
sired clustering and the use case (von Luxburg et al.,
2012). For this reason, apart from a wide variety of
distance or similarity functions, there exists a plethora
of clustering algorithms. These clustering algorithms
are designed to find vastly different clusterings. If we
focus just on the shapes of the resulting clusterings
(and ignore other areas of clustering like hierarchi-
cal structures (Ward Jr, 1963), fuzzy clusterings (Rus-
pini et al., 2019) or different subspaces (Parsons et al.,
2004)), we can see two different groups of clustering
algorithms.
The first group consists of algorithms creating
convex datasets, mostly by defining cluster medoids
and assigning objects to the cluster of the clos-
est medoid. This group contains important and
widely used algorithms like kMeans (McQueen,
1967), kMedoids (Kaufman and Rousseeuw, 1990)
or kCenter (Lim et al., 2005). These algorithms are
mostly fast and generate easily interpretable cluster-
ings, where each cluster is a convex region and can be
seen as a voronoi cell in the space of the used distance
metric. However, these algorithms struggle in finding
the correct clustering, if the clusters are not arranged
in convex forms like the datasets depicted in Figure 1.
This is the reason, a second group of clus-
tering algorithms exists, the density-connectivity
based algorithms like DBSCAN (Ester et al.,
1996), OPTICS (Ankerst et al., 1999) and HDB-
SCAN (Campello et al., 2015). These algorithms have
no fixed idea of the shape of the dataset, but are based
on a constant minimal density in the clusters. This
means, that every object in a cluster has at least a min-
imum amount of other elements of the same cluster in
the proximity. This way, these algorithms can gener-
ate clusters based on the dense regions and adapt to
their shape.
As different clustering algorithms and different
notions of clusterings exist, it is hard to compare the
“goodness” of two clustering solutions. However,
evaluating clusterings can be important in scenar-
ios like Automated Clustering (Schlake and Beecks,
2023; Schlake and Beecks, 2024a; Schlake et al.,
2024) or in different pipelines (von Luxburg et al.,
2012). For this reason, a number of Clustering Val-
idation Indices (CVIs) exists. Like the clustering al-
gorithms, these CVI have different notions of a good
clustering, so there cannot be one objectively best
CVI for any situation. While there exists a number of
surveys, these are either dated (Halkidi et al., 2001;
Deborah et al., 2010), done on a small set of datasets
and without in depth evaluation (Hassan et al., 2024)
or lack qualitative insight into different metafeatures
of datasets (Schlake and Beecks, 2024b).
This lack of qualitative studies might be connected
to the problem of generating non-convex high dimen-
sional datasets. While high-dimensional dataset gen-
erators are known and easily available (e.g. multiple
in scikit-learn (Pedregosa et al., 2011)), these gen-
erating arbitrary shaped clusters is seldom. While a
few generators can generate such datasets (Gan and
Tao, 2015; Li and Zhou, 2023), none of them can
guarantee that the datasets are not linearly separable.
For this reason, we make use of the dataset generator
Densired (Jahn et al., 2024), which complies with the
aforementioned properties.
Earlier in this section, we reckoned the similar-
ity or distance between objects to be of importance
when generating a clustering solution. While the
distance between objects like the points in Figure 1
can easily be represented by the Euclidean distance
or other Minkowski distances, more complex objects
required more intricate distances like the Signature
Quadratic Form Distances (Beecks et al., 2010) for
signatures or Dynamic Time Warping (Berndt and
Clifford, 1994) for time series. While these distances
enable the (dis)similarity quantification of complex
objects, there also exist distances to change the over-
all notion of a distance, like the DC-distance (Beer
et al., 2023). This distance is designed to incorpo-
rate the density-connectivity concept in the distance
computation, enabling different algorithms or CVI to
work with the notion of density-connectivity, even if
this is not part of their original design.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
364

Figure 2: Exemplary Silhouette computation of the light
yellow element. The (blue) distances between the light yel-
low element and the other yellow elements are used to com-
pute the Compactness, while the (red) distances to the ele-
ments of the green cluster are used for the Separation. The
elements of the purple cluster are not used for this Silhou-
ette.
3 METHODS
As we aim for an intuitive overview, we will describe
and (mostly) illustrate the intuition for the different
CVI. For mathematical details, we refer the reader to
the original papers or to our previous paper (Schlake
and Beecks, 2024b) for a survey of all these CVI
with consistent mathematical formulations and infor-
mation on the complexity of the approaches.
3.1 Reference CVI
We do not only investigate CVI for arbitrary shaped
clusters, but also a few “classical” CVIs as baseline
models. These are not designed for arbitrary shaped
clusters and will likely deliver worse results than spe-
cialised measures.
3.1.1 Silhouette Coefficient
A well known measure for clustering validation is the
Silhouette Coefficient or Silhouette Width Criterion
(SWC) (Rousseeuw, 1987). In this criterion, a Silhou-
ette is computed for every object. These silhouettes
are averaged to get a result for the complete dataset.
To get the silhouette of an object, the average distance
of said object to other objects of the same cluster is
used as Compactness, while the Separation is the av-
erage distance to objects in the next different cluster.
The difference between these values, normalized by
the bigger of both, is used as Silhouette. The values
for the SWC can range between -1 and 1, where a
high value means a good clustering, as the Separation
is much higher than the Compactness. An example of
the computation of the SWC can be seen in Figure 2.
3.1.2 VRC
The Variance Ratio Criterion or Calinski Harabasz
Index (Cali
´
nski and Harabasz, 1974) is also a well
known baseline index, where the dispersion between
Figure 3: Exemplary computation of the D bw for S Dbw.
The hatched yellow and green elements represent the
medoids of their corresponding clusters, while the red
hatched elements marks the mid between these points. The
circles around these elements have the radius of stdev.
groups and within groups is measured. The VRC can-
not be adapted to use any distance functions, so it is
only usable on a limited set of problems.
3.1.3 S Dbw
The last baseline CVI investigated in this paper is
Scattering-Density between (S Dbw) (Halkidi and
Vazirgiannis, 2001). In this approach, the density
between clusters (D bw) and the Scattering for the
whole clustering are added to generate a value, where
both values should be small. The Scattering is mea-
sured as the average standard deviation of all clus-
ters divided by the standard deviation of the complete
dataset, whereas the density between clusters is mea-
sured by dividing the density of objects in the mid-
point between two clusters by the higher density of
both clusters midpoints. The density of objects is in
this CVI the number of objects closer than stdev, the
average standard deviation of all clusters. An exam-
ple for this can be seen in Figure 3.
3.2 MST Based
Multiple CVI are generated using a Minimum Span-
ning Tree (MST) of the data. This MST is a graph
connecting all objects in the dataset while minimiz-
ing the total weight of the edges, where the weight of
an edge between two objects corresponds to their sim-
ilarity. An MST should - in case of a good clustering -
connect elements of the same cluster and should have
only very limited edges between objects of different
clusters (in the optimal case the number of clusters,
as all clusters need to be connected, but no edge be-
tween two clusters should be shorter than the shortest
edge in a cluster). An MST automatically adapts to
the shape of the dataset and hence is apt to help mea-
suring arbitrary shaped clusterings.
3.2.1 DBCV
The density-connectivity based method of this pa-
per is the Density Based Clustering Validation
(DBCV) (Moulavi et al., 2014). The first important
Arbitrary Shaped Clustering Validation on the Test Bench
365

Figure 4: An example for the MRD between the light yel-
low and the light purple object. The circles around the
object represent the core distance of each object based on
the (green) similarity to objects in the same cluster. As the
(red) distance between both objects is bigger, this distance
is dominating the MRD.
Figure 5: An example of the computation of the DBCV. The
Sparsity per cluster is depicted as the green edge in each
cluster. The light gray edges are not part of this computa-
tion, as they connect to border objects. The red edges rep-
resent the Separation between two clusters as the minimum
distance between objects of these. The lighter coloured ob-
jects are excluded as those are border objects.
step of this algorithm is to adjust the distance between
two objects by first assigning a core distance to each
object based on the density of other objects of the
same cluster and secondly replacing the “normal” dis-
tance between two objects by the maximum of each
objects core distance and the “normal” distance be-
tween those objects as Mutual Reachability Distance
(MRD). As the core distance will be high for an object
in a sparse region, this will raise the distance between
objects in sparse regions, whereas it will have little
effect in dense regions or for objects far apart. An
example of this computation can be seen in Figure 4.
Using this MRD, an MST is build in each cluster,
which is then pruned of its border object, which are
only connected by a single edge. Now, the DBCV for
each cluster is based on the Sparsity, the maximum
edge of these MSTs and the Separation, which is the
minimum distance of a non-border object to any non-
border object of a different cluster. An example of this
can be seen in Figure 5. To combine these, the Spar-
sity is deducted from the Separation and the result is
normalized by the bigger of these numbers. The result
for the complete clustering is the weighted average of
each cluster.
3.2.2 IC-av
Intracluster average gap (IC-av) (Bay
´
a and Granitto,
2013) is an approach penalizing clusterings with long
distances on the shortest path on an MST between
Figure 6: An example for the calculation for the path dis-
tance for IC-av of the light yellow and the dark yellow point.
The green and red edges comprise the path between those
objects. The red edge is used as distance between the ob-
jects, as it is the longest edge on the path. The weight of the
green edges is shorter, so those are not taken into account
for this distance.
two objects in the same cluster. The distance between
two objects is measured as the longest distance on the
shortest path between them. These distances are aver-
aged to get the IC-av.
3.2.3 DCVI
Figure 7: An example of the computation of the DCVI. The
Compactness resembles the green edges, whilst the Separa-
tion resembles the red edges.
Density-core-based clustering validation index
(DCVI) (Xie et al., 2020) is a similar measure to the
DBCV, but without using a sophisticated reachability
distance. The Separation of a cluster is measured as
the minimal distance of an object in the cluster to an
element in another cluster, whilst the Compactness
is measured as the maximum edge weight in the
MST of a cluster. The Compactness is divided by
the Separation to gain each clusters value, which is
averaged over all clusters.
3.2.4 CVDD
Another Approach is the Cluster Validity index
based on Density-involved Distance (CVDD) (Hu and
Zhong, 2019). Here, a density is generated based on
the k-nearest Neighbors of an object and a density-
connectivity distance is used. As this approach uses
a wide variety of correcting factors deduced from the
dataset, it is not possible for us to give more than the
crudest intuition of this method in our given space, so
we refer to the original paper for more information.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
366

Figure 8: The Separation of the light yellow objects re-
sembles the share of other clusters objects in its k nearest
Neighbors, whilst for the compactness, every distances two
objects in the same cluster (blue) are computed.
Figure 9: The local density of the light yellow objects for
the CDR is its closest distance to another object of the same
cluster, depicted by the green edge.
3.3 Other Methods
We use this umbrella subsection, to introduce a few
other, unique CVI, which do not use MSTs to approx-
imate the shape of the data.
3.3.1 CVNN
The Clustering validation index based on nearest
neighbours (CVNN) (Liu et al., 2013) uses Near-
est Neighbors for its notion of Separation, where the
maximum Separation of a cluster is the average num-
ber of other clusters elements in the k-Nearest Neigh-
bors of the clusters objects. The maximum Separation
of any cluster is used as Separation for the clustering.
The Compactness is measured as the average pairwise
distance between two objects of the same cluster. An
example for an object with k := 5 can be seen in Fig-
ure 8. In the end, Separation and Compactness of the
clustering are added to retrieve the value. A slight
variation of the weighting is found in (Halkidi et al.,
2015), which we also investigate as CVNN hal.
3.3.2 CDR
Another CVI is the Contiguous Density Region
(CDR) index (Rojas Thomas and Santos Pe
˜
nas,
2021). In this CVI, the local density is measured by
the distance to the next object in the same cluster. The
notion of the CDR is, that this local density should be
very close the average density of the same cluster.
3.3.3 VIASCKDE
The Validity Index for Arbitrary-Shaped Clusters
Based on the Kernel Density Estimation (VI-
Figure 10: Exemplary VIASCKDE computation of the light
yellow element. The (blue) distances between the light yel-
low element and the other yellow elements are used to com-
pute the Compactness, while the (red) distances to the ele-
ments of the green cluster are used for the Separation. The
elements of the purple cluster are not used for this Silhou-
ette. The purple lines represent the isolines of the KDE, so
that elements inside those lines will have a higher weight.
Frequency
Distance
Figure 11: An illustration of the DSI computation. The
(green) distances of the light yellow object to the other yel-
low objects are part of the inner cluster distances of the yel-
low cluster. The (red) distances to objects of the other clus-
ters are part of the between cluster distances of the yellow
cluster. The difference between these histograms (seen on
the right) is used to calculate the DSI for the cluster.
ASCKDE) (S¸ enol, 2022) is very similar to the SWC
(see subsubsection 3.1.1). However, each object is
valued by the weight of a Kernel Density Estimation
of the dataset, meaning objects in more populated re-
gions will have a higher weight.
3.3.4 DSI
In the Distance-based Separability Index (DSI) (Guan
and Loew, 2022), the distances inside one cluster are
compared to the distances to objects of other clusters.
Instead of directly using the distances in the computa-
tions, two sets of distances are generated, comprising
the inner-cluster and the between-cluster distances of
one cluster. Then the difference between those his-
tograms is computed to gain a value for this cluster.
These values get averaged to retrieve a value for the
complete cluster.
3.4 Used Distances
The notion of similarity used to compute the dis-
tance between two objects plays an important role
in the evaluation of a clustering. Almost all pre-
sented methods are able to cope with different dis-
tance metrics without any problem. The exception
are the VRC, which directly works on the vector rep-
resentation of the objects, the S Dbw, which needs a
Arbitrary Shaped Clustering Validation on the Test Bench
367

Figure 12: An illustration of the DC-distance computation
between the blue and the green object. The edges depict the
MST of the dataset. The gray edges are ignored, because
they are not part of the path between both objects. The black
edges are shorter than the red edge, which resembles the
distance between both objects.
midpoint of clusters and between two objects and the
VIASCKDE, which needs a KDE based on the dis-
tance between objects. We will investigate all other
methods not only using the Euclidean distance, but
also the DC-distance (Beer et al., 2023). The DC-
distance measures the distance between two objects
in a density-connectivity based fashion, so that us-
ing this distance function even classical CVI like the
SWC should be able to find arbitrary shaped cluster-
ings. Similar to the DBCV, every object is assigned
a core distance, which is the distance to its µ nearest
neighbor. It also uses a mutual reachability distance,
where the MRD between two objects is the maximum
of both objects core distance and their Euclidean dis-
tance. To find the DC-distance between two objects,
an MST is build on the dataset using the MRDs. The
distance is now the longest edge on the path between
two points on this MST.
4 EXPERIMENTS
In this section, we will describe our experiments.
1
We
will start with a description of the used datasets (sub-
section 4.1), before we will explain which algorithms
were used to generate clusterings (subsection 4.2). To
conclude this section, we will describe the setup used
to generate our results (subsection 4.3).
4.1 Datasets
In order to generate valuable and qualitative results,
we generate datasets using the Densired dataset gen-
erator (Jahn et al., 2024). This generator is capable of
generating clusterings of arbitrary shapes, which are
guaranteed to be density-connectivity separable and
allows for a variety of parameters to be tuned. In addi-
tion to the parameters discussed in the following para-
graphs, we set the parameters safety to False, which
allows noise points to be generated next to or inside
a cluster. For each parameterization, we created three
1
Our implementation of the CVI can be found under
https://github.com/g-schlake/ASCVI
datasets to prevent an influence of chance and to see
the variance of the CVI in similar datasets. The actual
values used for each test can be seen in Table 1.
Dimensionality. The first metafeature we tuned was
the dimensionality. It is wide known, that real datasets
can contain many dimensions. However, many ar-
bitrary shaped, synthetical datasets are only two- or
three-dimensional, so many evaluations of methods or
even surveys like (Schlake and Beecks, 2024b) might
be misled, if CVI perform especially well in the lower
dimensional space. We used 5 dimensions as standard
value in order to not prevent former mistakes of only
investigating a low dimensional space, but also to not
investigate too high dimensional spaces.
Number of Clusters. The number of clusters is an-
other interesting metafeature of datasets. While some
algorithms take information of all clusters into ac-
count, others only compare neighboring clusters. We
change the number of clusters in the clustering to see,
whether this leads to CVI scaling with the number of
clusters. Our default will be 10 clusters.
Overlap Factor. Overlaps between clusters have
been shown to be a problem for a number of density-
connectivity based CVI (Schlake and Beecks, 2024b).
In the generation of datasets, the separability between
two clusters is guaranteed based on the Overlap fac-
tor. By lowering this value below 1, overlaps between
two clusters can happen, which can lead to problems
for some CVI. Our standard value will be 1.1, to en-
sure no overlaps for our standard tests.
Number of Connections. Another way to generate
overlapping clusters is the existence of lower density
bridges between two clusters. With this parameter,
we can control, how many low linkage bridges exist
between different clusterings. As standard, we will
have no bridges between two clusters.
Noise Ration. Another very important factor is the
ability to handle noise. While noise is occurring in
almost all real world scenarios, many CVI are not able
to handle it properly. For this reason, our standard
noise ratio will be 0.
Number of Objects. Our last tests will be about the
number of objects. As datasets tend to vary in size,
we will use datasets in varying sizes. Our standard
datasets will have the size of 5.000.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
368

Table 1: The different metafeatures, their respective parameters and the investigated values. The default value is printed in
bold.
Metafeature Parameter Values
Dimensionality dim 2, 3, 5, 10, 20
Number of Clusters clunum 2, 3, 5, 10, 20
Overlap Factor min dist 0.5, 0.8, 0.9, 1, 1.1, 1.2, 1.3
Number of Connections connections 0, 1, 4, 8, 10
Noise Ration ratio noise 0, 0.05, 0.1, 0.2
Number of Objects data num 100, 500, 1.000, 5.000, 10.000
4.2 Used Algorithms
To generate a number of clusterings, we use three
different clustering algorithms, out of which one is
partition-based and two are density-based.
The kMeans (McQueen, 1967) algorithm is a very
wide known algorithm to assign clusters based on
centroids. This algorithm is not capable of finding
arbitrary shaped clusters, but it is important to also in-
clude a partition based algorithm, as otherwise a lack
of partition based results might lead to CVI choosing
density based results, because they lack higher valued
partition based alternatives. For the k Parameter, we
used the values ranging between
clunum
3
and 3· clunum
and the implementation from (Pedregosa et al., 2011),
so we had (mostly) 27 different values.
DBSCAN (Ester et al., 1996) is a widely known
algorithm for density-connectivity based clustering,
which is still in wide use today (Schubert et al., 2017).
In this algorithm, objects with enough other objects in
close proximity are regarded as core objects of a clus-
ter and objects, which are close to these are part of the
same cluster. We used the implementation from (Pe-
dregosa et al., 2011), where we set MinPts to the even
values in [4,20] and eps to 100 equidistant values
in the distances between the minimum and the max-
imum distance between two objects, resulting in the-
oretically up to 900 clusterings. However, as most
of these are identical due to the very minor changes,
only the differing ones (maximum 102 per Dataset)
are considered.
An updated approach is HDBSCAN (Campello
et al., 2015), which uses hierarchical properties to
make a decision for a data scientist easier. We used
the implementation in (McInnes et al., 2017), where
we only tuned the MinPts parameter, which we set to
the even values in [4,20], resulting in 8 clusterings per
dataset.
4.3 Experimental Setup
In order to evaluate our clusterings, we test how well
they select a clustering resembling the ground truth
of the dataset. Even though the clustering Problem
in general is ambiguous (von Luxburg et al., 2012),
we can assume that the ground truth of our generated
datasets is what we are looking for, as it was generated
and evaluated with the same notion of similarity and
“good” clusterings. For every test, we have generated
3 datasets for each test value, which are then clus-
tered using our three algorithms using the described
parameters. Following this, we select the best cluster
on each dataset for each CVI based on the value of the
CVI. As not every CVI can process noise objects, we
remove objects clustered as noise from the clustering
and multiply the result of the clustering with a penalty
term based on the share of noise in the dataset like de-
scribed in (Schlake and Beecks, 2024b). Where pos-
sible, we also evaluated the clusterings using the DC-
distance as distance function. As we have 3 datasets
per test and value, we get 3 results per test, value
and CVI, out of which we will mostly focus on the
median. We will evaluate the quality of each clus-
tering by measuring the Adjusted Mutual Information
(AMI) (Nguyen et al., 2009) with the ground truth.
As the AMI has no special considerations for noise,
we will assign every noise object to its own, singleton
cluster. This will prevent the AMI from confusing a
cluster in one clustering with similar noise objects in
the other clustering.
5 RESULTS
In the following section, we will present the results of
our 6 tests. Every subsection will show the results of
both used distances. We will start with the dimension-
ality (subsection 5.1) and number of clusters (subsec-
tion 5.2), before we will have a look at the overlap
factor (subsection 5.3) and the number of connections
(subsection 5.4). We will finish with the noise ratio
(subsection 5.5) and the number of objects (subsec-
tion 5.6).
5.1 Dimensionality
When looking at the results at different dimensional-
ities, we see widely varying results in many datasets
Arbitrary Shaped Clustering Validation on the Test Bench
369
2 3 5 10 20
Number of dimensions
0.0
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
2 3 5 10 20
Number of dimensions
0.0
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 13: AMI per CVI using datasets of different dimensionalities. The thick line represents the median value on three
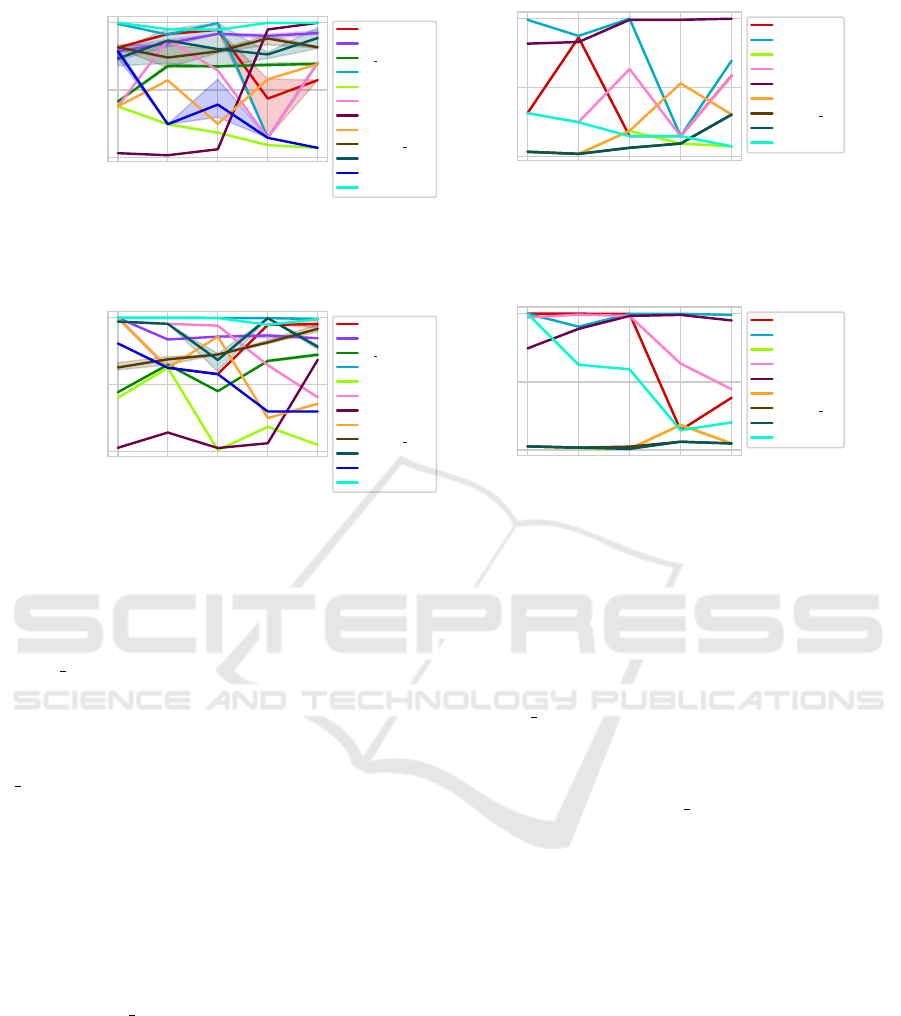
datasets, while the shaded area is based on maximum and minimum value.
2 3 5 10 20
Number of clusters
0.0
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
2 3 5 10 20
Number of clusters
0.0
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 14: AMI per CVI using datasets with a different number of clusters. The thick line represents the median value on
three datasets, while the shaded area is based on maximum and minimum value.
(see Figure 13). When focusing on the Euclidean dis-
tance (Figure 13a), it can be seen that DSI, VRC,
CVNN hal and CDR seem to find good clusterings
using all dimensionalities. IC-av and VIASCKDE
struggle to evaluate the clusterings correctly in higher
dimensional spaces. CVNN and DCVI have high
variations between the different dimensionalities. The
S Dbw struggles in the two-dimensional case, but is
constant in all the other dimensionalities. The SWC
and the DBCV seem to deliver good results, but both
have a negative outlier at 10 dimensions. It can be
seen, that VIASCKDE, DCVI, SWC and CDR have
different results for the different datasets and are quite
sensitive to minimal changes.
When looking at the results using the DC-distance
(Figure 13b), it can be seen that the CVDD delivers
good results using all dimensionalities, whilst IC-av,
CVNN, CVNN hal, DSI and have struggles to find
good clusterings. SWC, DBCV and DCVI have vary-
ing results.
5.2 Number of Clusters
When looking at the varying number of clusters us-
ing the Euclidean distance (Figure 14a), it can be seen
that DBCV, DSI and produce good clusterings regard-
less of the number of clusters. CVNN, VIASCKDE
and IC-av work well for 2 and 3 clusters, but strug-
gle to find good clusterings using more clusters. The
SWC works well only for a high number of clus-
ters. Similarly, the CVDD has low results on the most
numbers of clusters but works well using 20 clusters.
CVNN hal seems to find better clusterings, the more
clusters are in the ground truth.
Using the DC-dist, most algorithms seem to ei-
ther find very good (SWC, DCVI, DBCV, CVDD) or
very bad (CVNN, CVNN hal, CDR, IC-av) cluster-
ings with 5 or less clusters. The exception is DSI,
which only fingds good clusterings with 2 clusters and
struggles even from 3 on. The SWC and DCVI strug-
gle, when there are more than 5 clusters present.
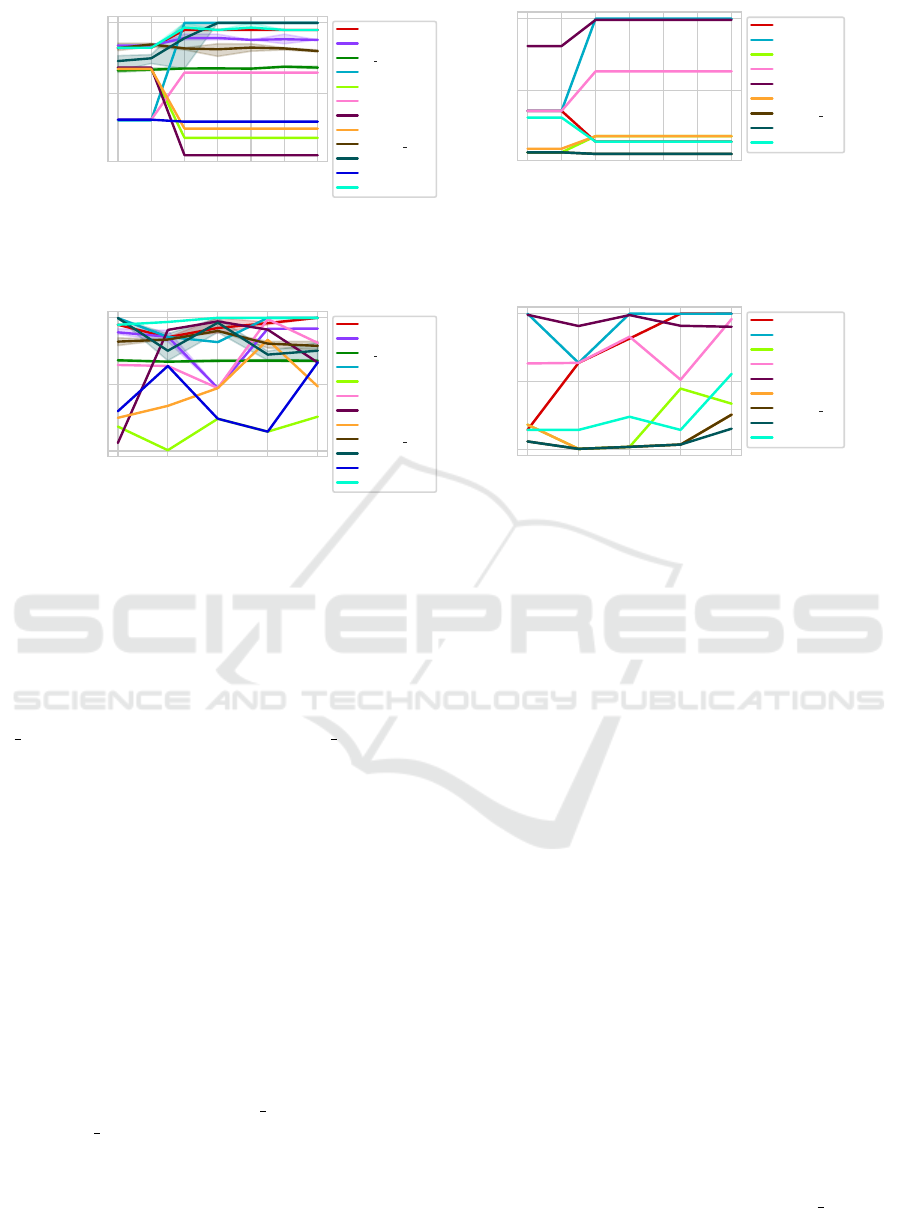
5.3 Overlap Factor
The only notable change in the CVI using the Overlap
happens between 0.8 and 0.9. DBCV, DCVI, SWC
and CDR produce much better results when there are
no overlaps. CVDD, CVNN and IC-av produce their
only good results if there are overlaps.
The trend is similar using the DC-distance. How-
ever, the SWC and CDR are producing better results
with overlaps using this distance.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
370
0.5 0.8 0.9 1 1.1 1.2 1.3
Overlap factor
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
0.5 0.8 0.9 1 1.1 1.2 1.3
Overlap factor
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 15: AMI per CVI using datasets with different overlap factors. The thick line represents the median value on three
datasets, while the shaded area is based on maximum and minimum value.
0 1 4 8 10
Number of connections between clusters
0.0
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
0 1 4 8 10
Number of connections between clusters
0.0
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 16: AMI per CVI using datasets with a different number of connections. The thick line represents the median value on
three datasets, while the shaded area is based on maximum and minimum value.
5.4 Number of Connections
When looking at the presence of connections between
different clusters using the Euclidean distance (Fig-
ure 16b), it can be seen that this has little effect on
S Dbw, DBCV, DSI, SWC and CVNN hal. How-
ever, more connections seem to lead to better results
for CVDD, CVNN and VIASCKDE. VRC and DCVI
have no general trend, but a relatively large drop at 4
connections.
Using the DC-distance (Figure 16a), you can see
that the effect of the number of connections is much
smaller. SWC, DCVI, DSI and IC-av have slight trend
to find better clusterings with a higher number of con-
nections between clusters. Apart from this, most CVI
have a stable trend with little outliers.
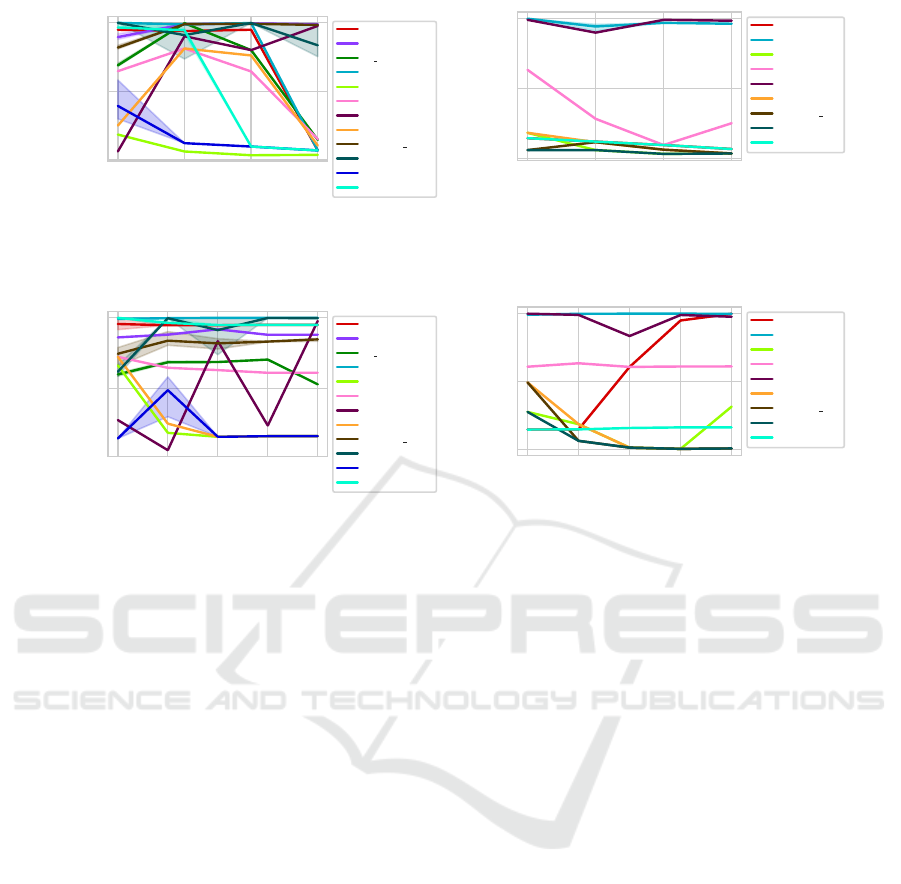
5.5 Noise Ratio
When looking at the noise ratio using the Euclidean
distance (Figure 17a), it can be seen that most algo-
rithms (all apart from IC-av, S Dbw, CVDD, VRC
and CVNN hal) have a massive drop in selecting the
correct clustering when too many outliers are present.
This drop happens earlier for the DSI than for the
other methods.
Using the DC-distance (Figure 17b), the noise ra-
tios chosen by us have little influence on the perfor-
mance of the CVI.
5.6 Number of Objects
When looking at the number of objects, you can see
that most CVI stay constant using the Euclidean dis-
tance (Figure 18a), IC-av and CVNN show much bet-
ter results using only a little number of object. The
performance of CVDD has a general positive trend,
but also has many negative outliers.
Using the DC-distance (Figure 14b), you can see
that the SWC works better, the more objects are
present.
6 DISCUSSION
In this section, we will discuss our results and con-
clude our findings. We have seen that some CVI like
CVDD work well in high dimensional cases or with
many clusters and might have struggles in “easier”
cases, which are similar to many well known bench-
mark datasets, explaining the poor results in previous
surveys (Schlake and Beecks, 2024b). Of all CVI, the
DBCV seems to work well in most scenarios, except
for a high number of outliers, where CVNN hal, CDR
and CVDD perform better. Interestingly, CVNN, IC-
av and CVDD perform better, when there are overlaps
Arbitrary Shaped Clustering Validation on the Test Bench
371
0 0.05 0.1 0.2
Noise ratio
0.0
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
0 0.05 0.1 0.2
Noise ratio
0.0
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 17: AMI per CVI using datasets with different noise ratios. The thick line represents the median value on three
datasets, while the shaded area is based on maximum and minimum value.
100 500 1000 5000 10000
Number of objects
0.5
1.0
AMI
SWC
VRC
S Dbw
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
VIASCKDE
DSI
(a) Euclidean.
100 500 1000 5000 10000
Number of objects
0.0
0.5
1.0
AMI
SWC
DBCV
IC-av
DCVI
CVDD
CVNN
CVNN hal
CDR
DSI
(b) DC-distance.
Figure 18: AMI per CVI using datasets of different sizes. The thick line represents the median value on three datasets, while
the shaded area is based on maximum and minimum value.
between clusters. This might be interesting for further
research. We also figured out that the combination
of DC-distance and DBCV or CVDD will aptly find
clusterings in all our scenarios.
However, our results have to be treated with some
caveats. While we did investigate all these different
factors in isolation, the combination of these factors
might lead to completely different results. An inves-
tigation of all combinations of these properties is not
possible due to their sheer numbers. The second big
caveat is that the quality of these results is depen-
dent on the quality of the Densired dataset generator.
While many CVI have trends, which are expected or
invariant and support the use of this dataset generator,
some results are unexpected and unintuitive. Obvi-
ously, this might be due to an interesting behaviour
of the corresponding CVI, but also some subopti-
mal generated datasets are possible. Another caveat
is, that there is no possibility to measure the “arbi-
trary” of a clusterings shape. It is possible that some
datasets actually contain partitioning based clusters,
which would explain the good performance of the
SWC. However, this unintuitively good performance
matches the results of (Schlake and Beecks, 2024b),
so that it could be, that there are some properties mak-
ing the SWC better suited for arbitrary shaped data
than known.
REFERENCES
Ankerst, M., Breunig, M. M., Kriegel, H., and Sander, J.
(1999). OPTICS: ordering points to identify the clus-
tering structure. In SIGMOD Conference, pages 49–
60. ACM Press.
Bay
´
a, A. E. and Granitto, P. M. (2013). How many clus-
ters: A validation index for arbitrary-shaped clus-
ters. IEEE ACM Trans. Comput. Biol. Bioinform.,
10(2):401–414.
Beecks, C., Uysal, M. S., and Seidl, T. (2010). Signature
quadratic form distance. In CIVR, pages 438–445.
ACM.
Beer, A., Draganov, A., Hohma, E., Jahn, P., Frey, C.
M. M., and Assent, I. (2023). Connecting the dots -
density-connectivity distance unifies dbscan, k-center
and spectral clustering. In KDD, pages 80–92. ACM.
Berndt, D. J. and Clifford, J. (1994). Using dynamic time
warping to find patterns in time series. In KDD Work-
shop, pages 359–370.
Cali
´
nski, T. and Harabasz, J. (1974). A dendrite method
for cluster analysis. Commun. Stat. - Theory Methods,
3(1):1–27.
Campello, R. J. G. B., Moulavi, D., Zimek, A., and Sander,
J. (2015). Hierarchical density estimates for data
clustering, visualization, and outlier detection. ACM
Trans. Knowl. Discov. Data, 10(1):5:1–5:51.
Deborah, L. J., Baskaran, R., and Kannan, A. (2010). A sur-
vey on internal validity measure for cluster validation.
IJCSES, 1(2):85–102.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
372

Ester, M., Kriegel, H., Sander, J., and Xu, X. (1996). A
density-based algorithm for discovering clusters in
large spatial databases with noise. In KDD, pages
226–231. AAAI Press.
Gan, J. and Tao, Y. (2015). DBSCAN revisited: Mis-claim,
un-fixability, and approximation. In SIGMOD Con-
ference, pages 519–530. ACM.
Guan, S. and Loew, M. H. (2022). A distance-based sepa-
rability measure for internal cluster validation. Int. J.
Artif. Intell. Tools, 31(7):2260005:1–2260005:23.
Halkidi, M., Batistakis, Y., and Vazirgiannis, M. (2001). On
clustering validation techniques. J. Intell. Inf. Syst.,
17:107–145.
Halkidi, M. and Vazirgiannis, M. (2001). Clustering valid-
ity assessment: Finding the optimal partitioning of a
data set. In ICDM, pages 187–194. IEEE Computer
Society.
Halkidi, M., Vazirgiannis, M., and Hennig, C. (2015).
Method-independent indices for cluster validation and
estimating the number of clusters. In Handbook
of cluster analysis, pages 616–639. Chapman and
Hall/CRC.
Hassan, B. A., Tayfor, N. B., Hassan, A. A., Ahmed, A. M.,
Rashid, T. A., and Abdalla, N. N. (2024). From a-to-z
review of clustering validation indices. Neurocomput-
ing, 601:128198.
Hu, L. and Zhong, C. (2019). An internal validity index
based on density-involved distance. IEEE Access,
7:40038–40051.
Jahn, P., Frey, C. M. M., Beer, A., Leiber, C., and Seidl, T.
(2024). Data with density-based clusters: A generator
for systematic evaluation of clustering algorithms. In
ECML/PKDD (7), volume 14947 of Lecture Notes in
Computer Science, pages 3–21. Springer.
Kaufman, L. and Rousseeuw, P. J. (1990). Partitioning
Around Medoids (Program PAM), chapter 2, pages
68–125. John Wiley & Sons, Ltd.
Li, W. and Zhou, Z. (2023). Ac: A data generator for eval-
uation of clustering. Authorea Preprints.
Lim, A., Rodrigues, B., Wang, F., and Xu, Z. (2005). k-
center problems with minimum coverage. Theo. Com-
put. Sci., 332(1-3):1–17.
Liu, Y., Li, Z., Xiong, H., Gao, X., Wu, J., and Wu, S.
(2013). Understanding and enhancement of internal
clustering validation measures. IEEE Trans. Cybern.,
43(3):982–994.
McInnes, L., Healy, J., and Astels, S. (2017). hdbscan: Hi-
erarchical density based clustering. J. Open Source
Softw., 2(11):205.
McQueen, J. (1967). Some methods for classification and
analysis of multivariate observations. In Proc. Fifth
Berkeley Symposium on Mathematical Statistics and
Probability, 1967, pages 281–297.
Moulavi, D., Jaskowiak, P. A., Campello, R. J. G. B.,
Zimek, A., and Sander, J. (2014). Density-based clus-
tering validation. In SDM, pages 839–847. SIAM.
Nguyen, X. V., Epps, J., and Bailey, J. (2009). Informa-
tion theoretic measures for clusterings comparison: is
a correction for chance necessary? In ICML, volume
382 of ACM International Conference Proceeding Se-
ries, pages 1073–1080. ACM.
Parsons, L., Haque, E., and Liu, H. (2004). Subspace clus-
tering for high dimensional data: a review. SIGKDD
Explor., 6(1):90–105.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V., VanderPlas, J., Passos, A.,
Cournapeau, D., Brucher, M., Perrot, M., and Duch-
esnay, E. (2011). Scikit-learn: Machine learning in
python. J. Mach. Learn. Res., 12:2825–2830.
Rojas Thomas, J. C. and Santos Pe
˜
nas, M. (2021). New
internal clustering validation measure for contigu-
ous arbitrary-shape clusters. Int. J. Intell. Syst.,
36(10):5506–5529.
Rousseeuw, P. J. (1987). Silhouettes: a graphical aid to
the interpretation and validation of cluster analysis. J.
Comput. Appl. Math., 20:53–65.
Ruspini, E. H., Bezdek, J. C., and Keller, J. M. (2019).
Fuzzy clustering: A historical perspective. IEEE
Comput. Intell. Mag., 14(1):45–55.
Schlake, G. S. and Beecks, C. (2023). Towards automated
clustering. In IEEE Big Data, pages 6268–6270.
IEEE.
Schlake, G. S. and Beecks, C. (2024a). The skyline operator
to find the needle in the haystack for automated clus-
tering. In IEEE Big Data, pages 6117–6122. [IEEE.
Schlake, G. S. and Beecks, C. (2024b). Validating arbi-
trary shaped clusters - A survey. In DSAA, pages 1–12.
IEEE.
Schlake, G. S., Pernklau, M., and Beecks, C. (2024). Au-
tomated exploratory clustering. In IEEE Big Data,
pages 5711–5720. IEEE.
Schubert, E., Sander, J., Ester, M., Kriegel, H. P., and Xu, X.
(2017). DBSCAN revisited, revisited: why and how
you should (still) use DBSCAN. TODS, 42(3):1–21.
S¸enol, A. (2022). VIASCKDE index: A novel internal clus-
ter validity index for arbitrary-shaped clusters based
on the kernel density estimation. Comput. Intell. Neu-
rosci., 2022.1:4059302.
von Luxburg, U., Williamson, R. C., and Guyon, I. (2012).
Clustering: Science or art? In ICML Unsupervised
and Transfer Learning, volume 27 of JMLR Proceed-
ings, pages 65–80. JMLR.org.
Ward Jr, J. H. (1963). Hierarchical grouping to optimize an
objective function. JASA, 58(301):236–244.
Xie, J., Xiong, Z., Dai, Q., Wang, X., and Zhang, Y. (2020).
A new internal index based on density core for clus-
tering validation. Inf. Sci., 506:346–365.
Arbitrary Shaped Clustering Validation on the Test Bench
373
