
Harnessing Diet and Gene Expression Insights Through a Centralized
Nutrigenomics Database to Improve Public Health
Shriya Samudrala
1
, Ijeoma Ezengwa
1
, Fahmida Hai
2
, Rubayat Khan
3
, Saif Nirzhor
4
and Don Roosan
5
1
College of Health Sciences, Western University of Health Sciences, Pomona, U.S.A.
2
Tekurai Inc., San Antonio, U.S.A.
3
University of Nebraska Medical Center, Omaha, U.S.A.
4
University of Texas Southwestern Medical Center, Dallas, U.S.A.
5
School of Engineering and Computational Sciences, Merrimack College, North Andover, U.S.A.
Keywords: Nutrigenomics, Nutrition, Public Health, Artificial Intelligence, Database Management.
Abstract: Nutrigenomics is an emerging field that explores the intricate interaction between genes and diet. This study
aimed to develop a comprehensive database to help clinicians and patients understand the connections
between genetic disorders, associated genes, and tailored nutritional recommendations. The database was built
through an extensive review of primary journal articles and includes detailed information on gene
characteristics, such as gene expression, location, descriptions, and their interactions with diseases and
nutrition. The data suggest that a patient's food intake can either increase or decrease the expression of genes
related to specific diseases. These findings underscore the potential of nutrition to modify gene expression
and reduce the risk of chronic diseases. The study highlights the transformative role nutrigenomics could play
in medicine by enabling clinicians to offer personalized dietary recommendations based on a patient’s genetic
profile. Future research should focus on validating the database in clinical counselling to further refine its
practical applications.
1 INTRODUCTION
The impact of nutrigenomics on daily life is profound
and far-reaching. Nutrigenomics, the study of the
relationship between diet and gene expression, is a
rapidly evolving field with tremendous potential for
growth, particularly through the use of advanced
databases. Given the current health challenges faced
by many communities, this area of research is
increasingly relevant, as nutrition plays a pivotal role
in the development and management of many chronic
disorders, such as colorectal cancer. Nutrigenomics
integrates diverse scientific and environmental
factors to explore the intricate connections between
nutrition and gene expression (Franzago et al, 2020).
The origins of nutrigenomics trace back to the
groundbreaking Human Genome Project of the late
20th century, which provided researchers with the
tools to investigate how diet influences gene activity
and contributes to the onset of diseases. As the field
continues to grow, nutrigenomics holds the promise
of advancing personalized nutrition and improving
public health outcomes.
Nutrigenomics offers valuable insights into how
individuals may respond differently to specific foods
or nutrients based on their genetic makeup. Food
serves as the primary source of nutrients essential for
the body to function effectively, supporting daily
activities and overall survival. Through
nutrigenomics, individuals can better understand how
modifying their diet can influence gene expression,
particularly in cases involving cancerous tissues.
The database empowers consumers by providing
knowledge to make informed dietary decisions that
can positively impact their health. Research has
identified numerous genes affected by dietary intake.
For instance, in colorectal cancer, consuming more
than 2 ounces of red meat per day has been linked to
the downregulation of COL1A1, a gene involved in
extracellular matrix (ECM) regulation and cell matrix
adhesion (Zeng et al, 2023; Aykan, 2015). In addition
to COL1A1, other genes such as the anti-metastasis
and angiogenesis-related gene COL4A2, the tumor
Samudrala, S., Ezengwa, I., Hai, F., Khan, R., Nirzhor, S., Roosan and D.
Harnessing Diet and Gene Expression Insights Through a Centralized Nutrigenomics Database to Improve Public Health.
DOI: 10.5220/0013457600003967
In Proceedings of the 14th International Conference on Data Science, Technology and Applications (DATA 2025), pages 291-298
ISBN: 978-989-758-758-0; ISSN: 2184-285X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
291

suppressor gene TP53, and the cytokine signaling
regulation gene IL22RA1 have been implicated in
cancer pathogenesis (Hall et al., 2016; Abo El-Ella &
Bishayee, 2019; Nasir et al, 2019). Developing a
deeper understanding of these interactions opens new
opportunities for personalized dietary strategies to
prevent and manage diseases, underscoring the
transformative role of nutrition in promoting health
and well-being.
Nutrigenomics also explores how nutrition
interacts with our genes to influence brain function
and mood, offering significant potential in the field of
mental health (Marcum, 2020). As an emerging
discipline, further research in nutrigenomics could
lead to innovative treatments for mental health
conditions, potentially minimizing the unpleasant
side effects and withdrawal symptoms often
associated with traditional medications. Key genes
implicated in depression and anxiety include the
serotonin transporter gene (SLC6A4/5HTT), the
serotonin transporter-linked promoter region (5-
HTTLPR), the serotonin receptor gene (HTR2A), and
brain-derived neurotrophic factor (BDNF). These
genes play critical roles in regulating mood and
emotional well-being (Birla et al., 2022). Notably, L-
tryptophan (TRP) supplements have shown promise
in improving affective states by promoting serotonin
synthesis, a neurotransmitter essential for mood
regulation, appetite control, and sleep. Serotonin is
synthesized from TRP, which can be obtained from
foods such as salmon, nuts and seeds, turkey and
poultry, and pineapple (Rodrigues et al., 2021).
Identifying the genetic pathways influenced by
nutrition, nutrigenomics leads to the potential to
revolutionize mental health care, paving the way for
personalized dietary strategies that enhance
emotional and cognitive well-being.
Consumers have access to various companies that
offer services to use genetic information to tailor
dietary recommendations to their unique genome.
However, the databases currently available tend to be
more generalized, offering insights into genes
associated with common diseases rather than
personalized, specific guidance (Jaskulski et al,
2023). Among these, NutrigenomeDB stands out as a
primary resource for finding nutrition-specific
articles related to various diseases. While other
databases exist, their focus is often limited to the
general connection between diet and genes in the field
of nutrigenomics. To improve the functionality and
precision of these databases, several key concepts
should be incorporated. First, databases should
specify the required intake amounts of specific foods
necessary to influence gene expression effectively.
Additionally, providing detailed descriptions of the
interactions between particular foods and genes,
along with the precise location of these genes, would
enhance their utility for consumers and researchers
alike (Jaskulski et al, 2023).
As a relatively new and emerging field,
nutrigenomics faces challenges in accessing
comprehensive information on the effects of nutrition
on gene expression. While existing studies have
explored a variety of nutrigenomic interactions, there
remains a lack of literature that delves deeply into
these interactions on a disease-specific level (Alegría-
Torres et al, 2011). Continued research is essential to
bridge this gap, as nutrigenomics has the potential to
significantly advance our understanding of these
mechanisms and contribute to the development of
targeted interventions and treatments. With increased
funding and resources, we anticipate a surge in
scientific studies and research in the near future,
enabling nutrigenomics to fulfill its potential as a
transformative tool in personalized medicine and
nutrition.
There is a pressing need for companies to provide
more detailed insights into how specific interactions
between food and genes result in positive health
outcomes. It is essential for consumers to have the
ability to track and understand the pathways involved
in these interactions, enhancing their knowledge of
how nutrigenomics works. Given the strong correlation
between nutrigenomics and diseases, understanding
the effects of excessive or insufficient intake of certain
foods is critical. For instance, increased consumption
of certain foods can lead to epigenetic changes, such as
methylation, which can alter gene sequences. An
example is the overconsumption of red meat, which
has been linked to the downregulation of the NCL gene
in colorectal cancer (Genkinger & Koushik, 2007).
Conversely, improper portion sizes or poor dietary
habits can lead to the overexpression or
underexpression of genes, further contributing to
disease risk (Zhang et al, 2019).
With the rising incidence of hospital admissions
related to poor nutrition and the rapid advancements
in nutrigenomics, there is hope that personalized
dietary strategies can help combat chronic diseases
linked to nutrition (Peña-Romero et al, 2017). By
emphasizing proper food intake and understanding
the genetic implications of diet, nutrigenomics has the
potential to transform healthcare and reduce the
burden of nutrition-related illnesses. Recognizing the
profound impact of nutrients on gene expression and
overall health has the potential to enhance the well-
being of entire communities, driving a new era of
precision health and disease prevention.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
292

2 METHODS
We identified a suitable database, Nutrigenetics, to
collect data on gene classification and the interaction
between genes and specific nutrients. The
Nutrigenetics database provides information on
various gene-food interactions and allowed us to
access relevant articles related to specific nutrients
(Martín-Hernández et al, 2019). During the selection
process, shown in Figure 1, we noted the database’s
limitations, including a lack of detailed explanations on
gene-nutrition interactions and recommended intake
amounts. These factors were considered as part of our
inclusion criteria. A total of 196 articles were
thoroughly reviewed to extract data on the effects of
nutrients on genes associated with chronic diseases.
We carefully examined the limitations and concerns
highlighted by researchers in the "gap" sections of
these articles, identifying commonalities among them.
To ensure the reliability of the findings, we analyzed
the methods and results sections of each study to
confirm the presence of robust and supporting data.
Based on the criteria listed in Table 1, the pool of
articles was narrowed down to eight. Two independent
reviewers conducted the data extraction process,
entered the extracted information into the database, and
cross-checked all inputs for accuracy. This rigorous
approach ensured the integrity and reliability of the
data collected for our study.
Gene location was included in the database to
provide patients with detailed information about
where each gene is positioned on their chromosomes.
This addition enhances patients’ understanding of
their genetic makeup. Gene descriptions were also
incorporated to explain each gene's function in its
natural state, uninfluenced by specific nutrients. This
information helps patients grasp the potential
mechanisms through which certain foods may
contribute to cancer development.
Figure 1: Flow chart illustrating the process of paper
selection for inclusion in the study.
Table 1: The inclusion and exclusion criteria were
implemented to create an adequate and proper
nutrigenomics database.
Inclusion Criteria Exclusion Criteria
Articles available in full
text
Articles is not available in
full text
Articles has correlation
between the nutrition
and the gene
Articles does not have
correlation between the
nutrition and the gene
Articles have measure of
consumption, whether
that be numerical intake
or frequency intake.
Articles does not have
measure of consumption,
whether that be numerical
intake or frequency intake.
Method and result
section of the paper has
proper and supporting
data
Method and result section
of the paper does not have
proper and supporting data
Food intake was measured using two different
approaches. The first method quantified the nutrient
intake in numeric values. The second method relied
on a Food Frequency Questionnaire (FFQ) to assess
consumption frequency. This questionnaire included
11 response categories, ranging from "never" to "2 or
more times a day," and reflected dietary habits from
the year prior to the study.
Data collected from various research articles and
studies were initially compiled into an Excel sheet
and subsequently transformed into a comprehensive
database. The database was implemented using
MySQL, a widely used relational database
management system, with data structured into tables
for genes, diseases, nutrients, and their interactions,
allowing for efficient querying and retrieval of
information.
3 RESULTS
The database results revealed critical insights into the
correlation between various nutritious foods and gene
expression. It offers a comprehensive depiction of
gene characteristics and their interactions with
specific foods. The data demonstrated that the intake
of certain foods can either upregulate or downregulate
the expression of genes associated with specific
diseases. The database is organized into ten sections:
disease, associated gene, P-value, gene expression,
location, species, description, function, interaction,
and intake amount. Each gene is linked to its
corresponding disease, along with the P-value that
quantifies the strength of its interaction with
particular foods. The inclusion of P-values provides a
clear understanding of the statistical significance of
these correlations.
Harnessing Diet and Gene Expression Insights Through a Centralized Nutrigenomics Database to Improve Public Health
293
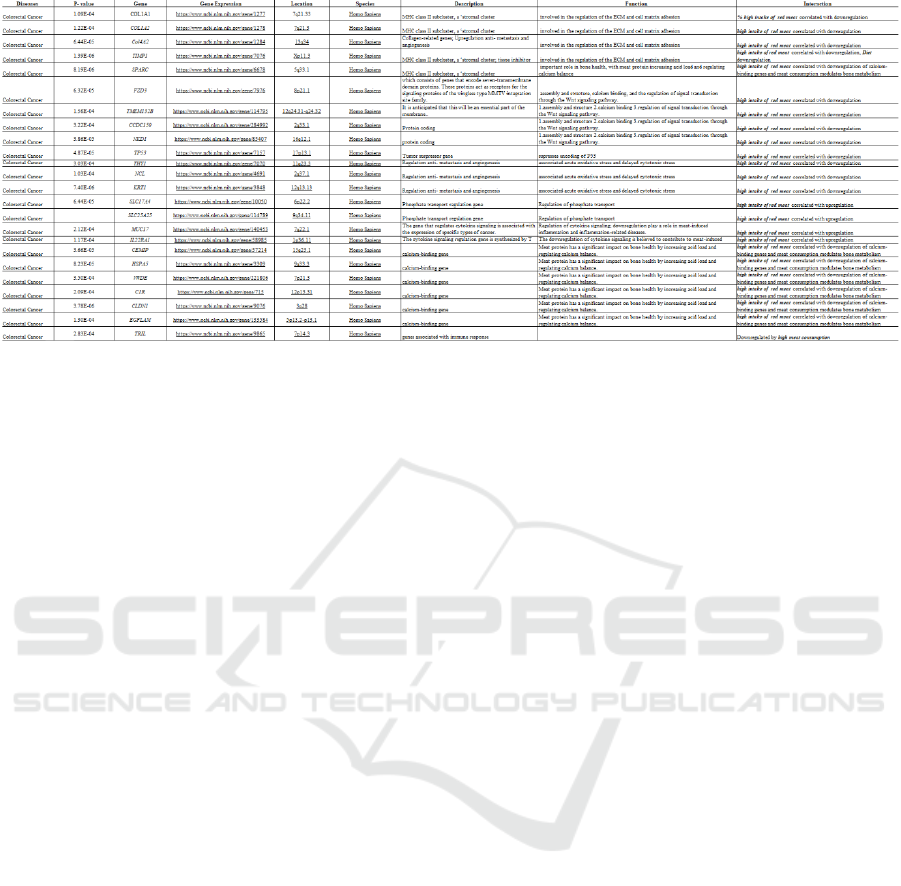
Figure 2: Snapshot of a section of the database, showcasing detailed information on the relationship between nutritious foods,
gene expression, and disease prevention. This resource offers valuable insights for both clinicians and patients.
The gene expression section highlights the gene's
characteristics, including its type and a summary of
how it is expressed. The location section specifies the
gene's position on the chromosome, with the first
number representing the chromosome, the "p"
indicating the short arm, and the "q" denoting the long
arm. The database also includes concise descriptions
of each gene and their biological functions.
The interaction section outlines how specific
foods interact with genes to influence their
expression, offering insights into the metabolic
pathways involved. The intake amount category
provides a quantitative measure of the required
nutrient consumption to induce changes in gene
expression.
By integrating these elements, the database equips
clinicians and patients with a comprehensive
understanding of the intricate relationships between
nutritious foods, gene expression, and disease
prevention. This resource has the potential to inform
personalized nutrition strategies and enhance efforts
to prevent and manage chronic diseases. To evaluate
the functionality and utility of our database, we
conducted several test queries; for example, querying
the database for genes associated with colorectal
cancer and their interaction with red meat
consumption returned results including the
downregulation of COL1A1 with a recommended
intake limit of less than 2 ounces per day. In contrast,
a similar query in NutrigenomeDB provided general
information on gene-diet interactions but lacked
specific intake recommendations, highlighting the
enhanced detail our database offers. To demonstrate
the database's implementation and functionality, we
present a sample query output in Figure 3, which
shows the detailed information retrieved for the gene
MTHFR and its interaction with folate, including the
gene's location, description, and the specific intake
amount that influences its expression.
4 DISCUSSIONS
Nutrigenomics is an emerging and rapidly evolving
field, but current research remains limited and
fragmented. While our database underscores the
significant impact of nutrition on gene expression and
its potential to influence disease risk, it is crucial to
acknowledge that nutrition is one of several factors
contributing to chronic disease management. Genetic
predispositions, medical treatments, and lifestyle
choices all play integral roles, and thus, the nutritional
recommendations provided should be considered as
part of a holistic healthcare approach. While valuable
information exists, compiling a cohesive and
comprehensive understanding of disorders, their
associated genes, and corresponding nutritional
recommendations often requires navigating multiple,
often inaccessible, sources. To address this gap, we
have developed a curated database that systematically
organizes diseases, associated genes, and specific
nutritional interventions that can upregulate or
downregulate these genes.
This database serves as a valuable resource for
nutrition counselors and healthcare clinicians,
offering evidence-based recommendations to support
patient health. Currently, our research focuses on
specific cancers and neuropsychiatric disorders—
relatively new areas of study within nutrigenomics.
As the field continues to grow, we anticipate an
expanding body of knowledge that will enable the
identification of additional targets and further
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
294

enhance personalized approaches to nutrition and
disease prevention.
The potential applications of this database extend
across various domains, including nutritional
counselling, informatics, and public health. Insights
derived from the database enable clinicians to offer
personalized, gene-based dietary recommendations
tailored to an individual’s unique genetic profile
(Agrawal et al, 2023; Mullins et al, 2020). Such
personalized nutrition plans not only enhance the
effectiveness of dietary interventions but also reduce
the risk of nutrition-related diseases, presenting a
transformative approach to preventive healthcare
(Meng et al., 2019).
Current research in nutrigenomics has significant
implications for informatics. The database serves as a
centralized, well-organized resource that researchers
can easily access to support the discovery of new
genes and dietary interventions. Moreover, the
nutrigenomics database can integrate with advanced
technologies, such as artificial intelligence (AI), to
unlock deeper insights from large datasets of genetic
and nutritional information. AI applications have the
potential to analyze these datasets, identify patterns
and associations, and develop predictive models to
assist healthcare clinicians in making informed
decisions about nutrition and health (Gao & Chen,
2017; Marcum, 2020). Currently, healthcare
applications utilizing AI, augmented and virtual
reality have been increasing rapidly. This technology
is being utilized for patient counselling, medication
reminders, and even to aide in surgery (Roosan, 2024;
Li et al., 2023). For example, AI could generate
personalized nutrition plans tailored to an
individual’s unique genetic profile that the provider
reviews (Roosan, 2024). By analyzing genetic data
and identifying variations that influence how an
individual metabolizes specific nutrients or responds
to certain diets, AI can provide highly targeted dietary
recommendations (Malle, 2021). This integration of
AI with nutrigenomics could revolutionize the field,
paving the way for more precise and effective
healthcare solutions.
Integrating the nutrigenomics database with
electronic health records (EHRs) could significantly
enhance its clinical utility. With access to
comprehensive patient data, including genetic
profiles and dietary habits, clinicians can develop
more precise and tailored nutrition recommendations
(Roosan et al, 2019). Additionally, using AI-powered
tools like ChatGPT alongside the database could
enable patients to monitor and refine their dietary
intake in real time, empowering them to achieve their
nutritional goals (Roosan et al, 2023; Roosan et al,
2024).
The applications of genomic knowledge extend
far beyond healthcare. Genomics forms the
foundation for “personalized medicine,” offering
unique, patient-specific clinical interventions that
improve outcomes. Beyond healthcare, genomics has
transformative potential in agriculture, environmental
science, forensic science, and drug development
(Senthil et al., 2019).
From a public health perspective, the database
could enhance the effectiveness of interventions by
providing tailored nutritional recommendations based
on genetic profiles and dietary needs (Alegría-Torres
et al, 2011; Roosan et al, 2024). Targeted strategies
could address specific subpopulations with unique
genetic traits and nutritional risk factors, enabling the
development of more effective public health policies
and programs (Wu et al., 2024; Roosan et al, 2024).
The database also provides valuable insights into the
relationships between genetics, nutrients, and health
outcomes, offering opportunities to identify
therapeutic targets and improve disease prevention at
the population level (Fenech et al., 2011).
There is growing evidence linking genetics, poor
nutrition, and disease risk. For example, individuals
with genetic variations affecting folate metabolism
are at an increased risk of colorectal cancer when
consuming low-folate diets (Kim, 2006; Zeng et al,
2023). Variations in the MTHFR gene can impair
folic acid metabolism, and excessive folic acid
consumption in these individuals may mask vitamin
B12 deficiency, leading to adverse health outcomes
(Birla et al., 2022). Diets high in processed and red
meat have similarly been associated with a
heightened risk of colorectal cancer, particularly in
individuals with genetic predispositions (Aykan,
2015; Bertucci et al, 2004). Additionally, genetic
variations have been shown to influence
susceptibility to mental health disorders such as
depression and anxiety, with poor dietary quality
exacerbating these conditions. These findings
highlight the intricate interplay between genetics,
nutrition, and health, emphasizing the importance of
personalized dietary strategies to mitigate risks and
improve outcomes.
The database is designed primarily for clinicians,
offering a robust tool to assist in creating personalized
treatment plans. By analyzing genetic profiles,
clinicians can identify variations that influence
nutrient metabolism and recommend tailored dietary
changes or supplements to optimize patient health.
Equally important is educating patients about the
benefits of the database, which can be achieved
Harnessing Diet and Gene Expression Insights Through a Centralized Nutrigenomics Database to Improve Public Health
295

through online resources and consultations with
healthcare professionals. Empowering patients with
this knowledge enables them to take an active role in
managing their health through informed dietary and
lifestyle changes (Roosan et al, 2022; Roosan, 2022).
It is important to note that while nutrigenomics
provides valuable insights into potential areas of
concern and personalized strategies, it is not a
diagnostic tool. Instead, it serves as a resource to
enhance understanding and promote proactive health
management. Although nutrigenomics is still in its
early stages, it represents the beginning of a deeper
understanding of the complex relationship between
genetics and nutrition. This field has already made
significant strides in identifying how genetic factors
and nutrients interact to impact health and reduce
disease risk. As research continues to advance, we
anticipate a growing body of knowledge that will
pave the way for more personalized approaches to
medicine, tailoring healthcare interventions to
individual genetic profiles. To ensure the
nutrigenomics database can be effectively integrated
into clinical workflows and patient care, it is crucial
to adhere to established health data standards (Roosan
et al., 2020b), explore the development of AI-
powered applications for personalized dietary
guidance (Roosan et al., 2020a), and consider
advanced data management solutions like blockchain
for secure and efficient access (Roosan et al., 2022).
While many companies currently focus on
genomic testing to raise awareness of genetic
diseases, there is a growing need for these companies
to provide customers with more comprehensive
information on the connection between genes and
nutrition. Enhancing access to such insights would
enable customers to develop a deeper understanding
of nutrigenomics. Achieving this goal may involve
initiatives such as education programs, accessible
genetic testing, consultations with healthcare
providers, and greater transparency from companies
(Guasch-Ferré et al., 2018). These studies
collectively highlight the multifaceted nature of
clinical decision-making, exploring cognitive
strategies, complexity measurement, expert
heuristics, and disease-specific reasoning to inform
the design of effective decision support systems
((Islam et al, 2014; Islam et al, 2015; Islam et al,
2016a; Islam et al, 2016b). By expanding awareness
and accessibility, nutrigenomics has the potential to
transform how individuals approach health and
nutrition, fostering a deeper understanding of
personalized wellness.
5 CONCLUSION
In conclusion, the nutrigenomics database offers a
comprehensive understanding of the relationship
between nutritious foods, gene expression, and
disease prevention. By cataloging diseases,
associated genes, and specific nutritional
interventions that can upregulate or downregulate
gene activity, this resource serves as a valuable tool
for nutrition counselors and healthcare clinicians
seeking evidence-based recommendations to support
their patients' health. The database has far-reaching
implications for nutritional counseling, informatics,
and public health. It holds the potential to enable the
creation of personalized nutrition plans tailored to an
individual’s genetic makeup, integrate seamlessly
with AI applications for data analysis and predictive
modeling, and enhance clinical practice through
integration with EHRs.
Furthermore, genomics knowledge provides
opportunities for personalized medicine, agriculture
environment, forensic science, and drug
development. The nutrigenomics database plays a
crucial role in preventing or managing disease
through diet and recognizing the significant impact of
nutrients on overall health, which can notably
enhance the health and well-being of the entire
community. Lastly, the nutrigenomics database could
help identify targeted interventions for subgroups of
the population with specific genes and risk factors of
nutritional needs and offer valuable insights into
disease pathogenesis and potential targets for
interventions, ultimately optimizing health in
individuals and communities through nutrigenomics.
ACKNOWLEDGEMENTS
We are grateful to Merrimack College for support.
REFERENCES
Abo El-Ella, D. M., & Bishayee, A. (2019). Chapter 6—
The Epigenetic Targets of Berry Anthocyanins in
Cancer Prevention. In A. Bishayee & D. Bhatia (Eds.),
Epigenetics of Cancer Prevention (Vol. 8, pp. 129–
148). Academic Press. https://doi.org/10.1016/B978-0-
12-812494-9.00006-8
Agrawal, P., Kaur, J., Singh, J., Rasane, P., Sharma, K.,
Bhadariya, V., Kaur, S., & Kumar, V. (2024). Genetics,
Nutrition, and Health: A New Frontier in Disease
Prevention. Journal of the American Nutrition
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
296

Association, 43(4), 326–338. https://doi.org/10.1080/
27697061.2023.2284997
Alegría-Torres, J. A., Baccarelli, A., & Bollati, V. (2011).
Epigenetics and Lifestyle. Epigenomics, 3(3), 267–277.
https://doi.org/10.2217/epi.11.22
Aykan N. F. (2015). Red Meat and Colorectal Cancer.
Oncology reviews, 9(1), 288. https://doi.org/10.4081/
oncol.2015.288
Bertucci, F., Salas, S., Eysteries, S., Nasser, V., Finetti, P.,
Ginestier, C., Charafe-Jauffret, E., Loriod, B.,
Bachelart, L., Montfort, J., Victorero, G., Viret, F.,
Ollendorff, V., Fert, V., Giovaninni, M., Delpero, J.-R.,
Nguyen, C., Viens, P., Monges, G., … Houlgatte, R.
(2004). Gene expression profiling of colon cancer by
DNA microarrays and correlation with histoclinical
parameters. Oncogene, 23(7), 1377–1391.
https://doi.org/10.1038/sj.onc.1207262
Birla, M., Choudhary, C., Singh, G., Gupta, S., Bhawana,
& Vavilala, P. (2022). The Advent of Nutrigenomics:
A Narrative Review with an Emphasis on
Psychological Disorders. Preventive nutrition and food
science, 27(2), 150–164. https://doi.org/10.3746/
pnf.2022.27.2.150
Fenech, M., El-Sohemy, A., Cahill, L., Ferguson, L. R.,
French, T. A., Tai, E. S., Milner, J., Koh, W. P., Xie, L.,
Zucker, M., Buckley, M., Cosgrove, L., Lockett, T.,
Fung, K. Y., & Head, R. (2011). Nutrigenetics and
nutrigenomics: viewpoints on the current status and
applications in nutrition research and practice. Journal
of nutrigenetics and nutrigenomics, 4(2), 69–89.
https://doi.org/10.1159/000327772
Franzago, M., Santurbano, D., Vitacolonna, E., & Stuppia,
L. (2020). Genes and Diet in the Prevention of Chronic
Diseases in Future Generations. International Journal
of Molecular Sciences, 21(7), 2633. https://doi.org/
10.3390/ijms21072633
Gao, Y., & Chen, J. (2017). Informatics for Nutritional
Genetics and Genomics. In B. Shen (Ed.), Translational
Informatics in Smart Healthcare (Vol. 1005, pp. 143–
166). Springer Singapore. https://doi.org/10.1007/978-
981-10-5717-5_7
Genkinger, J. M., & Koushik, A. (2007). Meat
Consumption and Cancer Risk. PLoS Medicine, 4(12),
e345. https://doi.org/10.1371/journal.pmed.0040345
Guasch-Ferré, M., Dashti, H. S., & Merino, J. (2018).
Nutritional Genomics and Direct-to-Consumer Genetic
Testing: An Overview. Advances in nutrition
(Bethesda, Md.), 9(2), 128–135. https://doi.org/10.10
93/advances/nmy001
Hall, J. E., do Carmo, J. M., da Silva, A. A., Wang, Z., &
Hall, M. E. (2016). Obesity, kidney dysfunction and
hypertension: mechanistic links. Nature Reviews
Nephrology, 12(5), 339-52. doi: 10.1038/nrneph.20
16.32
Islam, R., Roosan, M., Mayer, J., & Clutter, J. (2016).
Supporting novice clinicians cognitive strategies:
System design perspective. IEEE-EMBS International
Conference on Biomedical and Health Informatics,
2016, 509. https://doi.org/10.1109/BHI.2016.7455946
Islam, R., Weir, C., & Del Fiol, G. (2016). Clinical
complexity in medicine: A measurement model of task
and patient complexity. Methods in Information in
Medicine, 55, 14–22. https://doi.org/10.3414/ME15-
01-0031
Islam, R., Weir, C., & Del Fiol, G. (2014). Heuristics in
managing complex clinical decision tasks in experts’
decision making. Proceedings (IEEE International
Conference on Healthcare Informatics), 2014, 186–
193. https://doi.org/10.1109/ICHI.2014.32
Islam, R., Weir, C. R., Jones, M., Del Fiol, G., & Samore,
M. H. (2015). Understanding complex clinical
reasoning in infectious diseases for improving clinical
decision support design. BMC Medical Informatics and
Decision Making, 15, 101. https://doi.org/10.1186/
s12911-015-0221-z
Jaskulski, S., Nuszbaum, C., & Michels, K. B. (2023).
Components, prospects and challenges of personalized
prevention. Frontiers in Public Health, 11, 1075076.
https://doi.org/10.3389/fpubh.2023.1075076
Kim Y. I. (2006). Folate: a magic bullet or a double edged
sword for colorectal cancer prevention?. Gut, 55(10),
1387–1389. https://doi.org/10.1136/gut.2006.095463
Ma, Y., Liu, X., Zhuang, Y., & Yang, W. (2016). Dietary
intake alters gene expression in colon tissue: Possible
underlying mechanism for the influence of diet on
disease. Pharmacogenetics and Genomics, 26(6), 294-
306. doi: 10.1097/FPC.0000000000000217.
Malle, M. (2021). Ethical Aspects of Artificial Intelligence
and Robotics. In V. Taatgen & E. A. Groen (Eds.),
Handbook of Artificial Intelligence (pp. 1-10).
Helsinki: Finnish Center for Artificial Intelligence.
Retrieved from https://helda.helsinki.fi/bitstream/
handle/10138/340241/Artificial_intelligence.pdf?sequ
ence=1
Marcum, J. A. (2020). Nutrigenetics/Nutrigenomics,
Personalized Nutrition, and Precision Healthcare.
Current Nutrition Reports, 9(4), 338–345.
https://doi.org/10.1007/s13668-020-00327-z
Martín-Hernández, R., Reglero, G., Ordovás, J. M., &
Dávalos, A. (2019). NutriGenomeDB: A nutrigenomics
exploratory and analytical platform. Database, 2019,
baz097. https://doi.org/10.1093/database/baz097
Meng, H., Yue, P., Tao, L., Liu, Y., & Liu, Y. (2019).
Effects of Exercise on Sleep Quality in Older Adults: A
Systematic Review and Meta-Analysis. Journal of
Clinical Medicine, 8(10), 1489. doi: 10.3390/jcm8101
489.
Mullins, V. A., Bresette, W., Johnstone, L., Hallmark, B.,
& Chilton, F. H. (2020). Genomics in Personalized
Nutrition: Can You “Eat for Your Genes”? Nutrients,
12(10), 3118. https://doi.org/10.3390/nu12103118
Nasir, A., Bullo, Mir. M. H., Ahmed, Z., Imtiaz, A.,
Yaqoob, E., Safdar, M., Ahmed, H., Afreen, A., &
Yaqoob, S. (2020). Nutrigenomics: Epigenetics and
cancer prevention: A comprehensive review. Critical
Reviews in Food Science and Nutrition, 60(8), 1375–
1387. https://doi.org/10.1080/10408398.2019.1571480
Pellatt, A. J., Slattery, M. L., Mullany, L. E., Wolff, R. K.,
& Pellatt, D. F. (2016). Dietary intake alters gene
Harnessing Diet and Gene Expression Insights Through a Centralized Nutrigenomics Database to Improve Public Health
297

expression in colon tissue: possible underlying
mechanism for the influence of diet on disease.
Pharmacogenetics and genomics, 26(6), 294–306.
https://doi.org/10.1097/FPC.0000000000000217
Peña-Romero, A. C., Navas-Carrillo, D., Marín, F., &
Orenes-Piñero, E. (2018). The future of nutrition:
Nutrigenomics and nutrigenetics in obesity and
cardiovascular diseases. Critical Reviews in Food
Science and Nutrition, 58(17), 3030–3041.
https://doi.org/10.1080/10408398.2017.1349731
Rodrigues, J., Coelho, A., Figueiredo, D., Silva, C., &
Almeida, I. (2021). The Role of Virtual Reality in the
Assessment and Rehabilitation of Cognitive
Impairment in Older Adults. Frontiers in Psychology,
12, 627309. doi: 10.3389/fpsyg.2021.627309
Roosan, D., Chok, J., Karim, M., Law, A. V., Baskys, A.,
Hwang, A., & Roosan, M. R. (2020a). Artificial
intelligence–powered smartphone app to facilitate
medication adherence: Protocol for a human factors
design study. JMIR Research Protocols, 9(11), e21659.
Roosan, D., Hwang, A., Law, A. V., Chok, J., & Roosan,
M. R. (2020b). The inclusion of health data standards
in the implementation of pharmacogenomics systems:
A scoping review. Pharmacogenomics, 21(16), 1191–
1202.
Roosan, D., Wu, Y., Tatla, V., Li, Y., Kugler, A., Chok, J.,
& Roosan, M. R. (2022). Framework to enable
pharmacist access to health care data using Blockchain
technology and artificial intelligence. Journal of the
American Pharmacists Association, 62(4), 1124–1132.
Roosan, D. (2022). The Promise of Digital Health in
Healthcare Equity and Medication Adherence in the
Disadvantaged Dementia Population. Pharmaco-
genomics, 23(9), 505–508. https://doi.org/10.2217/pgs-
2022-0062
Roosan, D. (2024). Integrating Artificial Intelligence with
Mixed Reality to Optimize Health Care in the
Metaverse. In V. Geroimenko (Ed.), Augmented and
Virtual Reality in the Metaverse (pp. 247–264).
Springer Nature Switzerland. https://doi.org/10.1007/
978-3-031-57746-8_13
Roosan, D. (2024). Comprehensive guide and checklist for
clinicians to evaluate artificial intelligence and machine
learning methodological research. Journal Of Medical
Artificial Intelligence, 7. doi:10.21037/jmai-24-65
Roosan, D., Chok, J., Nersesian, T., Li, Y., Law, A., & Li,
Y. (2023). Development of a dashboard analytics
platform for dementia caregivers to understand
diagnostic test results. https://doi.org/10.21203/rs.3.rs-
3032181/v1
Roosan, D., Chok, J., Li, Y., & Khou, T. (2024). Utilizing
Quantum Computing-based Large Language
Transformer Models to Identify Social Determinants of
Health from Electronic Health Records. 2024
International Conference on Electrical, Computer and
Energy Technologies (ICECET, 1–6. https://doi.org/
10.1109/ICECET61485.2024.10698600
Roosan, D., Law, A. V., Karim, M., & Roosan, M. (2019).
Improving Team-Based Decision Making Using Data
Analytics and Informatics: Protocol for a Collaborative
Decision Support Design. JMIR Research Protocols,
8(11), e16047. https://doi.org/10.2196/16047
Roosan, D., Padua, P., Khan, R., Khan, H., Verzosa, C., &
Wu, Y. (2024). Effectiveness of ChatGPT in clinical
pharmacy and the role of artificial intelligence in
medication therapy management. Journal of the
American Pharmacists Association, 64(2), 422-428.e8.
https://doi.org/10.1016/j.japh.2023.11.023
Roosan, D., Wu, Y., Tran, M., Huang, Y., Baskys, A., &
Roosan, M. R. (2023). Opportunities to integrate
nutrigenomics into clinical practice and patient
counseling. European Journal of Clinical Nutrition,
77(1), 36–44. https://doi.org/10.1038/s41430-022-
01146-x
Senthil, R. A., Suresh, S., & Anandh, G. (2019). Leveraging
the power of artificial intelligence in healthcare:
Exploring the issues, opportunities and challenges.
Journal of Medical Systems, 43(8), 233.
https://doi.org/10.1007/s10916-019-1386-3
Tharmaratnam, T., Koczwara, A., Schneider, E., & Palmer,
C. (2021). Mental health impacts of the COVID-19
pandemic on international students: A systematic
review. BMC Public Health, 21(1), 1–18.
https://doi.org/10.1186/s12889-021-10672-2
Xiang Zeng, Guoxue Chen, Dunwei Guo, Yong Chen, &
Xinpeng Hu. (2023). Expression, prognostic value and
potential immunotherapeutic target of COL1A1 in
colon cancer. Cellular and Molecular Biology, 69(15),
120–125. https://doi.org/10.14715/cmb/2023.69.15.21
Zhang, F. F., Cudhea, F., Shan, Z., Michaud, D. S.,
Imamura, F., Eom, H., Ruan, M., Rehm, C. D., Liu, J.,
Du, M., Kim, D., Lizewski, L., Wilde, P., &
Mozaffarian, D. (2019). Preventable Cancer Burden
Associated With Poor Diet in the United States. JNCI
Cancer Spectrum, 3(2), pkz034. https://doi.org/
10.1093/jncics/pkz034
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
298
