
A Multi-Scale Feature Fusion Network for Detecting and Classifying
Apple Leaf Diseases
Assad S. Doutoum
1 a
, Recep Eryigit
2 b
and Bulent Tugrul
3 c
1
Computer Science Department, University of Ndjamena, Farcha, Ndjamena, Chad
2
Ankara University, Department of Computer Engineering, Ankara, Turkey
3
Ankara University, Department of Computer Science, Ankara, Turkey
Keywords:
Deep Learning, Multi-Scale CNN, Apple Leaf Diseases.
Abstract:
Early detection and identification of leaf diseases reduce expenses and increase profits. Thus, it is essential
for producers to be aware of the symptoms and indications of these leaf diseases and take the necessary pre-
ventative measures. Early diagnosis and treatment can also help prevent the disease from spreading to healthy
plants. For successful disease control, regular inspections of orchards are essential. As well as being costly and
time-consuming, traditional methods require a great deal of labor. However, the use of modern technologies
and methods such as computer vision will both increase successes and reduce costs. Deep learning methods
can be used to detect and classify diseases, as well as predict the likelihood of them occurring. Though, a
particular CNN architecture may focus on a subset of features, while another may discover other additional
features not extracted from the dataset. Robust classification models should be developed that perform con-
sistently well when different environmental factors such as light, angle, background and noise vary. To solve
these challenges, this study proposes a multi-scale feature fusion network (MFFN) that combines features
from different scales or levels of detail in an image to improve the performance and robustness of classifica-
tion models. The proposed method is evaluated on a publicly available dataset and is shown to improve the
performance of the original models. Four branches applied to CNN architectures were simultaneously trained
and merged to accurately classify and predict infected apple leaves. The merged model was able to detect
infected leaves with a high degree of accuracy, significantly through the combined models. The merged model
was able to accurately predict the unhealthy apple leaves with a 99.36% on the training accuracy, 98.90% on
the validation accuracy, and 98.28% on the test accuracy. The results show that combining the models is an
effective way to increase the accuracy of predictions under volatile conditions.
1 INTRODUCTION
Plant leaf diseases severely affect the quality and pro-
ductivity of agricultural products. The agricultural in-
dustry loses millions of dollars every year due to yield
loss and unnecessary or misuse of pesticides (Alsayed
et al., 2021). It is therefore crucial to detect and diag-
nose plant diseases early to prevent their spread and
minimize their impacts. Several pathogens such as
Black Rot disease, Cedar Rust disease, and Scab dis-
ease may affect apple leaves (Su et al., 2022; Nand-
hini and Ashokkumar, 2022). Traditional methods
of detecting plant diseases require specialized knowl-
a
https://orcid.org/0000-0003-4822-1876
b
https://orcid.org/0000-0002-4282-6340
c
https://orcid.org/0000-0003-4719-4298
edge and expertise in plant pathology. These meth-
ods are often time-consuming and require the use of
expensive equipment, making them inaccessible to
those without adequate budgets. These limitations
hinder the ability to detect and monitor diseases on
small farms and in low-income countries where re-
sources and experts are scarce. Consequently, ad-
vanced technologies and automated systems are re-
quired to detect plant sicknesses at an early stage.
With the help of such technologies it is possible
to detect, classify and diagnose plant diseases more
quickly and accurately. Thanks to these methods,
agricultural enterprises can save time and money and
gain a competitive advantages over their competitors
on both national and international markets.
Various machine learning and deep learning meth-
ods have been used to detect and classify plant ill-
Doutoum, A. S., Eryigit, R., Tugrul and B.
A Multi-Scale Feature Fusion Network for Detecting and Classifying Apple Leaf Diseases.
DOI: 10.5220/0013447700003967
In Proceedings of the 14th International Conference on Data Science, Technology and Applications (DATA 2025), pages 13-20
ISBN: 978-989-758-758-0; ISSN: 2184-285X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
13

nesses (Barbedo, 2018; Doutoum and Tugrul, 2023).
It is suggested that deep learning methods shorten
the time-consuming and cost-efficient process of fea-
ture extraction in image processing (Yan et al., 2020).
The models obtained through deep learning meth-
ods demonstrate significantly improved accuracy and
robustness compared to traditional machine learning
methods. In traditional machine learning, prediction
or classification models are trained using manually
extracted features. In contrast, deep learning mod-
els have the ability to automatically extract features
from images and build the model on them. As a re-
sult, manual feature extraction is no longer necessary,
increasing the efficiency and reducing the cost of clas-
sification methods. Furthermore, deep-learning algo-
rithms have the potential to recognize complex corre-
lations and patterns as well as various high-level ele-
ments that are invisible to humans. As a result, they
are more suitable for tasks that require more domain-
specific knowledge and experience.
Feature fusion uses multiple scales or levels of de-
tail in computer vision and image processing to in-
tegrate facts into feature representation to enable a
better understanding of visual elements (Elizar et al.,
2022). As a result, they are more resistant to changes
in object size, orientation, and scale, because charac-
teristics at various sizes can give complementing in-
formation. This may rise item recognition and clas-
sification accuracy, as well as the capability to dis-
tinguish things in a more problematic context. There
are many ways of accomplishing multi-scale feature
fusion in computer vision. One possible approach is
to use distinct Convolutional Neural Network (CNN)
architectures to extract features of varying sizes and
aspects and then efficiently integrate them. Integrat-
ing detailed features from small scales with contex-
tual knowledge from large scales yields more com-
prehensive and robust models.
The contributions of this article can be summa-
rized as follows: By proposing an architecture based
on multi-scale CNN for apple leaf disease classifica-
tion and prediction, we improve the overall perfor-
mance of the model by allowing it to better under-
stand the spatial structure of the images, leading to
more accurate predictions. Our proposed model is
based on parallel training of big datasets, is less com-
plex, and has accurate results compared with other
deep learning models (Yan et al., 2020; G
¨
und
¨
uz and
Yılmaz G
¨
und
¨
uz, 2022). According to the compari-
son dataset, our model achieved the highest accuracy
result of 98.28% on the test set.
2 METHODS AND MATERIALS
The following section provides detailed information
about the Apple data source. Additionally, the pre-
processing procedures applied to the dataset and the
training process of the proposed multi-scale feature
fusion network (MFFN) will be explained.
2.1 Dataset
The dataset utilized in this study consists of images
with healthy and unhealthy apple leaves. As can be
seen from Table 1, there are four main classes within
the dataset. The first class contains leaf images of
Black Rot disease. The second class contains images
of Cedar Rust disease. The third and fourth classes
contain images of healthy leaves and Scab disease re-
spectively. In total, there are 10,173 images of apple
leaves in the dataset (Hughes and Salath
´
e, 2015). The
class distribution of the apple dataset is shown in Ta-
ble 1.
2.2 Data Pre-Processing
Image pre-processing and validation is an essential
step to calibrate the images before being trained by
the deep learning model (Divakar et al., 2021). Deep
learning classifiers require huge datasets to overcome
over-fitting problems and obtain better precision level
of achievement. It is therefore necessary to perform
an augmentation techniques in order to expand the
dataset from the one that currently exists. This re-
quires changes such as resizing, zooming, rotating, or
adjusting the color of the images. Nevertheless, it is
necessary to split the dataset into training, validation,
and testing sets (G
¨
und
¨
uz and Yılmaz G
¨
und
¨
uz, 2022).
Using the image data augmentation techniques, we
horizontally shift the dataset’s images.
2.3 Dataset Augmentation Techniques
Common data augmentation techniques include flip-
ping, rotating, and scaling images to create new vari-
ations. Other methods involve adjusting brightness,
contrast, and adding noise to enhance the dataset fur-
ther. By expanding the dataset, it allows models to
learn features more robustly and generalize better to
unseen data. Additionally, it introduces variability
that can make the model more resilient to noise and
variations in real-world data. This leads to improved
accuracy and performance across diverse scenarios.
Common data augmentation techniques include flip-
ping, rotating, and scaling images to create new vari-
ations. Other methods involve adjusting brightness,
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
14

Table 1: Distribution of the apple dataset.
Disease type Black Rot Cedar Rust Healthy leaf Scab Total
Number of images 2352 2792 2510 2519 10173
Figure 1: Sample of images after the augmentation pro-
cesses.
contrast, and adding noise to enhance the dataset fur-
ther (Baranwal et al., 2019; Su et al., 2022). Table 2
displays the augmentation details. Figure 1 shows a
sample of images after the augmentation processes.
Table 2: Augmentation parameters.
Parameter Value
Rotation 40
Width 0.2
Height 0.2
Horizontal flipping True
Vertical flipping True
Shear 0.2
Zoom 0.2
2.4 Data Splitting
The dataset used in this study contains a total of
10173 images. The dataset is divided into three fold-
ers: the training and the validation sets contain 6353
and 2014 images respectively. The remaining 1806
images are assigned to the test set. The validation
set is allocated to assess the model’s generalization
ability. If over-fitting is observed, hyper-parameters
such as learning rate, epoch size, or number of lay-
ers can be fine-tuned to enhance the model’s perfor-
mance. This process of fine-tuning hyper-parameters
based on validation results help to identify the opti-
mal configuration that maximizes the model’s perfor-
mance on unseen test data. The apple dataset is di-
vided into four classes, and each class contains one
apple disease (Yadav et al., 2022).
2.5 Multi-Scale CNN Model Structure
The improved model architecture is based on the
multi-scale CNN model, which is shown in Figure 2.
Our network consists of one input image shape with a
size of 224×224×3 divided into four branches. Each
CNN branch has one convolution layer (64×64) fol-
lowed by Batch Normalization layers (BN) and Max-
Pooling layers. After each Batch Normalization we
use ReLu as an activation function. We add separa-
ble layers before concatenating the four branches net-
works. Following the merging of the CNN networks,
two dense layers are added to the network. Table 3
displays the description of the Multi-scale CNN net-
work architecture.
Table 3: Description of MFNN architecture.
Layer CNN (Input=224 × 224 × 3)
Conv1 1 × 1
64 × 64
MaxPool 2 × 2
Conv2 1 × 1
64 × 64
MaxPool 2 × 2
Conv3 1 × 1
64 × 64
MaxPool 2 × 2
Conv4 1 × 1
64 × 64
MaxPool 2 × 2
Concatenate conv1, conv2 , conv3 , conv4
Conv5 3 × 3
64 × 64
MaxPool 2 × 2
Conv6 3 × 3
64 × 64
MaxPool 2 × 2
Conv7 3 × 3
128 × 128
MaxPool 2 × 2
Dense -
2.6 Experimental Environment and
Parameters
For the purpose of training, our proposed model for
classification and detection of Apple leaf diseases,
used TensorFlow/Keras framework, and Python 3.11
A Multi-Scale Feature Fusion Network for Detecting and Classifying Apple Leaf Diseases
15
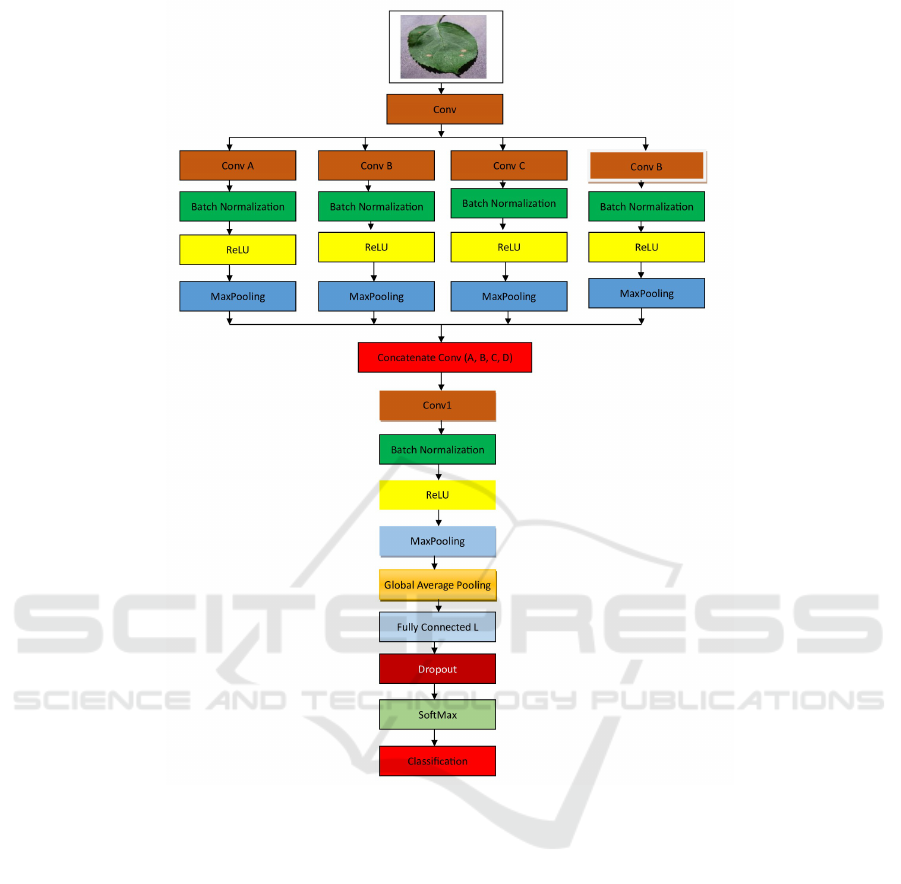
Figure 2: MFFN architecture.
version. Table 4 shows the hardware and software
details of the experimental environment. The Apple
dataset was divided into three parts: training, valida-
tion, and testing sets. There are multiple parameters
applied to the training and validation dataset. Table
5 shown the parameters utilized during training and
validation.
3 EXPERIMENTAL RESULTS
AND ANALYSIS
This section evaluates the performance of our pro-
posed MFFN model in the classification and detection
of apple leaf diseases. The experimental evaluates and
analyzes confusion metrics and compares the module
with other existing models. To evaluate the efficiency
of the proposed model we trained on the apple leaf
dataset, which consists of four distinct classes. The
percentage of true guesses in prediction results com-
pared to all guesses is known as accuracy rate. Recall
rate is the percentage that a category is accurately pre-
dicted in all actual values, whereas precision rate is
the percentage that a category is correctly forecasted
in all prediction outcomes.
3.1 Evaluation Metrics
The confusion matrix is used to evaluate the achieve-
ments of our model. A confusion matrix is presented
in this section for a thorough analysis. The confu-
sion matrix’s rows correspond to the actual category
labels, while its columns correspond to the labels that
the model predicted. Through the number of cases of
True Positive (TP), False Positive (FP), True Negative
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
16

Table 4: Experimental hardware and software.
Name Model
Framework TensorFlow/Keras
GPU NVIDIA RTX A4000
CPU Intel(R) Xeon(R) W-2235 CPU @ 3.80GHz, 3792 MHz
OS Win 11 Pro
Ram 32 GB
Python 3.11
Table 5: Experimental parameters setting.
Parameter Name Parameter setting
Learning rate 0.001
Batch size 32
Optimizer Adam
Activation function SoftMax
Beta1 0.9
Beta2 0.999
Amsgrad False
Iteration epochs 100
(TN), and False Negative (FN), the confusion matrix
produces four assessment metrics: accuracy, recall,
specificity, and F1 score. The formula for each metric
is given below.
Precision =
T P
T P + FP
(1)
Recall =
T P
T P + FN
(2)
F1 − score = 2 ×
Precision × Recall
Precision + Recall
(3)
It is evident from the confusion matrix displayed
in Figure 3 that the majority of classification errors
occur in scenarios where there is a distinction be-
tween cedar rust and scab. One of the main reasons
for this is that the complex background context of the
data set facilitates errors in judgment. The early dis-
ease spots in the Scab and Cedar rust dataset are pri-
marily small-scale features with intricate background
settings. Another reason for that is the texture of
the affected areas might appear irregular or spotted,
which is a common feature between the two diseases.
Additionally, the impact of nearby leaves that fall into
different health categories results in classification er-
rors and insufficient reliable information about the fi-
nal marks that the model obtains. Nonetheless, the
model’s overall effect still complies with the require-
ments of the leaf disease detection scenario. Figure
4 displays precision, recall, and F1 scores for each
class.
Figure 3: Confusion matrix of the MFFN.
Figure 4: Classification performance of the MFFN.
Table 6: Training and validation results.
Metrics Training Validation
Accuracy 99.36% 98.90%
Loss 0.1072 0.1140
3.2 Experimental Results
The MFFN model was trained with an apple dataset
for four classes. These classes contain different types
A Multi-Scale Feature Fusion Network for Detecting and Classifying Apple Leaf Diseases
17
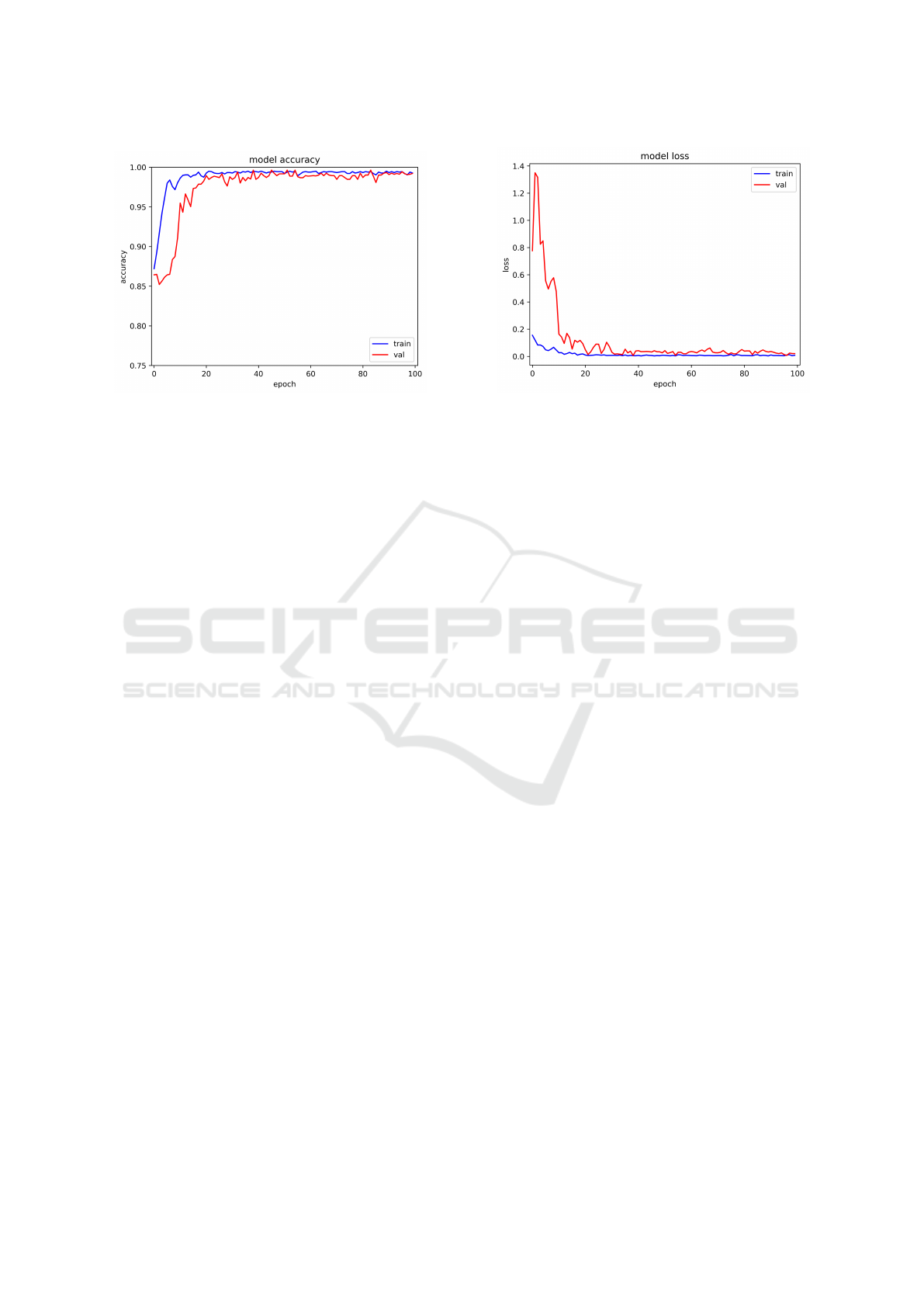
(a) Training and validation accuracy (b) Training and validation loss
Figure 5: Training and validation results.
of apple leaf diseases including Black Rot disease,
Cedar Rust disease, and Scab disease. Our model was
trained for 100 epochs to achieve the best accuracy in
training and validation. The outcomes of the assess-
ment highlighted areas in need of development, of-
fered insightful information about the model’s func-
tionality, and improved it even further. Finally, the
performance review showed that our suggested model
is a strong solution in its field and very successful at
accomplishing the aims it set out to do.
The preciseness of the training and validation of
the suggested model demonstrated remarkable accu-
racy. The model performed well, as evidenced by the
average training and validation accuracy of 99.36%
and 98.90%, respectively. Furthermore, it was noted
that the model’s thoroughness was further supported
by the training and validation losses, which were
0.1072 and 0.1140, respectively. Figure 5 and Table
6 indicate that our proposed model achieved the best
performance during training on the apple plant village
dataset for apple diseases classification.
3.3 Model Performance
The efficiency of the proposed MFFN model has been
compared with other multi-scale CNN models to con-
firm its performance. Each model is trained while
maintaining the aforementioned experimental proce-
dure and experimental setting, and the outcomes are
shown in Table 7. Our model demonstrates an excel-
lent stability, as demonstrated by its high initial iden-
tification accuracy, low error rate, and rapid conver-
gence throughout both training and validation.
Improvements have been observed in the disease
classification of Apple leaves using deep learning
techniques. The suggested approach represents a ma-
jor advancement in the detection and classification of
Apple leaf diseases, enabling precise and effective re-
sults. It is worthwhile to note that the recommended
model can be made even better by adding more data
sources, adjusting hyperparameters, and investigating
cutting-edge deep learning architectures.
3.4 Comparison Between Different
Approaches and Our Proposed
Model
To examine the effectiveness and performance of the
proposed MFFN, we compare our model with 10
other techniques that are used to classify Apple leaf
diseases. The outcomes are contrasted with those of
other pre-trained models, such as the built exclusively
model. Table 8 provides an overview of the findings.
From the table we find out that our suggested method
performs significantly better than pre-trained mod-
els and built models. From the comparison between
the techniques and MFFN in Table 8, we observed
that our provided MFFN model has an accuracy rate
higher than the other architectures in the field of Ap-
ple leaf diseases classification.
4 CONCLUSION AND FUTURE
WORKS
Apple leaf diseases can significantly affect produc-
tivity and quality by causing early fruit drops and
compromising the appearance, size, and flavor of the
fruit. This can lead to financial losses for farmers
and a reduction in the overall quality of apple con-
sumption. As a result, it can lead to the economic
decline of the agricultural industry. Therefore, early
diagnosis of apple leaf disease and preventative mea-
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
18

Table 7: MFFN performance compared with various multi-scale CNN.
Reference Plant Methodology Accuracy
Ren et al. (Ren et al., 2023) Apple Multi-scale parallel fusion 97.20%
Ahmed et al. (Ahmed et al., 2022) Apple Multi-Contextual Feature Fusion Network 90.86%
Luo et al. (Luo et al., 2021) Apple Multi-scale extraction of disease features 94.23%
Suo et al. (Suo et al., 2022) Grape Multi-scale fusion module 95.95%
Tian et al. (Tian et al., 2022) Apple Multi-scale feature extraction 83.19%
Proposed MFFN Apple Our proposed model 98.28%
Table 8: Comparison between different approaches and our proposed model on Apple leaf diseases.
Reference Methodology Plant Accuracy
Zhang et al. (Zhang et al., 2021) ResNet34 Apple 93.76%
Sangeetha et al. (Sangeetha et al., 2022) VGG16 Apple 93.30%
Zhao et al. (Zhao and Huang, 2023) CBAM-ENetV2 Network Apple 97.49%
Alsayed et al. (Alsayed et al., 2021) ResNetV2 Apple 94.00%
Sheng et al. (Sheng et al., 2022) MobileViT-based Apple 96.76%
Rehman et al. (Rehman et al., 2021) Mask RCNN Apple 96.60%
Chakraborty et al. (Chakraborty et al., 2021) SVM Apple 96.00%
Assad et al. (Assad et al., 2023) AppleNet Apple 96.00%
Chen et al. (Chen et al., 2023) ResNet Apple 97.78%
Yu et al. (Yu et al., 2022) ResNet-50 Apple 95.70%
Proposed MFFN MFFN Apple 98.28%
sures can reduce the economic loss. Multiscale CNNs
can enhance apple leaf disease classification by ana-
lyzing local and global features. They accommodate
size variability in lesions, handle texture changes, and
make the model more robust to real-world variations.
Therefore, we utilized deep learning based on a multi-
scale CNN approach to identify apple leaf diseases
with a high test accuracy rate of 98.28%. The demon-
strated method has shown significant and robust re-
sults in evaluation metrics such as precision, recall,
and F1-score. After applying a deep learning model
to the apple leaf dataset to detect apple leaf diseases
in their earlier stages and achieving promising re-
sults, this simulation can be deployed as a web- or
mobile-based application. Such applications will help
farmers detect apple leaf diseases in real life. In fu-
ture works, our proposed multi-scale CNN technique
can be implemented to classify different plant species.
Besides, it could be employed to identify other apple
leaf diseases, making it easier for farmers and produc-
ers to categorize multiple diseases. Overall, the im-
plementation of the proposed model may help farmers
make better-informed decisions.
REFERENCES
Ahmed, M. R., Ashrafi, A. F., Ahmed, R. U., and Ahmed,
T. (2022). Mcffa-net: Multi-contextual feature fusion
and attention guided network for apple foliar disease
classification. In 2022 25th International Conference
on Computer and Information Technology (ICCIT),
pages 757–762. IEEE.
Alsayed, A., Alsabei, A., and Arif, M. (2021). Classifica-
tion of apple tree leaves diseases using deep learning
methods. International Journal of Computer Science
& Network Security, 21(7):324–330.
Assad, A., Bhat, M. R., Bhat, Z., Ahanger, A. N., Kun-
droo, M., Dar, R. A., Ahanger, A. B., Dar, B., et al.
(2023). Apple diseases: detection and classification
using transfer learning. Quality Assurance and Safety
of Crops & Foods, 15(SP1):27–37.
Baranwal, S., Khandelwal, S., and Arora, A. (2019). Deep
learning convolutional neural network for apple leaves
disease detection. In Proceedings of International
Conference on Sustainable Computing in Science,
Technology & Management (SUSCOM-2019), pages
260–267.
Barbedo, J. G. (2018). Factors influencing the use of deep
learning for plant disease recognition. Biosystems En-
gineering, 172:84–91.
Chakraborty, S., Paul, S., and Rahat-uz Zaman, M. (2021).
Prediction of apple leaf diseases using multiclass sup-
port vector machine. In 2nd International Conference
on Robotics, Electrical and Signal Processing Tech-
niques (ICREST), pages 147–151.
Chen, Y., Pan, J., and Wu, Q. (2023). Apple leaf disease
identification via improved cyclegan and convolu-
tional neural network. Soft Computing, 27(14):9773–
9786.
Divakar, S., Bhattacharjee, A., and Priyadarshini, R. (2021).
Smote-dl: A deep learning based plant disease detec-
tion method. In 6th International Conference for Con-
vergence in Technology (I2CT), pages 1–6.
A Multi-Scale Feature Fusion Network for Detecting and Classifying Apple Leaf Diseases
19

Doutoum, A. S. and Tugrul, B. (2023). A review of leaf
diseases detection and classification by deep learning.
IEEE Access, 11:119219–119230.
Elizar, E., Zulkifley, M. A., Muharar, R., Zaman, M. H. M.,
and Mustaza, S. M. (2022). A review on multiscale-
deep-learning applications. Sensors, 22(19):7384.
G
¨
und
¨
uz, H. and Yılmaz G
¨
und
¨
uz, S. (2022). Plant disease
classification using ensemble deep learning. In 2022
30th Signal Processing and Communications Applica-
tions Conference (SIU), pages 1–4.
Hughes, D. P. and Salath
´
e, M. (2015). An open ac-
cess repository of images on plant health to en-
able the development of mobile disease diagnostics
through machine learning and crowdsourcing. CoRR,
abs/1511.08060.
Luo, Y., Sun, J., Shen, J., Wu, X., Wang, L., and Zhu, W.
(2021). Apple leaf disease recognition and sub-class
categorization based on improved multi-scale feature
fusion network. IEEE Access, 9:95517–95527.
Nandhini, S. and Ashokkumar, K. (2022). An automatic
plant leaf disease identification using densenet-121 ar-
chitecture with a mutation-based henry gas solubility
optimization algorithm. Neural Computing and Ap-
plications, 34(7):5513–5534.
Rehman, Z. U., Khan, M. A., Ahmed, F., Dama
ˇ
sevi
ˇ
cius, R.,
Naqvi, S. R., Nisar, W., and Javed, K. (2021). Recog-
nizing apple leaf diseases using a novel parallel real-
time processing framework based on mask rcnn and
transfer learning: An application for smart agriculture.
IET Image Processing, 15(10):2157–2168.
Ren, H., Chen, W., Liu, P., Tang, R., Wang, J., and Ge, C.
(2023). Multi-scale parallel fusion convolution net-
work for crop disease identification. In 2023 8th In-
ternational Conference on Cloud Computing and Big
Data Analytics (ICCCBDA), pages 481–485. IEEE.
Sangeetha, K., Rima, P., Kumar, P., Preethees, S., et al.
(2022). Apple leaf disease detection using deep learn-
ing. In 6th International Conference on Computing
Methodologies and Communication (ICCMC), pages
1063–1067.
Sheng, X., Wang, F., Ruan, H., Fan, Y., Zheng, J., Zhang,
Y., and Lyu, C. (2022). Disease diagnostic method
based on cascade backbone network for apple leaf
disease classification. Frontiers in Plant Science,
13:994227.
Su, J., Zhang, M., and Yu, W. (2022). An identification
method of apple leaf disease based on transfer learn-
ing. In 7th International Conference on Cloud Com-
puting and Big Data Analytics (ICCCBDA), pages
478–482.
Suo, J., Zhan, J., Zhou, G., Chen, A., Hu, Y., Huang, W.,
Cai, W., Hu, Y., and Li, L. (2022). Casm-amfmnet: A
network based on coordinate attention shuffle mech-
anism and asymmetric multi-scale fusion module for
classification of grape leaf diseases. Frontiers in Plant
Science, 13:846767.
Tian, L., Zhang, H., Liu, B., Zhang, J., Duan, N., Yuan, A.,
and Huo, Y. (2022). Vmf-ssd: A novel v-space based
multi-scale feature fusion ssd for apple leaf disease
detection. IEEE/ACM Transactions on Computational
Biology and Bioinformatics.
Yadav, A., Thakur, U., Saxena, R., Pal, V., Bhateja, V.,
and Lin, J. C.-W. (2022). Afd-net: Apple foliar dis-
ease multi classification using deep learning on plant
pathology dataset. Plant and Soil, 477(1):595–611.
Yan, Q., Yang, B., Wang, W., Wang, B., Chen, P., and
Zhang, J. (2020). Apple leaf diseases recognition
based on an improved convolutional neural network.
Sensors, 20(12):3535.
Yu, H., Cheng, X., Chen, C., Heidari, A. A., Liu, J., Cai, Z.,
and Chen, H. (2022). Apple leaf disease recognition
method with improved residual network. Multimedia
Tools and Applications, 81(6):7759–7782.
Zhang, D., Yang, H., and Cao, J. (2021). Identify apple leaf
diseases using deep learning algorithm. arXiv preprint
arXiv:2107.12598, abs/2107.12598.
Zhao, G. and Huang, X. (2023). Apple leaf disease recog-
nition based on improved convolutional neural net-
work with an attention mechanism. In 5th Interna-
tional Conference on Natural Language Processing
(ICNLP), pages 98–102.
DATA 2025 - 14th International Conference on Data Science, Technology and Applications
20
