
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with
Split Neural Networks for Distributed Patient Data
Bashair Alrashed
1 a
, Priyadarsi Nanda
1 b
, Hoang Dinh
1 c
, Amani Aldahiri
2
, Hadeel Alhosaini
2
and Nojood Alghamdi
2
1
Faculty of Engineering and Information Technology, University of Technology Sydney, Sydney, Australia
2
Faculty of Computing and Information Technology, University of Jeddah, Jeddah, Saudi Arabia
Keywords:
Privacy-Preserving, Vertical Federated Learning, Split Neural Networks, Patient Data.
Abstract:
Medical data privacy regulations pose significant challenges for sharing raw data between healthcare institu-
tions. These challenges are particularly critical when the data is vertically partitioned. In such scenarios, each
healthcare provider holds unique but complementary patient information. This makes collaborative learning
challenging while protecting patient privacy. As a result, developing effective machine learning models that
require integrated data becomes unfeasible. This leads to fragmented analyses and less effective patient care.
To address this issue, we developed a vertical federated learning framework using split neural networks to
enable secure collaboration while preserving privacy. The framework comprises three main stages: generating
symmetric keys to establish secure communication, aligning overlapping patient records across institutions
using a privacy-preserving record linkage algorithm, and collaboratively training a global machine learning
model without revealing patient privacy. We evaluated the framework on three well-known medical datasets.
Our evaluation focused on two critical scenarios: varying degrees of overlap in patient records and differing
feature distributions. The proposed framework ensures patient privacy and compliance with strict regulations,
providing a scalable and practical solution for real-world healthcare networks. It effectively addresses key
challenges in privacy-preserving collaborative machine learning.
1 INTRODUCTION
Over the past decade, the rapid digitization of health
systems and the exponential growth of digital med-
ical data have transformed the healthcare landscape.
This evolution offers new opportunities to revolution-
ize medical research and improve patient care deliv-
ery. Machine learning (ML) algorithms provide re-
searchers with new ways to efficiently analyze and
manage medical data. These advances drive innova-
tions that improve outcomes and streamline health-
care processes. Such algorithms enable predictive,
personalized, and cost-effective data management.
They can analyze medical images and patient records
to predict diseases and help healthcare providers de-
velop effective treatment plans. Moreover, they help
prevent complications by enabling early disease de-
tection through advanced medical systems. For ex-
a
https://orcid.org/0000-0002-7393-5355
b
https://orcid.org/0000-0002-5748-155X
c
https://orcid.org/0000-0002-9528-0863
ample, (Riedel et al., 2023) employed ResNetFed, a
modified ResNet50 model, to detect COVID-19 pneu-
monia on chest radiographs. Similarly, (Mali et al.,
2023) used artificial neural models to predict heart
disease.
The integration of ML algorithms into medical
systems delivers significant benefits to healthcare
providers, patients, and society. Despite their poten-
tial, ML applications in medical research face signif-
icant challenges. One key challenge is the distribu-
tion of medical data. Privacy regulations, such as
HIPAA and GDPR, restrict data sharing across health-
care providers, preventing the creation of centralized
repositories (Antunes et al., 2022). Hence, the data
remain within organizational boundaries.
In fact, patient records are distributed across mul-
tiple healthcare providers or institutions rather than
centralized in a single repository (Allaart et al., 2022).
(Allaart et al., 2022) also believed that the distribu-
tion of medical data generally follows two patterns:
horizontal or vertical, as shown in Figure 1. The hor-
izontal distribution involves sharing similar features
Alrashed, B., Nanda, P., Dinh, H., Aldahiri, A., Alhosaini, H., Alghamdi and N.
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data.
DOI: 10.5220/0013445300003979
In Proceedings of the 22nd International Conference on Security and Cryptography (SECRYPT 2025), pages 13-24
ISBN: 978-989-758-760-3; ISSN: 2184-7711
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
13

(a) Horizontal.
(b) Vertical.
Figure 1: Data Distribution Types.
across different population groups. In contrast, the
vertical distribution involves sharing data about the
same individuals, but with different attributes. For
example, in the vertical distribution, a data provider
might store attributes such as patient ID, gender, and
heart attack status. Another might have details such
as gender, age, and ST slope. This distributed nature
of medical data highlights the need for a decentral-
ized ML framework that enables collaborative model
training without sharing raw data. This framework
ensures the privacy of sensitive medical information.
Therefore, (McMahan et al., 2017) introduced a
new decentralized approach to collaboratively train
the global model without compromising data privacy,
known as federated learning (FL). FL has two main
categories based on the data distribution: horizontal
federated learning (HFL) and vertical federated learn-
ing (VFL). In HFL, each training sample shares the
same feature space. As a result, each data provider
creates its own local model independently based on its
local training samples. These local models are then
used to iteratively train a global model. This kind
of learning improves the performance of the global
model and resolves the data-shortage problem when
the data size is limited. Since HFL requires all data
providers to access the same feature space, it cannot
be directly applied to vertically distributed data.
To address the limitations of HFL for vertically
partitioned data, a VFL was introduced. Unlike HFL,
VFL enables collaboration between institutions that
hold different but complementary feature sets. To fa-
cilitate deep learning in VFL, split neural networks
were introduced (Vepakomma et al., 2018). This ar-
chitecture partitions the neural network layers among
participants, ensuring privacy by exchanging only in-
termediate outputs and gradients instead of full neu-
ral network updates. Since data providers possess
distinct feature sets, this method allows collabora-
tive model training without exposing raw data. Re-
cent research has widely adopted this architecture as
a baseline for VFL frameworks. For example, (Sun
et al., 2023) optimized communication efficiency in
split learning, while (Anees et al., 2024) explored its
application in scenarios with limited overlap between
participants, addressing real-world data sharing chal-
lenges.
These studies often ignore practical implementa-
tion details. They also fail to evaluate split learning
performance on diverse datasets, feature distributions,
and overlap conditions, leaving key VFL challenges
unresolved. Data heterogeneity is one of the main
challenges in the VFL framework. It requires han-
dling diverse feature distributions, which can degrade
model performance. In addition, incomplete overlap
between healthcare providers complicates the record
linking and training process in the VFL framework.
Moreover, none of the existing studies provides sys-
tematic evaluations across diverse datasets and real-
world scenarios.
Therefore, to address these limitations, we pro-
pose a novel privacy-preserving VFL framework us-
ing split neural networks (PPVFL-SplitNN). It is
designed to enable secure and efficient collabora-
tion among healthcare providers while preserving pa-
tient privacy. PPVFL-SplitNN incorporates three
key stages. First, symmetric key generation estab-
lishes a secure communication channel among par-
ticipants, preventing unauthorized data access during
the linking and training process. Next, record link-
age uses privacy-preserving algorithms to accurately
align overlapping patient records across institutions
while enabling error-tolerant comparisons. Finally,
split model training exchanges intermediate embed-
dings and gradients instead of raw data to collabora-
tively train a global model. These components ad-
dress data heterogeneity, limited participant overlap,
and strict privacy constraints, offering a practical so-
lution for vertically partitioned medical data.
The proposed framework has been evaluated on
three diverse medical datasets under varying over-
lap percentages and feature distributions. The results
show that the framework achieves predictive perfor-
mance comparable to centralized learning (CL) while
preserving privacy. This makes it a robust and secure
solution for collaborative model training. To the best
of our knowledge, this is the first work to systemat-
ically evaluate split learning across a broad range of
distributed patient data scenarios. It highlights the po-
tential of split learning to enable effective collabora-
tive learning in real-world healthcare networks. The
main contributions of this paper are summarized as
follows:
• Development of a Privacy-Preserving VFL
Framework. The proposed framework trains
split neural networks that are distributed among a
server and a number of healthcare providers. This
framework is significant as it enables collabora-
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
14

tive model training on vertically partitioned med-
ical data while preserving patient privacy. It en-
sures compliance with data protection regulations
and addresses challenges such as incomplete over-
lap and data heterogeneity.
• Implementation of Key Stages. The framework
includes three core stages: symmetric key gen-
eration, record linkage, and training split learn-
ing model. These stages ensure secure commu-
nication, accurate alignment of overlapping pa-
tient records, and collaborative training without
sharing raw data. Together, they address critical
challenges such as privacy preservation, data het-
erogeneity, and limited overlap. This enables se-
cure and efficient model training on vertically par-
titioned datasets.
• Comprehensive Evaluation and Identification
of Challenges. We evaluated the framework’s
predictive performance on three diverse medical
datasets with varying overlap percentages and fea-
ture distributions. The results demonstrate its ro-
bustness in real-world conditions. The results
show that the framework achieves predictive ac-
curacy and F1 scores comparable to CL while
preserving privacy. However, the evaluation re-
veals key challenges in current VFL frameworks,
including communication overhead, suboptimal
performance under limited overlap, and sensitiv-
ity to heterogeneous feature distributions. These
findings highlight areas for future research, such
as improving record linkage algorithms, optimiz-
ing communication efficiency, and enhancing ro-
bustness against feature heterogeneity.
2 SYSTEM OVERVIEW
2.1 System Design
This study proposes a privacy-preserving VFL frame-
work designed to address the challenges of training
ML models on vertically partitioned medical data.
The system consists of two types of entities: a cen-
tral server and multiple clients (healthcare providers),
as illustrated in Figure 2a.
A. Server. In a medical context, the server acts as a
central authority, such as a hospital group or an-
alytics provider. It ensures data privacy during
collaborative training. Figure 2a highlights the
server’s primary roles including:
• Symmetric Key Generation. It is respon-
sible for generating and distributing crypto-
(a) Entities Types.
(b) Architecture.
Figure 2: Framework Overview.
graphic keys for secure communication during
the record linkage and training process.
• Record Linkage. The server identifies over-
lapping patients across participating hospitals
using their identifier attributes. Techniques
like the Bloom filter are used to align patient
records while safeguarding privacy.
• Training and Updating Global Model. It has
the capability to store all the labels and the
global model, as shown in Figure 2b. In ad-
dition, it has the computing power to train and
analyze the distributed model and make pre-
dictions by aggregating the embeddings from
healthcare providers. The server is also respon-
sible for updating the global model and calcu-
lating the gradients to update the local model.
B. Client. Each client corresponds to a healthcare
provider, such as hospitals and laboratories, that
holds complementary patient attributes. For ex-
ample, in Figure 2a, Client 1 is a Cardiology
Clinic holding data on patient’s heart and blood
vessels. Client m stores radiological images, like
chest X-rays and MRIs, to help diagnose diseases
and plan treatments. Each healthcare provider can
usually store a large number of training samples
that can be used to train the ML model locally, as
shown in Figure 2b. As feedback, each client re-
ceives from the server the indices of the overlap
records and the gradients to shuffle its local data
and update the local model, respectively. Note
that any client can act as a “VFL server” if it has
the labels. We commonly refer to it as an ac-
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data
15
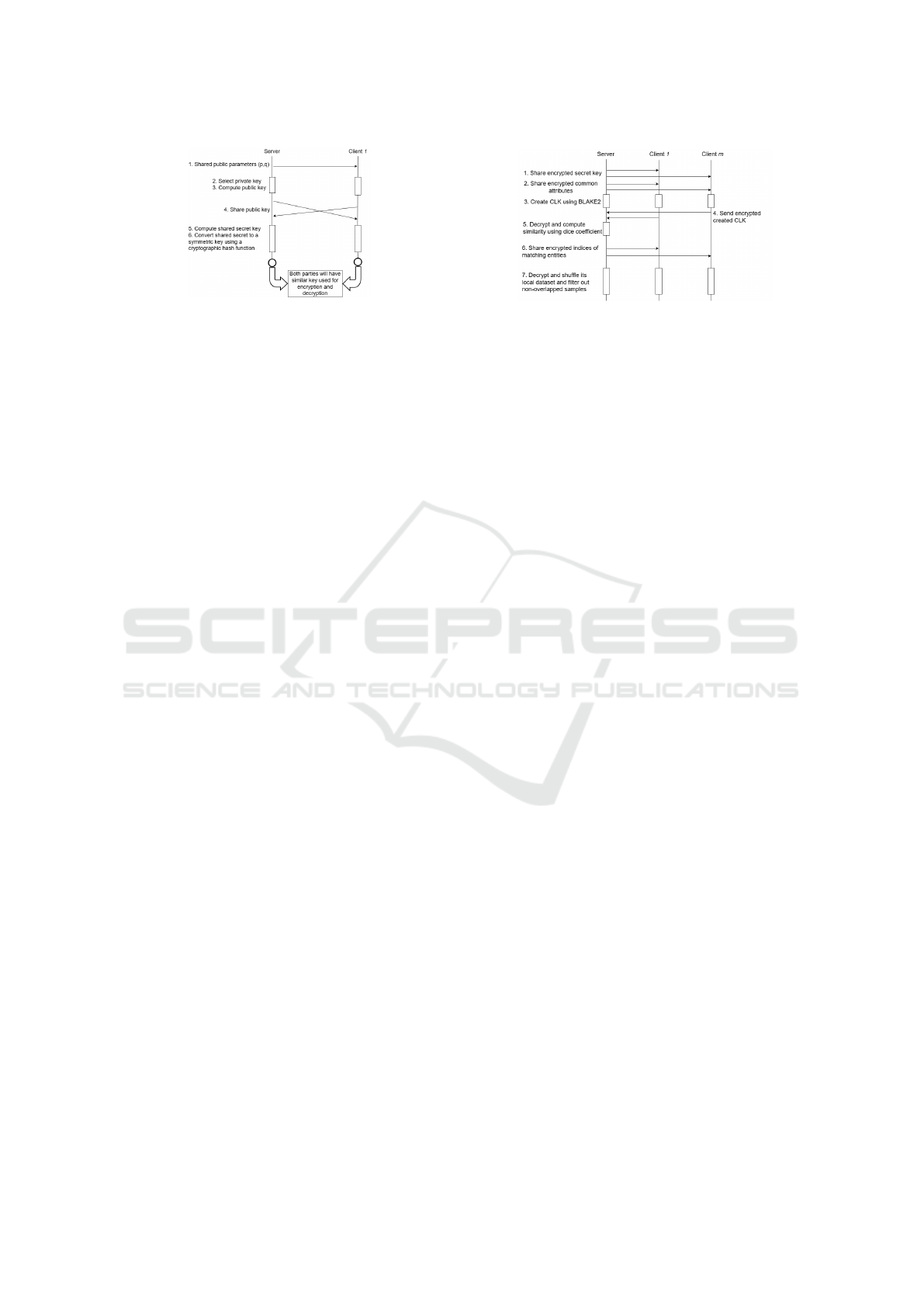
Figure 3: Symmetric Key Generation.
tive party, while a passive party refers to the client
holding features only.
In this paper, we adopt a semi-honest model in
which all entities honestly follow the protocol but ex-
ploit any opportunity to extract private data from in-
termediate results generated during the execution of
the proposed system. Therefore, each client cannot
interact with each other directly.
2.2 Proposed Framework
The proposed framework has been divided into three
main stages: symmetric key generation, record link-
age, and training and updating a global model. Each
stage involves the roles of the server and the clients.
• Stage 1: Symmetric Key Generation. The
server agrees first on two public parameters (g
and p) where p should be a large prime number
and g is a primitive root modulo p (Rodriguez-
Henriquez et al., 2007), as follows:
g
k
mod p, ∀k ∈ {1, 2, . . . , p −1}. (1)
Then, each participant chooses a secret key and
exchanges its public key with the server only.
Next, each participant computes the shared secret
key using the shared public key. The secret key
should be the same for both parties (server and
healthcare provider). Then, each participant con-
verts the shared secret key to a symmetric key us-
ing a secure cryptographic hash function such as
SHA-256.
Figure 3 shows the complete process of the sym-
metric key generation. After generating the sym-
metric key, all messages between the server and
the client will be encrypted and decrypted using
the generated key. Adding this stage to the pro-
posed system helps enhance its security level dur-
ing the record linkage and model training.
• Stage 2: Record Linkage. The first preprocess-
ing step in VFL to start a collaboration training
process is to link distributed records that belong to
the same sample ID anonymously using a privacy-
preserving algorithm. This algorithm is called a
Figure 4: Record Linkage Process.
long-term cryptographic key (CLK) and was pro-
posed in (Hardy et al., 2017) and (Nock et al.,
2018). It uses Bloom filters to preserve the pri-
vacy of identifier attributes while enabling error-
tolerant comparisons.
At this stage, each participant must securely re-
ceive a hashing secret key from the server, along
with essential identifier attributes that uniquely
distinguish each patient. These attributes, in-
cluding patient ID, age, and gender, are critical
for ensuring accurate and reliable identification
throughout the process. The server encrypts these
information using the generated symmetric key
and shares them with all participants. Each client
decrypts these information using the same sym-
metric key and starts clustering the training sam-
ple using the K-means algorithm to minimize the
mean distances between the user data points and
their closest cluster centers. Followed by creating
a set of CLKs for each entity using a BLAKE2
hash function. Then, each client encrypts and ex-
changes the created CLKs with the server only.
The server decrypts and computes the similarity
between the three sets of CLKs using a dice co-
efficient. It then extracts the indices of all possi-
ble pairs above the given threshold. After aligning
the overlapping patient records using the privacy-
preserving algorithm, the server encrypts the in-
dices of the matched records and securely shares
them with all clients. This ensures that each client
can identify and use only the aligned records for
collaborative training without compromising pa-
tient privacy.
Figure 4 shows the complete process of the record
linkage between three parties, a server and two
clients, in order to find matching records without
revealing patient privacy. It is important to note
that all participants must formalize a combination
of personal characteristics, such as age and gen-
der, in the same data formats and presentations
before starting the linking process. The data for-
malization process ensures that similar records are
matched accurately while maintaining the confi-
dentiality of the data involved.
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
16

Figure 5: Model Overview.
• Stage 3: Training Split Learning Model. Due
to the medical data heterogeneity problem in
VFL, split learning is used to enable collaborative
model training while preserving patient privacy.
This approach offers several advantages, includ-
ing enhanced privacy, efficient communication,
and adaptability to heterogeneous data distribu-
tions. At this stage, each client trains the bottom
model of the global model using the overlapped
samples only. Then, each client shares only the
output of the trained model, known as embed-
dings, with the server instead of sharing the com-
plete model parameters or the patient raw data in
order to preserve the patient’s privacy, as shown in
Figure 5. The server concatenates all the received
embeddings and feeds them as input to the server-
side top model. It completes the training process,
makes predictions, and computes the gradients
required to update the global model. To main-
tain the privacy-preserving advantage, the server
splits the computed gradients for each client and
shares them individually. Each client then up-
dates its local model using its respective gradients.
This process ensures that raw data and client-side
model details remain private throughout the train-
ing process. Split learning reduces communica-
tion overhead by avoiding the need to share com-
plete model parameters. It also works well with
vertically partitioned datasets, where healthcare
providers hold different features.
Figure 6 illustrates the complete process of train-
ing and updating the global model that splits into
two sub-models: the bottom and the top mod-
els located at the clients and the server side, re-
spectively. This training process iterates until the
model converges or a maximum number of itera-
tions is met.
Figure 6: Training Split Learning Model.
3 PRIVACY-PRESERVING
ALGORITHM
This section presents the formalization process of the
privacy-preserving record linkage algorithm and the
practical implementation of the split learning algo-
rithm.
3.1 Record Linkage Algorithm
Several solutions have been proposed to link the med-
ical record, including traditional merging techniques,
record linkage toolkit (De Bruin, 2019), dedupe
(Gregg and Eder, 2022), and splink (Linacre et al.,
2022). However, these solutions do not guarantee
the preservation of patient privacy when perform-
ing the record linkage process. Therefore, to ad-
dress this issue, we select the CLK algorithm to
link matching records without compromising individ-
ual privacy. This method was introduced by (Hardy
et al., 2017; Nock et al., 2018) to link related records
anonymously. It encodes identifier attributes using
BLAKE2, which is a family of hash functions (Au-
masson et al., 2014). This method also uses Bloom
filters to construct a set of CLKs as follows:
clk =
k
∑
j=1
(l
j
), (2)
where k represents the number of different indepen-
dent hash functions to compute the indices for an en-
try, and l is the length of the bit array. Using the
BLAKE2 hash function to encode identifier attributes,
the possibility of a collision attack is minimized (Au-
masson et al., 2014).
Next, the constructed CLKs are used by the server
to assess the similarity between the two clients (A and
B) as follows:
m
i
=
1 i f D
clk
A
∼ D
clk
B
, and
0 otherwise
, (3)
where the operator ∼ can be interpreted as “the most
likely match”.
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data
17

Figure 7: Compute the Similarity between Two sets of
CLKs.
More precisely, the server uses the Dice coeffi-
cient algorithm to compare between bit strings as fol-
lows:
D
A,B
=
2h
a + b
, (4)
where h represents the number of bit positions that are
set to 1 in both bit strings, a denotes the number of bit
positions set to 1 in A, and b denotes the number of
bit positions set to 1 in B (Schnell et al., 2009).
Figure 7 shows how the identifier attributes in
client 1 are hashed using the BLAKE2 hash function
along with the secret hash key to add an additional
level of security to the record linking algorithm. Af-
ter creating the CLKs, each participant sends its own
set of CLKs to the server. The server then measures
the similarity between the two sets of CLKs to extract
the indices of the overlapped samples x
m
o
.
In this paper, we consider that there are com-
mon identifier attributes shared between all clients
to uniquely identify the same sample ID using the
method proposed in (Hardy et al., 2017; Nock et al.,
2018). This assumption is often considered in real-
world healthcare systems to match similar records
across institutions accurately (Sun et al., 2022).
Therefore, the training sample set of each client is
divided into overlapped samples X
m
o
∈ R
N
m
o
×d
m
and
non-overlapped samples X
m
n
∈ R
N
m
n
×d
m
, where N
m
o
and N
m
n
represent the number of training samples
in the two datasets, respectively N
m
= N
m
o
+ N
m
n
.
In addition, the server stores the ground truth labels
Y
o
∈ 0, 1
N
o
×C
for the overlapped samples, where C
represents the possible number of classes.
Algorithm 1 describes a general procedure of
record linkage using CLK. The server first generates
the secret hash key and calculates the encryption pa-
rameters, which are distributed to all clients, as shown
in Figure 4. The server also selects and specifies iden-
tifier attributes to uniquely identify each sample and
shares the encrypted attribute with the participants.
Then, using the K-means algorithm, each participant
clusters its own data to minimize the mean distances
between the user data points and their closest cluster
centers. Next, each client uses a BLAKE2 hash
Input: hashing secret key (HSKey) and identifier attributes
(IDAttr)
Output: X
m
o
∈ R
N
m
o
×d
m
Server:;
∥HSKey∥, ∥IDAttr∥ ← encrypt(HSKey, IDAttr);
Send ∥HSKey∥ and ∥IDAttr∥ to all clients;
for each client m = 1, 2, . . . , M in parallel do
HSKey, IDAttr = decrypt(∥HSKey∥, ∥IDAttr∥);
clusterData f rame =
∑
n
i=0
min(||x
i
− µ
j
||
2
);
clks =
∑
k
j=1
l
j
;
Send clks to the server;
end
Server:;
X
m
o
=
2h
a+b
← Equation (4) ;
Send the index of X
m
o
to all clients;
for each client m = 1, 2, . . . , M in parallel do
X
m
o
← reshiftDataFrame(X
m
o
);
end
Algorithm 1: Privacy Preserving Record Linkage Algo-
rithm.
function to implement Bloom filters and create the set
of CLKs with Equation (2) and sends it to the server.
The server computes the similarity between the three
sets of CLKs using the dice coefficient method ac-
cording to Equation (4) and returns the results X
m
o
to
all participating clients to re-shift their local data us-
ing reshiftDataFrame function.
After the record Linkage process, each participant
has to delete the non-overlap samples X
m
n
∈ R
N
m
n
×d
m
and uses only the overlapped samples X
m
o
∈ R
N
m
o
×d
m
to train the local neural network. Finally, using this
algorithm, which combines personally identifiable at-
tributes, the proposed system is able to link individual
records and extract machining records while preserv-
ing patient privacy.
3.2 Split Learning Algorithm
Each client at this stage can start training the split
learning model using the overlapped samples X
m
o
only in the privacy-preserving setting. The server ini-
tializes the training parameters θ
s
and sends them to
all clients. Each client trains the local model h
m
o
with
parameters θ
m
o
={x
i
m
o
, b
i
m
o
}. The output of the local
model train is called embedding or feature embed-
ding, which represents the data patterns within each
client. h
m
o
is defined as follows:
u
0
m
o
= x
m
o
,
u
i
m
o
= σ
i
w
i
m
o
u
i−1
+ b
i
m
o
, i ∈ {1, 2, . . . , I},
h
m
o
= u
I
m
o
,
(5)
where σ⊙ is a linear function and u
i
m
o
is the ith
layer of the neural network (Li et al., 2023). The
server receives and concatenates all client embedding
vectors in a weighted manner (i.e. w=[h
1
⊙;...; h
m
⊙]).
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
18

Concatenated embedding vectors act as input to the
server top model θ
0
that is connected to the interac-
tive layer to predict ˆy
n
o
. The server then calculates the
loss function L ⊙ as follows:
f (Θ) = L (h
s
(θ
0
, w); y
n
),
with w =
h
1
(θ
1
;d
1
)
.
.
.
h
m
(θ
m
;d
m
)
, m ∈ {1, 2, . . . , M},
(6)
where Θ = θ
M
i=0
represents the global model that con-
tains M local models (θ
1
, ..., θ
M
) and the top model θ
0
(Li et al., 2023).
The server calculates the gradients of the global
model
αι
αΘ
(
t)
to update the global model Θ
t+1
. It also
computes the gradients for each client
αι
αh
m
o
and sends
them back to all the clients. Then, each client per-
forms a backward propagation and updates its local
model as follows:
∇
θ
m
ι =
∂ι
∂θ
m
=
∑
i
∂ι
∂h
i−1
m
∂h
i−1
m
∂θ
m
, (7)
Next, each client obtains the new h
t+1
m
o
and sends
it to the server. The server repeats the process until
the global model converges or the maximum number
of iterations is met.
Algorithm 2 describes the pressure for standard
VFL training based on split neural networks using
adaptive moment estimation (ADAM). The server
first initializes the top model θ
0
and sends the ini-
tialization parameters to all clients. Each client then
trains and computes the local model output h
m
o
=
σ(θ
m
, x
m
o
) in a mini-batch β of samples X
m
o
and sends
h
m
o
to the server. With all {h
m
o
}
M
(m=1)
, the server
concatenates the embeddings in a weighted manner
(w = [h
1
⊙;...; h
m
⊙]) and computes the loss function
following Equation (6). It also updates its global
model Θ using the calculated gradients
αι
αΘ
. Next, the
server computes the gradients
αι
αh
m
for each client and
sends them back to all clients. Finally, each client
computes and updates its local model θ
m
with Equa-
tion (7). This procedure iterates until the global model
Θ converges or the maximum number of iterations is
met.
4 PERFORMANCE EVALUATION
The effectiveness of the proposed framework is eval-
uated on three well-known medical datasets. The first
section describes the simulation setups, including the
setting of three datasets and training parameters. We
Input: Feature data {X
m
}
M
m=1
, learning rate η, batch size β,
number of rounds T
Output: Global model parameters Θ
Server: Initialize top model parameters Θ
(0)
and send Θ
(0)
to all
clients;
for each round t = 0, 1, . . . , T − 1 do
for each client m = 1, 2, . . . , M in parallel do
Client m computes h
m
= σ(θ
m
· X
m
);
Client m sends h
m
to the server;
end
Server: w = {h
m
}
M
m=1
;
L
(t)
= L( f (Θ
(t)
, w), y);
Θ
(t+1)
= Θ
(t)
− η∇
Θ
L
(t)
;
∇
h
m
L
(t)
for each client’s output;
Send ∇
h
m
L
(t)
to all clients;
for each client m = 1, 2, . . . , M in parallel do
Client m ∇
θ
m
L
(t)
;
Client m θ
(t+1)
m
= θ
(t)
m
− η∇
θ
m
L
(t)
;
end
end
Algorithm 2: Training Split Learning Model.
then discuss the obtained results in detail. All exper-
iments are performed on a single machine using an
Intel (R) Core (TM) i7-8565U CPU.
4.1 Experimental Setups
4.1.1 Datasets
General information is provided on the three datasets
that were used to train and test the split learning
model: Diabetes Prediction Dataset (Mustafa, 2023),
Breast Cancer (Wolberg, 1990) and Gliomas (Tasci,
Erdal et al., 2022). The description of each dataset is
as follows.
• Diabetes Prediction. This dataset is a public col-
lection of medical and demographic data from the
Kaggle website. It is used to predict the possi-
bility of developing diabetes in patients based on
their medical history and demographic records.
It initially contains 100,000 records, each with
eight features along with the patient’s diabetes sta-
tus that is categorized as “Yes” and “No” indi-
cating the presence or absence of diabetes. For
this dataset, the learning rate is set to 0.001, and
the batch size is 256. Furthermore, the dataset
is significantly imbalanced, which can lead the
model to disproportionately favor the majority
class (e.g., non-diabetic cases) during training. To
address this issue and ensure fair representation, it
is crucial to reduce the volume of data in the ma-
jority class, thereby achieving a balanced distribu-
tion with the minority class (e.g., diabetic cases).
• Breast Cancer. This database was obtained from
the University of Wisconsin Hospitals in 1992. It
is used to classify the cell nuclei of breast masses
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data
19

as malignant or benign. Initially, it contains 699
records, each with nine multivariate attributes,
along with the target value that describes whether
breast cancer is benign or malignant. For this
dataset, the learning rate is set to 0.001, and the
batch size is 32.
• Glioma. This dataset represents the histological
medical records of patients with brain tumor (i.e.,
glioma) grading. It initially contains 839 records,
each with 20 mutated genes and three clinical fea-
tures along with the grade value that determines
whether a patient is a lower grade glioma or a mul-
tiforme glioblastoma. For this dataset, the learn-
ing rate is set to 0.001, and the batch size is 32.
To train the ML model efficiently, the quality of the
input data must be maintained because it significantly
impacts the output results. Therefore, it is essen-
tial to preprocess the selected datasets before making
predictions by cleaning the data, checking for miss-
ing data and duplicate records. There are no missing
data for the diabetes prediction dataset. However, it
has 3854 duplicated records. Therefore, duplicated
records have been deleted. Furthermore, records that
do not provide valuable information, such as a person
with unclear gender information or records with “no
information” in the smoking history variable, were
not included. However, the breast cancer dataset has
some missing data and duplications. Due to the lim-
itation of the dataset size, the missing values were
filled in with the mean, and duplicate records were
kept the same as in the Glioma dataset. Finally, the
datasets are divided into two portions: training and
testing using the following ratio 8:2.
4.1.2 Training Details
The proposed framework is designed to handle verti-
cally partitioned data efficiently while ensuring data
privacy. In this setup, the training features are split
vertically between two healthcare providers and the
labels are stored exclusively on the server side. Each
participant is equipped with specific neural network
components tailored to the dataset being trained.
For the Diabetes Prediction and Breast Cancer
datasets, each healthcare provider trains a bottom
model consisting of two fully connected layers with
Linear-ReLU activations. The server holds the top
model, which comprises a single Linear-Sigmoid
layer. The units of these fully connected layers are
16, 8 and 1, respectively.
For the Glioma dataset, which has a higher feature
dimensionality, the architecture is expanded to ac-
commodate the complexity of the data. Each client’s
bottom model consists of two fully connected Linear-
ReLU layers, while the server’s top model includes
three Linear-ReLU layers followed by a sigmoid out-
put layer. The units of these fully connected layers
are 32, 16, 16, 8, and 1, respectively.
The models are initialized with random weights
using PyTorch’s default initialization method to en-
sure consistent starting conditions across all training
rounds. The ADAM optimizer is used for training,
with a learning rate of 0.001 to balance convergence
speed and stability. Binary cross-entropy is used as
the loss function to handle binary classification tasks
effectively.
The training process involves 200 rounds of com-
munication between the clients and the server. During
each round, the clients process their local data and
compute intermediate embeddings, which are sent to
the server. The server concatenates these embeddings,
trains the top model, and computes gradients that are
propagated back to update global and local models.
This iterative process ensures the privacy of raw data
while enabling collaborative model training.
4.2 Performance Metrics
The performance of the proposed system is evaluated
using three key metrics: Accuracy, F1 Score, and the
Confusion Matrix.
1. Accuracy. Measures the proportion of correctly
predicted samples to the total predictions, reflect-
ing the overall performance of the model.
2. F1 Score. It captures the model’s ability to bal-
ance precision and recall, minimizing false posi-
tives (FP) and false negatives (FN). This is crucial
in healthcare applications due to the challenges
posed by imbalanced datasets.
3. Confusion Matrix. Visualizes the performance
of the classification model, showing the distribu-
tion of true positives (TP), true negatives (TN),
FP, and FN.
4.3 Evaluating Split Learning
Algorithm
The performance of the proposed PPVFL-SplitNN
framework is evaluated against four scenarios:
• Baseline (CL). In this scenario, all data are inte-
grated into a single repository to train a CL model
and achieve optimal performance. However, this
approach violates privacy regulations by requiring
raw data sharing (Blue line in the plots).
• PPVFL-SplitNN (Stander). The proposed
framework incorporates privacy-preserving tech-
niques such as symmetric key generation, record
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
20
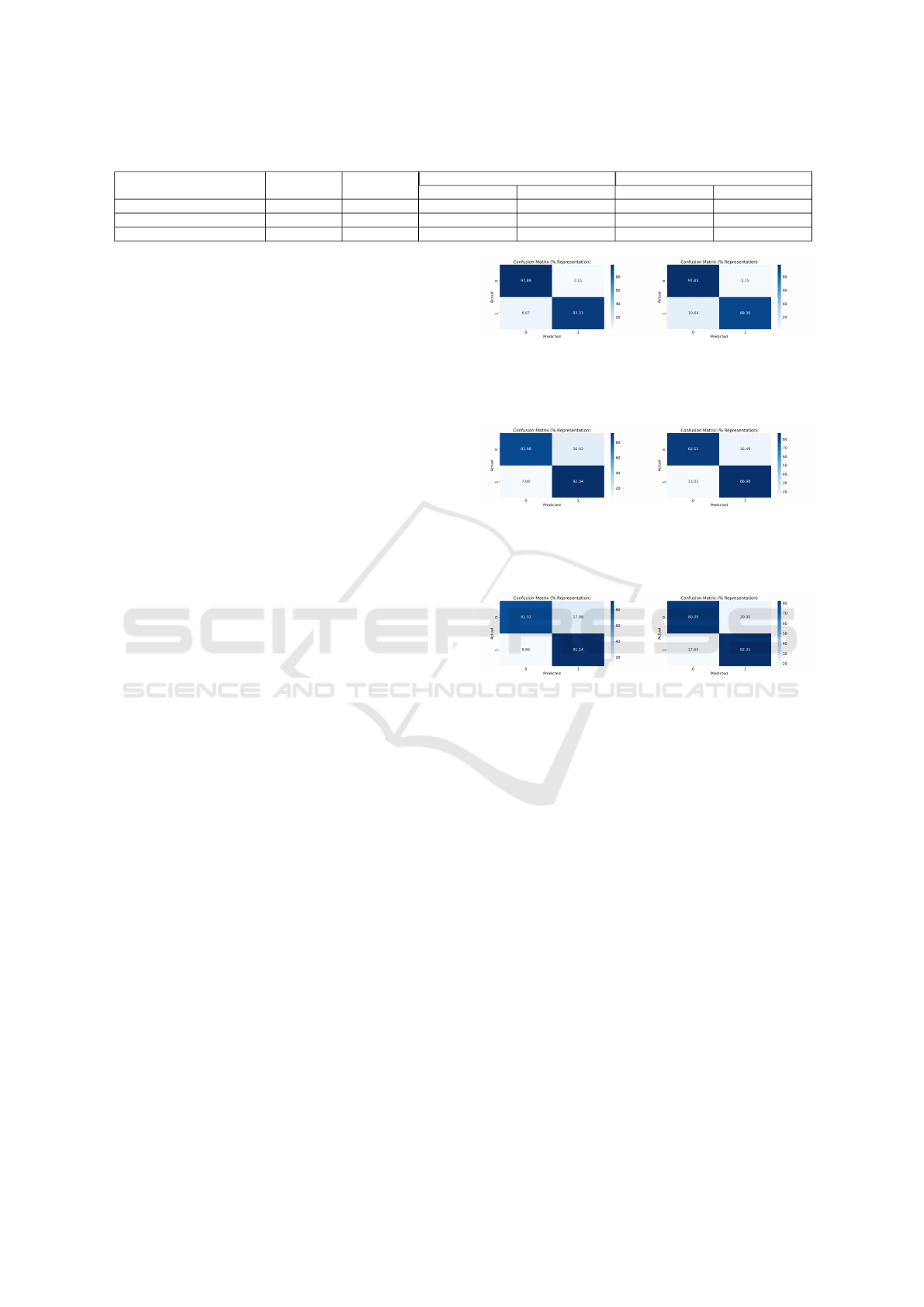
Table 1: Evaluation Metrics in Comparison between the Centralize and the PPVFL-SplitNN framework.
Database Name
Training
Samples
Testing
Samples
Centralize PPVFL-SplitNN
Accuracy (%) F1 Score (%) Accuracy (%) F1 Score (%)
Diabetes Prediction 5908 1478 88.70 90.22 85.38 86.55
Breast Cancer Wisconsin 559 140 96.42 94.38 95 92.30
Gliomas 671 168 86.30 86.22 80.95 77.77
linkage, and split learning. It represents the ideal
scenario with fully overlapping records and con-
sistent feature distributions (Orange line in the
plots).
• PPVFL-SplitNN + Varying Overlap Percent-
age. Evaluates the framework in conditions with
limited overlap among participants, simulating
real-world healthcare challenges (Green line in
the plots).
• PPVFL-SplitNN + Differ Feature Distribution.
Tests the framework’s ability to handle data het-
erogeneity, where participants hold diverse fea-
ture distributions (Red line in the plots).
Table 1 presents the evaluation metrics of the model
trained with the PPVFL-SplitNN framework com-
pared to the CL, while Figures 8 to 10 visualize the
confusion matrices for each dataset, showcasing the
number of TP, TN, FP and FN. These results demon-
strate that our framework achieves comparable perfor-
mance to the CL framework while preserving patient
privacy.
However, a slight performance gap is observed
between the two approaches. This gap can be pri-
marily attributed to the communication overhead in
split learning, which introduces latency and slows
down convergence due to the exchange of interme-
diate embeddings and gradients between the server
and clients. In contrast, CL benefits from seamless
data integration and optimization within a single en-
vironment. Additionally, split learning suffers from
the lack of end-to-end gradient optimization across all
model layers. Since only partial gradients are visible
during training, updates to the bottom and top models
may become suboptimal, especially when the client
data features are highly heterogeneous. Despite these
challenges, the confusion matrices in Figures 8 to 10
indicate that our framework achieves similar TP and
TN values as CL, demonstrating its effectiveness for
binary classification tasks.
In addition, Table 2 shows that our work achieves
comparable results to those reported by (Guo et al.,
2020), (Tasci et al., 2022) and (Fadillah et al., 2023).
However, there are significant differences in the
methodology and the system design. Specifically,
(Tasci et al., 2022; Fadillah et al., 2023) relied on the
CL approach, which requires the aggregation of raw
data from all healthcare providers in a single repos-
(a) CL. (b) PPVFL-SplitNN.
Figure 8: Show a Comparison between the CL, and the
Model Trained with PPVFL-SplitNN Framework when us-
ing the Breast Cancer.
(a) CL. (b) PPVFL-SplitNN.
Figure 9: Show a Comparison between the CL, and the
Model Trained with PPVFL-SplitNN Framework when us-
ing the Diabetes Prediction.
(a) CL. (b) PPVFL-SplitNN.
Figure 10: Show a Comparison between the CL, and the
Model Trained with PPVFL-SplitNN Framework when us-
ing the Gliomas Prediction.
itory. Although their methods achieved high predic-
tive performance, CL raises substantial privacy con-
cerns, particularly in healthcare, where data sensitiv-
ity is critical and regulations restrict data sharing. On
the other hand, (Guo et al., 2020) adopted an HFL
approach, achieving an F1 score of 0.88 and an ac-
curacy of 0.91, results close to those obtained by our
framework. However, HFL assumes horizontally par-
titioned data, where data providers share the same fea-
tures but for different patients. This assumption limits
the applicability of their method in vertically parti-
tioned settings, where different providers hold com-
plementary features for the same individuals.
Our work addresses this limitation by imple-
menting a PPVFL-SplitNN framework, which en-
ables collaborative training across vertically parti-
tioned datasets while preserving privacy. Importantly,
our method achieves predictive performance compa-
rable to (Guo et al., 2020) while operating under
stricter data constraints. By exchanging only inter-
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data
21
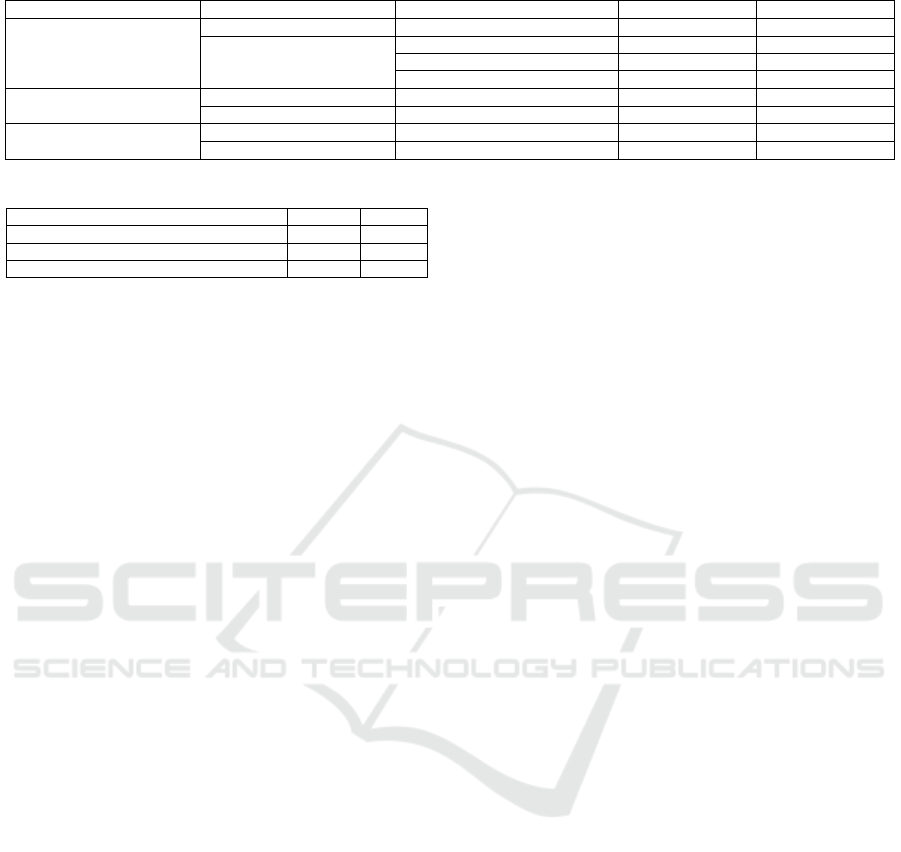
Table 2: Evaluation Metrics of the CL.
Database Name Reference Model Type Accuracy (%) F1 Score (%)
Diabetes Prediction
PPVFL-SplitNN NN 88.70 90.22
(Fadillah et al., 2023)
K-Nearest Neighbors 87 86.64
Random Forest 90.70 90.41
Logistic Regression 88.64 88.45
Breast Cancer Wisconsin
PPVFL-SplitNN NN 96.42 94.38
(Guo et al., 2020) NN 95 92
Gliomas
PPVFL-SplitNN NN 86.30 86.22
(Tasci et al., 2022) SVM + RF + AdaBoost 86.4 84.2
Table 3: Training Accuracy (%) of Split Learning Model.
Reference MNIST Titanic
PPVFL-SplitNN 81.16 68.16
Flower (Beutel et al., 2020) - 65
PyVertical (Romanini et al., 2021) 91.982 -
mediate embeddings and gradients, our framework
ensures compliance with privacy regulations and en-
hances suitability for real-world healthcare applica-
tions by eliminating the need to share raw data.
On the other hand, in VFL, we compare the train-
ing accuracy of our framework with existing frame-
works such as Flower (Beutel et al., 2020) and PyVer-
tical (Romanini et al., 2021) using two well-known
datasets: MNIST and Titanic. As shown in Table
3, our results are comparable to these frameworks,
with a slight improvement in training accuracy. In
comparison, Flower provides a general purpose FL
framework. However, it does not explicitly focus
on vertical partitioning, which is essential in medical
datasets where data are distributed between health-
care providers with complementary features. Simi-
larly, PyVertical focuses on vertical data. However, it
does not implement advanced techniques for privacy
record linkage or consider varying overlap percent-
ages, which are critical factors affecting performance
in real-world scenarios.
By addressing these limitations, our framework
ensures better alignment of shared records and im-
proved training efficiency, leading to a slight yet con-
sistent improvement in training accuracy. These re-
sults demonstrate the practical applicability of our
framework for vertically partitioned medical data,
where privacy and performance must be balanced si-
multaneously. The effectiveness of our framework
is further evaluated in two distinct scenarios to ana-
lyze its performance under realistic medical data chal-
lenges. These evaluations highlight the framework’s
robustness in handling varying overlap percentages
and feature distributions, reflecting the complexities
of real-world healthcare applications.
4.3.1 Impact of Overlap Percentage
In this scenario, we consider the effect of incomplete
overlap, where fewer than 100% of patient records
are shared across participating hospitals. This sit-
uation reflects real-world challenges, such as frag-
mented healthcare systems where not all patients
have records in every hospital, particularly in ru-
ral or under-resourced areas. As shown in Table 4,
lower overlap percentages (e.g., 60%) lead to a slight
degradation in model accuracy and F1 scores due
to the reduced number of shared samples available
for training. This limits the server’s ability to ag-
gregate meaningful embeddings across participants,
impacting global model performance. However, the
degradation is not dramatic, demonstrating the robust-
ness of our framework under incomplete data con-
ditions. These findings highlight the importance of
robust record linkage techniques to maximize shared
sample alignment and suggest opportunities to lever-
age non-overlapping samples for better data utiliza-
tion in future work.
4.3.2 Impact of Feature Distribution
In this section, we investigate the effect of the fea-
ture distribution on the model performance using the
three medical datasets. Randomized feature distri-
butions introduce redundancy, imbalance, and noise,
which degrade accuracy and F1 scores compared to
manually engineered distributions. As shown in Ta-
ble 5, this degradation underscores the critical role of
feature engineering in VFL. For instance, integrating
heterogeneous data sources, such as imaging labora-
tories (holding radiology data) and clinical databases
(storing demographic and test results), requires care-
ful feature selection to ensure meaningful contribu-
tions from all participants. Thus, performing fea-
ture engineering or feature selection within the VFL
framework becomes essential to maintain model per-
formance.
4.4 Performance Analysis
The testing accuracy and F1 scores for the evaluated
scenarios are presented in Figures 11 to 13. The re-
sults confirm that the CL achieves higher and more
stable performance due to seamless data integration
and full gradient optimization. However, CL is im-
practical for healthcare applications because of pri-
vacy regulations and the sensitive nature of patient
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
22
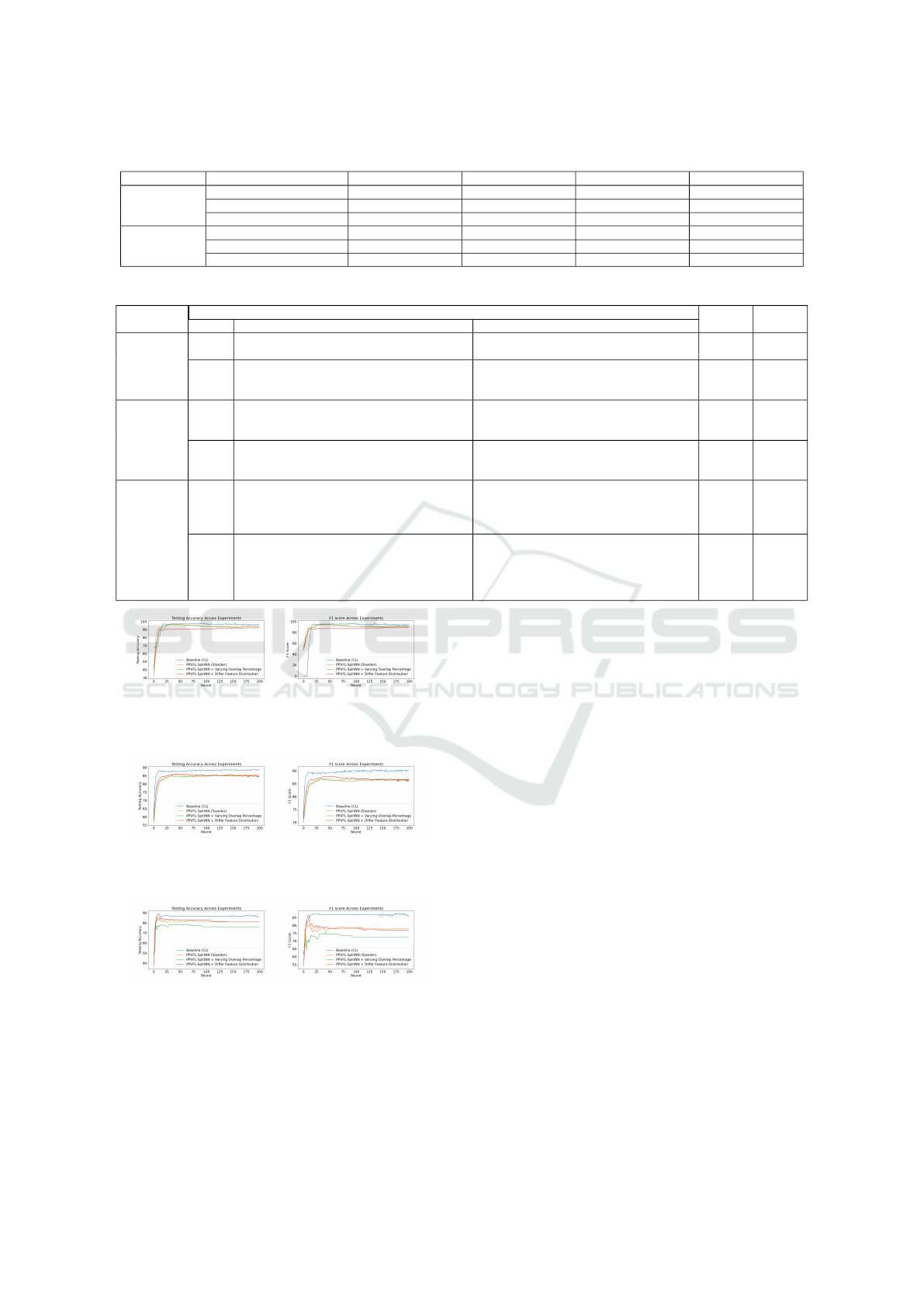
Table 4: Evaluation Metrics of Our Framework in Case if the Number of Overlap Records is Different.
Overlap (%) Database Name Training Samples Testing Samples Accuracy (%) F1 Score (%)
100
Diabetes Prediction 5908 1478 85.38 86.55
Breast Cancer Wisconsin 559 140 95 92.30
Gliomas 671 168 80.95 77.77
60
Diabetes Prediction 3544 887 84.44 85.91
Breast Cancer Wisconsin 335 84 94.04 91.22
Gliomas 402 101 76.23 72.72
Table 5: Evaluation Metrics of Our Framework based on the Feature Distribution.
Database
Name
Feature Distribution Accuracy
(%)
F1 Score
(%)Type Client 1 Client 2
Diabetes
Prediction
manual New ID, gender, age, hypertension, heart dis-
ease
New ID, gender, age, smoking history, bmi,
HbA1c level, blood glucose level
85.38 86.55
random New ID, gender, age, hypertension, blood glu-
cose level, HbA1c level, BMI, smoking history
(e.g., ever, current)
New ID, gender, age, heart disease, smoking
history (e.g., never, not current, former)
84.57 86.11
Breast Cancer
Wisconsin
manual New ID, Clump thickness, Uniformity of cell
size, Uniformity of cell shape, Marginal adhe-
sion
New ID, Single epithelial cell size, Bare nu-
clei, Bland chromatin, Normal nucleoli, Mi-
toses
95 92.30
random New ID, Bland chromatin, Uniformity of cell
shape, Uniformity of cell size, Normal nucle-
oli, Single epithelial cell size
New ID, Bare nuclei, Mitoses, Marginal ad-
hesion, Clump thickness
92.14 89.32
Gliomas
manual New ID, Gender, Age at diagnosis, IDH1,
TP53, ATRX, PTEN, EGFR, CIC, MUC16,
PIK3CA, NF1, PIK3R1, Race
New ID, Gender, Age at diagnosis,
FUBP1, RB1, NOTCH1, BCOR, CSMD3,
SMARCA4, GRIN2A, IDH2, FAT4,
PDGFRA
80.95 77.77
random New ID, Gender, Age at diagnosis, FUBP1,
NF1, ATRX, BCOR, PDGFRA, PTEN,
MUC16, TP53, GRIN2A, EGFR, RB1,
NOTCH1, Race (e.g., american indian or
alaska native, white)
New ID, Gender, Age at diagnosis,
PIK3CA, IDH2, CIC, PIK3R1, FAT4,
CSMD3, IDH1, SMARCA4, Race (e.g.,
black or african American, asian)
80.95 77.14
(a) Testing Accuracy. (b) F1-Score.
Figure 11: Show the Model Performance under the Evalu-
ated Scenarios when using Breast Cancer.
(a) Testing Accuracy. (b) F1-Score.
Figure 12: Show the Model Performance under the Evalu-
ated Scenarios when using the Diabetes Prediction.
(a) Testing Accuracy. (b) F1-Score.
Figure 13: Show the Model Performance under the Evalu-
ated Scenarios when using the Gliomas Prediction.
data.
In contrast, our proposed framework preserves
medical data privacy while achieving performance
comparable to CL. This demonstrates its practical-
ity for collaborative model training in healthcare net-
works. However, careful data utilization is critical
to avoid model degradation. Scenarios with reduced
overlap or randomized feature distributions highlight
the need for robust record linkage and feature engi-
neering techniques to maintain model performance.
5 CONCLUSIONS AND FUTURE
WORKS
In this paper, we proposed a privacy-preserving VFL
framework that uses split learning to address chal-
lenges in the training of ML models on vertically par-
titioned data. Our framework ensures privacy preser-
vation, data security, and collaboration among health-
care providers in real-world scenarios. Evaluations on
three medical datasets show that the proposed frame-
work achieves a performance comparable to CL while
preserving patient privacy. It demonstrates robust-
ness in handling incomplete overlap and diverse fea-
ture distributions, offering a practical solution for sen-
sitive healthcare networks. These findings highlight
the potential of our frameworks for advancing med-
ical research and patient care while maintaining pri-
vacy. Future work should focus on optimizing record
linkage and reducing communication overhead to im-
prove scalability and efficiency in large-scale settings.
PPVFL-SplitNN: Privacy-Preserving Vertical Federated Learning with Split Neural Networks for Distributed Patient Data
23

REFERENCES
Allaart, C. G., Keyser, B., Bal, H., and Van Halteren, A.
(2022). Vertical split learning-an exploration of pre-
dictive performance in medical and other use cases. In
2022 International Joint Conference on Neural Net-
works (IJCNN), pages 1–8. IEEE.
Anees, A., Field, M., and Holloway, L. (2024). A neural
network-based vertical federated learning framework
with server integration. Engineering Applications of
Artificial Intelligence, 138:109276.
Antunes, R. S., Andr
´
e da Costa, C., K
¨
uderle, A., Yari,
I. A., and Eskofier, B. (2022). Federated learning for
healthcare: Systematic review and architecture pro-
posal. ACM Transactions on Intelligent Systems and
Technology (TIST), 13(4):1–23.
Aumasson, J.-P., Meier, W., Phan, R. C.-W., Henzen, L.,
Aumasson, J.-P., Meier, W., Phan, R. C.-W., and Hen-
zen, L. (2014). Blake2. The Hash Function BLAKE,
pages 165–183.
Beutel, D. J., Topal, T., Mathur, A., Qiu, X., Fernandez-
Marques, J., Gao, Y., Sani, L., Kwing, H. L., Par-
collet, T., Gusm
˜
ao, P. P. d., and Lane, N. D. (2020).
Flower: A friendly federated learning research frame-
work. arXiv preprint arXiv:2007.14390.
De Bruin, J. (2019). Python Record Linkage Toolkit: A
toolkit for record linkage and duplicate detection in
Python.
Fadillah, M. I., Aminuddin, A., Rahardi, M., Abdulloh,
F. F., Hartatik, H., and Asaddulloh, B. P. (2023). Dia-
betes diagnosis and prediction using data mining and
machine learning techniques. In 2023 International
Workshop on Artificial Intelligence and Image Pro-
cessing (IWAIIP), pages 110–115. IEEE.
Gregg, F. and Eder, D. (2022). Dedupe. https://github.com/
dedupeio/dedupe.
Guo, Y., Liu, F., Cai, Z., Chen, L., and Xiao, N. (2020).
Feel: A federated edge learning system for efficient
and privacy-preserving mobile healthcare. In Pro-
ceedings of the 49th International Conference on Par-
allel Processing, pages 1–11.
Hardy, S., Henecka, W., Ivey-Law, H., Nock, R., Patrini, G.,
Smith, G., and Thorne, B. (2017). Private federated
learning on vertically partitioned data via entity reso-
lution and additively homomorphic encryption. ArXiv
Preprint ArXiv:1711.10677.
Li, A., Huang, J., Jia, J., Peng, H., Zhang, L., Tuan, L. A.,
Yu, H., and Li, X.-Y. (2023). Efficient and privacy-
preserving feature importance-based vertical feder-
ated learning. IEEE Transactions on Mobile Comput-
ing.
Linacre, R., Lindsay, S., Manassis, T., Slade, Z., Hep-
worth, T., Kennedy, R., and Bond, A. (2022). Splink:
Free software for probabilistic record linkage at scale.
International Journal of Population Data Science,
7(3):1794.
Mali, B., Saha, S., Brahma, D., Pinninti, R., and Singh, P. K.
(2023). Towards building a global robust model for
heart disease detection. SN Computer Science, 4(5):1–
12.
McMahan, B., Moore, E., Ramage, D., Hampson, S., and
y Arcas, B. A. (2017). Communication-efficient learn-
ing of deep networks from decentralized data. In Ar-
tificial intelligence and statistics, pages 1273–1282.
PMLR.
Mustafa, M. (2023). Diabetes prediction dataset.
https://www.kaggle.com/datasets/iammustafatz/
diabetes-prediction-dataset.
Nock, R., Hardy, S., Henecka, W., Ivey-Law, H., Patrini,
G., Smith, G., and Thorne, B. (2018). Entity resolu-
tion and federated learning get a federated resolution.
ArXiv Preprint ArXiv:1803.04035.
Riedel, P., von Schwerin, R., Schaudt, D., Hafner, A., and
Sp
¨
ate, C. (2023). ResNetFed: Federated deep learning
architecture for privacy-preserving pneumonia detec-
tion from COVID-19 chest radiographs. Journal of
Healthcare Informatics Research, pages 1–22.
Rodriguez-Henriquez, F., Perez, A. D., Saqib, N. A., and
Koc, C. K. (2007). A brief introduction to modern
cryptography. Cryptographic Algorithms on Recon-
figurable Hardware, pages 7–33.
Romanini, D., Hall, A. J., Papadopoulos, P., Titcombe, T.,
Ismail, A., Cebere, T., Sandmann, R., Roehm, R., and
Hoeh, M. A. (2021). Pyvertical: A vertical federated
learning framework for multi-headed splitnn. arXiv
preprint arXiv:2104.00489.
Schnell, R., Bachteler, T., and Reiher, J. (2009). Privacy-
preserving record linkage using bloom filters. BMC
medical informatics and decision making, 9:1–11.
Sun, C., van Soest, J., Koster, A., Eussen, S. J., Schram,
M. T., Stehouwer, C. D., Dagnelie, P. C., and Dumon-
tier, M. (2022). Studying the association of diabetes
and healthcare cost on distributed data from the maas-
tricht study and statistics netherlands using a privacy-
preserving federated learning infrastructure. Journal
of Biomedical Informatics, 134:104194.
Sun, J., Xu, Z., Yang, D., Nath, V., Li, W., Zhao, C., Xu, D.,
Chen, Y., and Roth, H. R. (2023). Communication-
efficient vertical federated learning with limited over-
lapping samples. In Proceedings of the IEEE/CVF
International Conference on Computer Vision, pages
5203–5212.
Tasci, E., Zhuge, Y., Kaur, H., Camphausen, K., and
Krauze, A. V. (2022). Hierarchical voting-based fea-
ture selection and ensemble learning model scheme
for glioma grading with clinical and molecular charac-
teristics. International Journal of Molecular Sciences,
23(22):14155.
Tasci, Erdal, Camphausen, Kevin, Krauze, Andra Valentina,
and Zhuge, Ying (2022). Glioma Grading Clinical and
Mutation Features. UCI Machine Learning Reposi-
tory. DOI: https://doi.org/10.24432/C5R62J.
Vepakomma, P., Gupta, O., Swedish, T., and Raskar, R.
(2018). Split learning for health: Distributed deep
learning without sharing raw patient data. arXiv
preprint arXiv:1812.00564.
Wolberg, W. (1990). Breast Cancer Wisconsin (Orig-
inal). UCI Machine Learning Repository. DOI:
https://doi.org/10.24432/C5HP4Z.
SECRYPT 2025 - 22nd International Conference on Security and Cryptography
24
