
Proposal for a Common Conceptual Schema for Metadata Integration
and Reuse Across Diverse Scientific Domains
Marcos Sfair Sunye
1 a
, Karolayne Costa Rodrigues de Lima
1 b
, Alberto Abell
´
o
2 c
and Elisabete Ferreira
1 d
1
Department of Informatics, Universidade Federal do Paran
´
a, Centro Polit
´
ecnico, Curitiba, Brazil
2
ESSI, Universitat Polit
`
ecnica de Catalunya, Carrer de Jordi Girona, Barcelona, Spain
Keywords:
Metadata Integration, Scientific Data, Metadata Standards, Conceptual Model.
Abstract:
Addresses challenges in reusing scientific data, highlighting the necessity for enhanced metadata standards to
facilitate data integration across various scientific domains. The primary contribution is a standardized concep-
tual schema for metadata, designed to improve data integration and reuse. This schema outlines the phenomena
and methods in scientific research to enable better identification and integration of datasets. The schema was
synthesized by analyzing four well-established metadata standards: DataCite, Darwin Core, Ecological Meta-
data Language, and Dublin Core. This analysis aimed to identify semantic correspondences among these stan-
dards to create a unified conceptual schema. The analysis validated the practicality of the proposed schema.
More than 11% of the 926 metadata elements analyzed were successfully integrated, demonstrating significant
potential to improve data integration across different scientific domains. The proposed common conceptual
schema for metadata improves data integration and reuse in scientific research. By introducing structured de-
scriptions of phenomena and methods, it facilitates the discovery of relationships between datasets, promoting
integrated and interdisciplinary reuse without additional metadata creation costs.
1 INTRODUCTION
The growth of specialized data repositories has in-
creased metadata standards across disciplines, en-
hancing data organization but complicating integra-
tion. Multidisciplinary repositories face challenges
in interoperability, which is critical for reusing and
integrating data across fields. Integrating datasets
has shown the potential to generate new knowledge
(Zimmermann, 2008), supported by initiatives like
the Data Documentation Initiative (Center, 2020) and
FAIR (Framework, 2020).
Despite advancements in metadata standards, data
reuse remains limited, with studies showing a small
percentage of resources in repositories being effec-
tively cited (Koesten et al., 2020; Wallis et al., 2011;
He and Nahar, 2016). This limited reuse is often
restricted to contextually relevant datasets with reli-
able descriptive attributes (Zimmerman, 2003; Zim-
a
https://orcid.org/0000-0002-2568-5697
b
https://orcid.org/0000-0002-6311-8482
c
https://orcid.org/0000-0002-3223-2186
d
https://orcid.org/0000-0002-8405-7906
mermann, 2008). Although most repositories adopt
metadata standards, fields such as title or date are
insufficient for precise searches, necessitating costly
manual interventions (Loffler et al., 2021; Park and
Tosaka, 2010). Fragmentation and inconsistencies in
metadata further hinder cross-disciplinary integration
efforts, highlighting the need for improved metadata
integration strategies (Gregory et al., 2020; Mutschke
et al., 2020).
We propose a generic conceptual model, the ”Inte-
grated Context Metadata Core,” focusing on scientific
elements: phenomena and methods. This core builds
on existing standards, establishing correlations and
facilitating reuse. By defining phenomena and meth-
ods (inquiry process), this model bridges datasets
through explicit descriptions (Porto and Spaccapietra,
2011). Although phenomena and methods exist in
current standards, they are often dispersed, especially
in multidisciplinary frameworks.
Our contributions include introducing the ”Inte-
grated Context Metadata Core” to consolidate criti-
cal elements for reuse and emphasizing the role of
metadata describing phenomena and methods. This
innovation supports better integration and interdisci-
278
Sunye, M. S., Lima, K. C. R., Abelló, A. and Ferreira, E.
Proposal for a Common Conceptual Schema for Metadata Integration and Reuse Across Diverse Scientific Domains.
DOI: 10.5220/0013291500003929
In Proceedings of the 27th International Conference on Enterprise Information Systems (ICEIS 2025) - Volume 1, pages 278-285
ISBN: 978-989-758-749-8; ISSN: 2184-4992
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

plinary collaboration.
The article organizes its content into five main sec-
tions. The introduction outlines the challenges and
motivations for metadata integration. In the Back-
ground section, the authors discuss fundamental con-
cepts and review existing metadata standards, includ-
ing DataCite, Dublin Core, EML, and Darwin Core.
The Methods section explains the methodology, de-
tailing the analysis of metadata standards and the ap-
plication of the Metadata Crosswalk method to iden-
tify semantic correspondences. The Results section
highlights the findings, focusing on the Integrated
Context Metadata Core (ICMC) proposal, a novel ty-
pology that consolidates key elements for describing
scientific phenomena and methods. Finally, the au-
thors summarize the study’s contributions, emphasiz-
ing the importance of ICMC for scientific data inte-
gration and reuse and proposing directions for future
research.
2 BACKGROUND
The benefits of data reuse are varied, highlight-
ing the advantages that involve maximizing the effi-
ciency of invested research resources, promoting in-
novation, and fostering interdisciplinary collabora-
tion among researchers and institutions, thereby pro-
moting broader and more diverse research networks
(Curty et al., 2017; Silva, 2019). With access to
data from various fields, researchers can perform
more complex and integrative analyses, crossing dis-
ciplinary boundaries that would otherwise be limited
by the lack of specific data. This approach facilitates
new discoveries and broadens the understanding that
data can be applied in practical contexts, contributing
to innovative solutions to complex problems.
In this context, metadata plays a crucial role in de-
scribing research data, thereby facilitating easier un-
derstanding, access, and effective utilization. Meta-
data are defined as ”data about data” (Borgman,
2015), and ”a statement about a potentially informa-
tive object” (Pomerantz, 2015, p. 73). These defi-
nitions are essential for understanding how data are
generated, used, and restricted, and for providing key
details about the author, location, access, and usage
rights.
Metadata standards like Dublin Core (DC), Eco-
logical Metadata Language (EML), DataCite, and
Darwin Core, among others, define common sets
of metadata elements and their encoding formats to
promote interoperability across various systems and
fields. These standards are tailored to accommodate a
range of requirements and scenarios, from simple bib-
liographic descriptions to the long-term preservation
of digital assets (Pomerantz, 2015).
Integrating metadata standards is a strategic ap-
proach to developing a unified conceptual framework.
This process includes the alignment of various stan-
dards to build a stronger metadata infrastructure that
supports a diverse range of disciplinary and institu-
tional needs. The effective integration of metadata
standards not only enhances the exchange and inter-
operability between different systems and communi-
ties but also increases the utility of scientific data by
making them more accessible and understandable to
a wider audience (Park and Tosaka, 2010).
2.1 Metadata Schema
A metadata schema comprises two components: se-
mantics, which define the meaning and refinements
of elements, and content, which specifies how values
should be assigned (Chan and Zeng, 2006). Schemas
establish rules for describing information, including
field definitions, data types, and required content.
A metadata standard expands on schemas by adding
guidelines for interoperability, consistency, and qual-
ity, ensuring usability of metadata across systems
((NISO), 2004). In this study, the ”metadata stan-
dard” refers to the comprehensive framework for or-
ganizing metadata and schemas.
Scientific metadata is categorized into three main
types: administrative, descriptive, and provenance
((NISO), 2004; Formenton et al., 2017). Admin-
istrative metadata details dataset management, such
as unique identifiers, responsible individuals, access
policies, and creation or update dates. Descriptive
metadata explains dataset content and purpose, em-
ploying keywords, controlled vocabularies, and struc-
tural information to aid in searching and understand-
ing data collection methods. Provenance metadata
documents data origins and transformations, provid-
ing transparency and reliability through records of
sources, processing history, and tools used.
2.2 Metadata Integration Based on
Crossing Metadata Standards
Metadata interoperability, a core principle of meta-
data, enables systems to exchange information effi-
ciently (Chan and Zeng, 2006; (NISO), 2004). Meta-
data integration is key to achieving this, aligning
and consolidating descriptive data to create a consis-
tent, interoperable framework across platforms (Chan
and Zeng, 2006). Techniques include standardized
schemas, metadata mapping, and interoperability-
enhancing protocols, aiming to unify metadata for ef-
Proposal for a Common Conceptual Schema for Metadata Integration and Reuse Across Diverse Scientific Domains
279

ficient data management, discovery, and utilization
across diverse ecosystems.
Integration approaches vary by context, data
sources, and desired interoperability levels, but the
principles of metadata interoperability are crucial for
standards like DataCite, Darwin Core, EML, and
Dublin Core. These standards address the unique
needs of scientific and cultural domains, improving
accessibility, citability, and reusability. By adopting
such standards, organizations ensure compliance with
international protocols, fostering seamless data shar-
ing, collaborative research, and knowledge discovery
within a connected data ecosystem. This practical
application of interoperability principles drives data-
driven advancements across fields.
The metadata standards analyzed include Dat-
aCite (version 4.4, 2021), a schema developed to en-
hance the citation and accessibility of research data by
assigning persistent digital object identifiers (DOI).
It supports proper attribution, access, and traceabil-
ity of data, with key metadata elements such as cre-
ators, titles, publication dates, and rights (DataCite
Metadata Working Group, 2021). The Darwin Core
standard (2009 version) (DwC), widely used in mu-
seums, herbaria, and biological studies, facilitates
communication about taxonomy, geographic distribu-
tion, and ecological contexts of species. It is piv-
otal in global conservation and ecological research
(Darwin Core Maintenance Interest Group, 2023).
The Ecological Metadata Language (version 2.2.0,
2019) (EML) focuses on ecological and environmen-
tal datasets, providing detailed descriptions of data
collection, methodologies, and quality, ensuring in-
tegrity for analysis and processing (Jones et al., 2019).
Lastly, Dublin Core (2020-01-20 version) is a sim-
ple yet versatile standard with 15 core elements, such
as title, author, subject, and description. It pro-
motes interoperability across information systems and
is particularly suited for libraries and archives (Dublin
Core, 2020).
3 METHODS
In terms of nature, the research adopts a qualita-
tive approach, and in terms of its purpose, it is ex-
ploratory and descriptive in nature. The study ana-
lyzed elements from four widely used metadata stan-
dards—DataCite, Dublin Core, EML, and Darwin
Core using the Metadata Crosswalk method to iden-
tify semantic correspondences and validate the feasi-
bility of an integrated conceptual schema. We found
156 metadata elements in DataCite, 15 in Dublin
Core, 493 in EML, and 262 in Darwin Core.
With elements collected from the standards, each
metadata item was analyzed to determine its defini-
tion, core type, obligation level, repeatability, and se-
mantic correspondence. The research aims to verify
metadata correspondence rather than propose a new
standard.
The selected standards are the most widely used
among repositories listed in Re3data
1
. The selection
of the DataCite, Dublin Core, EML, and Darwin Core
standards is justified by their broad adoption and rel-
evance across diverse contexts. DataCite and Dublin
Core were chosen for their wide applicability across
multiple domains and for providing foundational ele-
ments for describing and citing scientific data. EML
and Darwin Core, on the other hand, were selected for
their specialized focus on specific domains, making
them particularly valuable for addressing disciplinary
challenges in metadata integration. These standards
incorporate various descriptive elements, enabling the
analysis of semantic correspondences and supporting
the creation of a unified typology, such as the Inte-
grated Context Metadata Core (ICMC) proposed in
the article. Combining multidisciplinary and domain-
specific standards highlights the need to address gen-
eral interoperability and the unique requirements of
different scientific contexts.
Metadata collection was done manually via main-
tainers’ websites, and elements were categorized as
administrative, descriptive, or scientific. The Meta-
data Crosswalk method identified semantic and syn-
tactic correspondences to develop a generic concep-
tual model (St. Pierre et al., 1999).
Metadata Crosswalk maps and aligns metadata
across different standards to ensure interoperability
and consistency. The process involves five steps
(St. Pierre et al., 1999; Specka et al., 2019): (1) defin-
ing a shared terminology to unify content and ele-
ments across different standards; (2) identifying and
generalizing similar concepts, such as unique identi-
fiers or multiplicity, despite differing names; (3) de-
termining semantic mappings between equivalent el-
ements in the standards; (4) ensuring that mapped el-
ements also align in properties like obligation level
or multiplicity; and (5) converting element content to
accommodate restrictions like data type, value ranges,
or controlled vocabularies.
This method facilitates interoperability among
systems using different metadata standards, enabling
the aggregation and unified interpretation of descrip-
tive information in diverse environments. Its success
depends on accurately establishing correspondences
and ensuring the schema’s flexibility to accommodate
variations.
1
https://www.re3data.org
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
280

The metadata crosswalk steps were completed,
producing a corresponding metadata table
2
, which
outlines the semantic and syntactic correspondences
among the DataCite, Darwin Core, EML, and Dublin
Core schemes. Elements were collected from the
maintaining entities’ websites, focusing on integrat-
ing existing metadata rather than creating new ele-
ments.
4 RESULTS
The analysis identified 104 elements (11.23% of
926 total) with semantic correspondences across four
metadata standards, leading to the proposal of an In-
tegrated Context Metadata Core (ICMC) that consoli-
dates administrative, descriptive, and scientific meta-
data into a unified schema.
When establishing the semantic correspondence
between metadata, we classified the elements accord-
ing to their typology and identified a significant set
of metadata whose attributes represent the ”scientific”
context of the data. This set goes beyond the typology
of descriptive metadata, as its elements objectively
represent the specifics of the research phenomenon,
the methods used for acquisition, collection, process-
ing, and analysis, as well as geographic and temporal
coverage metadata.
Therefore, we assigned a new typology to this
group of metadata: Integrated Context Core Meta-
data. The proposed typology encompasses properties
and attributes that represent the subject in both quali-
tative and quantitative dimensions, the intrinsic nature
of the data, and its contextual elements. This typology
is distinct from the descriptive category, as scientific
metadata tends to focus on specific aspects related to
the methodology and underlying science of the data.
In contrast, descriptive metadata is more general, pro-
viding information to facilitate the identification and
organization of the data.
Our proposal to include the Integrated Context
Metadata Core in the current metadata typology com-
plements it by consolidating the fundamental scien-
tific properties of the data. Within this integrated
context core, two subcategories are developed to rep-
resent metadata related to the phenomenon and the
method.
We posit that to make the data integration process
sustainable, it is imperative that standards instantiate
fundamental integrated context metadata to improve
data representation and facilitate subsequent reuse.
Below we highlight the metadata of the standards ac-
2
http://dx.doi.org/10.5380/bdc/94
cording to their representative function within each
scheme.
After the matching process, the integrated meta-
data were classified into Administrative, Descriptive
and Integrated Context categories, containing 24, 17,
and 63 metadata in each category, respectively.
4.1 Administrative Core
The administrative core (Table 1) includes 24 meta-
data elements obtained through a crosswalk, covering
key aspects of data administration, management, and
use. It includes metadata for copyrights, licensing,
submission dates, public availability, data updates,
funding entities, and project-related awards, derived
from Darwin Core and EML standards.
Notably, <relatedMetadata> identifies relation-
ships between interconnected datasets, aiding navi-
gation and understanding in complex systems, while
<metadataProvider> details metadata sources, such
as software or organizations, supporting validation
and effective use.
Table 1: Administrative Core metadata.
Administrative Core
language
rights
rightsHolder
accessRight
date
dateSubmitted
dataIssued
dateUpdated
bibliographicCitation
relatedMetadata metadataProvider
project
title
personel
abstract
studyAreaDescription
designDescription
funding
funderName
funderIdentifier
awardTitle
awardNumber
awardURI
awardURL
4.2 Descriptive Core
The descriptive core (Table 2) is characterized by
the presence of 17 metadata elements that specify
the essence and content of a resource, contextualiz-
ing it and clarifying the nature and scope of this re-
source without the need to access it directly. In the
EML standard, this metadata is represented by the de-
scriptor <datasetType>, which can be complemented
Proposal for a Common Conceptual Schema for Metadata Integration and Reuse Across Diverse Scientific Domains
281

with the addition of other descriptive modules of the
EML to provide a more detailed representation of the
dataset (eml-resource, eml-methods, eml-project, and
the eml-literature modules). Another metadata found
in this core is <relationType> which connects a re-
source to its component parts (whether it is a new re-
source, derivative, or part of another). The presence
of this metadata across all standards provides context
to the data, enabling researchers to identify other re-
sources related to the data.
Table 2: Descriptive Core metadata.
Descriptive Core
title
identifier
creator
contributor
editor
publisher
distributor
description
abstract
source
series associeatedSequences
resourceType
relationType
isNewVersionOf
isPartOf
isReferencedBy
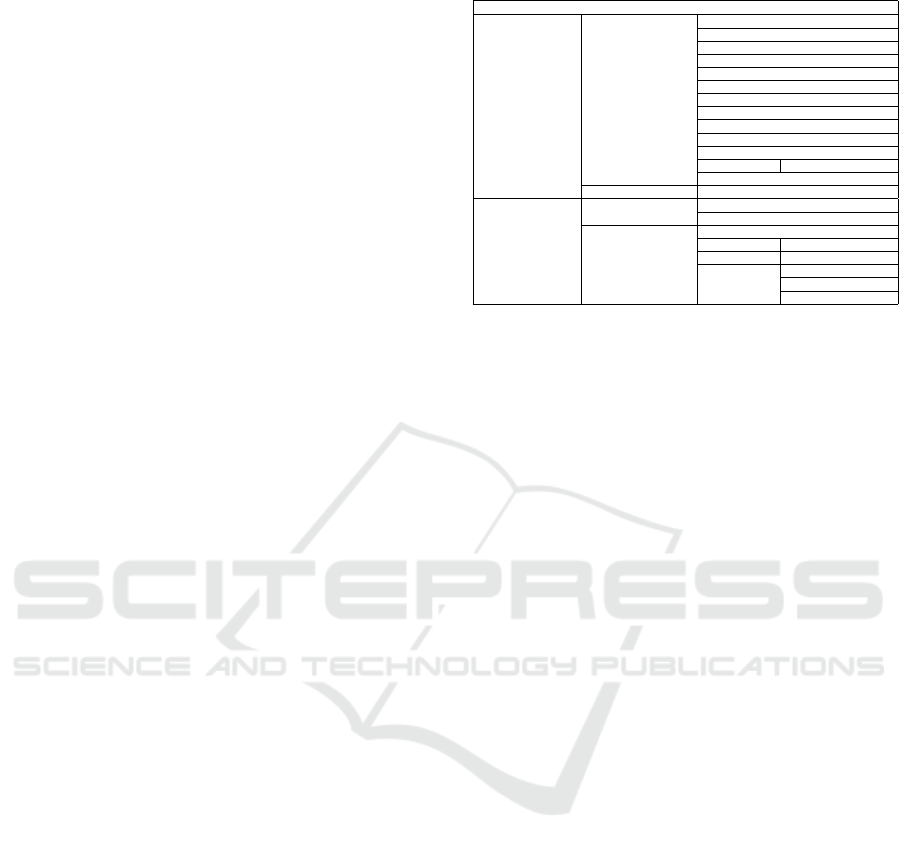
4.3 Integrated Context Metadata Core
As a contribution of this research, the metadata from
the Integrated Context Metadata Core consists of a
collection of metadata that represents the research ob-
ject supported by the data in its uniqueness. This
means it is an aggregation of descriptors that specifi-
cally detail the object (what we call the phenomenon)
and the methods of collection and analysis of this ob-
ject (what we call the method). The presence of In-
tegrated Context Metadata in a description is relevant
and significant for data reuse, as the absence or poor
completion of this information about the data impacts
the possibilities of recovery and reuse (Zimmermann,
2008).
The Integrated Context Metadata Core (Table 3)
includes 63 elements describing research subjects in
terms of size, extent, format, scientific and taxonomic
names, as well as geographic and temporal coverage,
which specify the object’s occurrence and data collec-
tion periods.
This metadata classification provides detailed de-
scriptions of research objects and methods, including
theoretical justification, specific methodologies, in-
Table 3: Integrated Context Core metadata.
Integrated Context
subject
scientificName
scientificNameAuthorship
vernacularName
acceptedNameUsage
taxonID
size
extent
format
procedutalStep
instrumentation
software
methods
methodStep
instrumentation
software
dataSource
sampling
studyExtent
coverage
description
samplingDescription
spatialDescription
spatialSamplingUnits referenceEntity
geographicCoverage
geoLocation
westBoundLongitude
eastBoundLongitude
northBoundLatitude
southBoundLatitude
decimalLongitude
decimalLatitude
footPrintSRS
footPrintWKT
country
locality
municipality
islandGroup island
stateProvince
geoReferenceProtocol geoReferenceSources
temporalCoverage
dateCollected
earliestDateCollected
latestDateCollected
time
singleDateTime
calendarDate dateyear
beginDate endDate
timeScaleTime
timeScaleAgeEstimate
timeScaleUncertainly
timeScaleCitation
struments, and analysis procedures. It enhances study
replicability and understanding, allowing researchers
to access data while comprehending its scientific con-
text, thus improving data integrity and fostering col-
laboration.
The analysis confirmed that the Integrated Context
Metadata Core contains descriptors for both phenom-
ena and methods, though these are scattered across
existing cores. To address this, the metadata were
divided into two subgroups: phenomena-related and
method-related, represented in Table 3.
4.3.1 Phenomenon Description
The metadata categorized as descriptors of the ”Phe-
nomenon” represent all aspects that provide coverage
and context to the research object. These include de-
scriptors that name the phenomenon, classify it taxo-
nomically, detail its metric attributes such as size, ex-
tent, and shape, and elaborate on the phenomenon’s
samples in relation to its field of study, research de-
sign, thematic and descriptive coverage. Addition-
ally, they include quality control metadata that inform
about the instrumental apparatus, including software,
used in data processing.
Regarding the <referenceEntity> element, the
EML standard includes a module called ”eml-Entity”
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
282
that details the data structure, its attributes, and identi-
fication information, facilitating the identification and
understanding of the data entity. This module serves
as a complementary resource to provide a more de-
tailed description of research data in the field of ecol-
ogy.
4.3.2 Method Description
The metadata representing the research method are in-
formational elements used to describe, document, and
record the procedures and techniques employed in the
collection, processing, and analysis of data within a
research project. These metadata are crucial for un-
derstanding how the data were generated, allowing for
the replication of methods and validation of results by
other researchers.
Information about the methods used in the gener-
ation, collection, processing, and analysis of data is
extremely relevant in the process of descriptive repre-
sentation. Just as in scientific research, understanding
the methodological approach is mandatory and essen-
tial for validating scientific activities. Therefore, the
same attention must be given to data description. A
lack of methodological detail can lead to a misunder-
standing of the results and, consequently, reduce the
possibilities for data reuse.
4.3.3 Geographic and Temporal Coverage
Metadata
Geographic and temporal metadata are classified
within the integrated context metadata core. Anal-
ysis of these elements reveals their versatile nature:
they not only provide information about the location
and period in which a phenomenon occurs but also
record the place and time of data collection, process-
ing, and analysis. These metadata play a crucial role
in describing the methods of data collection, treat-
ment, and analysis, thus serving two sub-cores of the
scientific core. This versatility enables a comprehen-
sive representation of both the observed phenomenon
and the employed methods. In other words, any event,
whether it be the observation of a species, a simula-
tion, or a physical phenomenon, occurs in a specific
location and period. Similarly, the methods used for
data collection, processing, and analysis are defined
by a specific spatial and temporal context, such as the
collection of a sample at a determined location, on a
specific day, during a particular time period.
Table 4 illustrates the geographic and temporal
coverage metadata obtained through the metadata
crosswalk process.
Table 4: Geographic and Temporal Coverage.
Geographic and Temporal Coverage
geographicCoverage
geoLocation
westBoundLongitude
eastBoundLongitude
northBoundLatitude
southBoundLatitude
decimalLongitude
decimalLatitude
footPrintSRS
footPrintWKT
country
locality
municipality
islandGroup island
stateProvince
geoReferenceProtocol geoReferenceSources
temporalCoverage
dateCollected
earliestDateCollected
latestDateCollected
time
singleDateTime
calendarDate dateyear
beginDate endDate
timeScaleTime
timeScaleAgeEstimate
timeScaleUncertainly
timeScaleCitation
4.4 A Common Conceptual Schema for
Scientific Core
Data repositories serve as a community infrastructure
for sharing knowledge, organizing data into generic
patterns to define ”scientific data.” While the FAIR
Principles (Wilkinson et al., 2016) advocate for ho-
mogenized data descriptions, no standard concep-
tual model exists, as seen in the genome project
(Bernasconi et al., 2022; Bernasconi et al., 2017).
Research communities vary in methods and data
types, requiring curation services to address di-
verse sub-disciplinary characteristics, a costly task
for ”small science” repositories (Cragin et al., 2010;
Thomer, 2022). High metadata quality is essential for
reuse, as data that are easy to find, access, and inter-
operate become more reusable (Brandizi et al., 2022).
The Integrated Context Metadata Core addresses
scattered and non-mandatory information in multi-
disciplinary standards and the overwhelming volume
in disciplinary standards. Properties like ”Temporal
Coverage” and ”Geographic Coverage” are common
across standards, aiding dataset correspondence. Sim-
ilarly, ”Method” properties frequently describe anal-
ysis techniques. Though limited to four standards,
this proposal highlights common properties beyond
administrative attributes like title and author.
Phenomenon and method metadata often appear
as free text, complicating dataset interoperability.
Structuring these concepts through frequent proper-
ties simplifies dataset correspondence and automates
data transformations (e.g., unit harmonization, out-
lier detection). Controlled vocabularies, such as for
equipment identification, further enhance metadata
quality. This proposal identifies generic concepts in
DataCite, Darwin Core, EML, and Dublin Core stan-
dards (Figure 1).
This Integrated Context Metadata Core proposal
encompasses metadata that at least represents a re-
Proposal for a Common Conceptual Schema for Metadata Integration and Reuse Across Diverse Scientific Domains
283
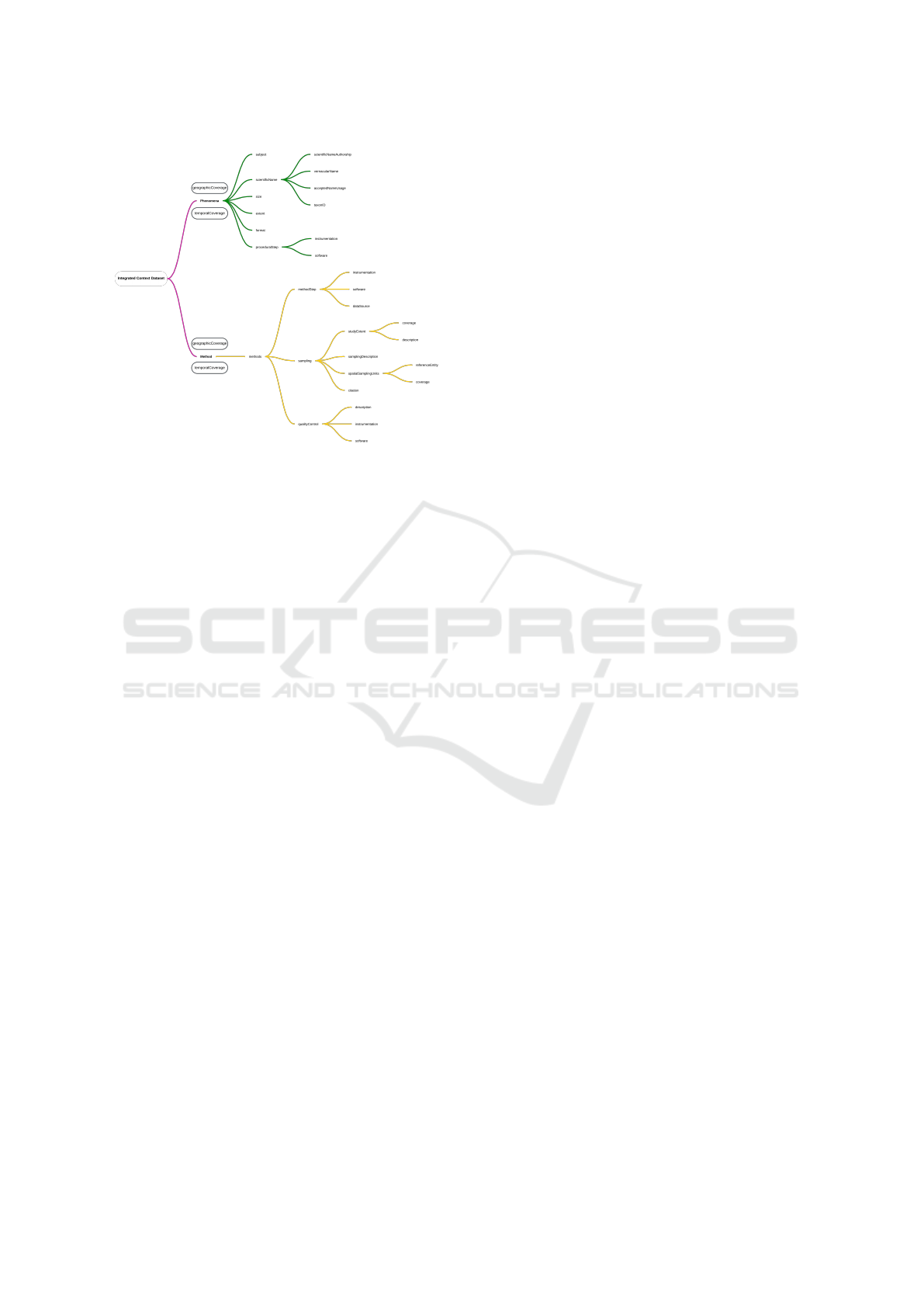
Figure 1: Conceptual description of scientific dataset in sci-
entific repositories metadata.
search phenomenon, characterizing it by its common
and scientific names and properties, as well as the op-
tional adoption of protocols according to the scien-
tific domain area. Similarly, the metadata represent-
ing the method is covered by metadata representing
the processes of data collection, treatment, and anal-
ysis, including detailed information about the instru-
ments and software used. The geographic and tempo-
ral coverage (as detailed in Section 4.3.2) serves the
core in a modular fashion and is a mandatory param-
eter in the description.
The Integrated Context Metadata Core (ICMC) is
a model designed to address the shortcomings of the
DataCite, Dublin Core, EML, and Darwin Core stan-
dards. Through an integrated and flexible approach,
the ICMC seeks to enhance the management of sci-
entific metadata, with a focus on interoperability and
reuse.
DataCite and Dublin Core are widely used in vari-
ous fields, but provide only basic elements, such as ti-
tle, authorship, and date, for the registration and cita-
tion of data. For more specific elements, such as study
phenomena or scientific methods, these standards of-
fer little or no semantic support. In contrast, EML
and Darwin Core offer sufficient semantic depth for
describing specific domains, such as ecology and bio-
diversity, but are limited to those fields. By combin-
ing elements of these standards, the ICMC addresses
both challenges.
Interoperability is another key feature. By in-
tegrating and merging elements from different stan-
dards, the ICMC overcomes the inflexibility of ex-
isting frameworks, which are typically designed to
meet isolated needs. Standardized metadata frame-
works that emphasize semantic relationships enhance
the accuracy of comparisons and make the data easier
for machines to process.
The ICMC is designed for interdisciplinary re-
search. Given the breadth and diversity of scientific
fields, as well as the depth of variety within these do-
mains, generalist standards are often too generic to be
effective, while specialist standards can be overly spe-
cific. The ICMC bridges this gap by accommodating
both general and domain-specific needs.
5 CONCLUSIONS
The analysis of metadata revealed a significant subset,
termed the Integrated Context Metadata Core, which
goes beyond descriptive metadata by capturing essen-
tial scientific attributes, such as the specifics of the
research phenomenon, methods used, and geographic
and temporal coverage. This new typology enhances
data reuse by providing detailed descriptions of phe-
nomena and methods, ensuring better recovery, inter-
operability, and integration of datasets.
The proposal consolidates existing metadata el-
ements into two subcategories: phenomenon and
method. These Integrated Context metadata address
the challenges of dispersed information in existing
schemas, enabling precise cross-dataset correspon-
dences and automating processes like normalization
and error detection. The inclusion of controlled
vocabularies for attributes like equipment improves
metadata quality and usability.
The study demonstrates that the Integrated Con-
text Metadata Core (ICMC) enhances interoperabil-
ity and reuse by consolidating metadata elements that
describe research phenomena and methods. This ap-
proach addresses challenges in scattered metadata and
supports cross-disciplinary integration, aligning with
FAIR principles and promoting broader scientific col-
laboration.
ACKNOWLEDGEMENT
Coordination for the Improvement of Higher Educa-
tion Personnel (CAPES) - Program of Academic Ex-
cellence (PROEX).
REFERENCES
Bernasconi, A., Ceri, S., , Campi, A., and Masseroli, M.
(2017). Conceptual modeling for genomics: Building
an integrated repository of open data. In Mayr, H. C.,
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
284

Guizzardi, G., Ma, H., and Pastor, O., editors, Inter-
national Conference on Conceptual Modeling, pages
325–339, Cham. Springer International Publishing.
Bernasconi, A., Ceri, S., Canakoglu, A., and Masseroli, M.
(2022). Meta-base: A novel architecture for large-
scale genomic metadata integration. In Mayr, H. C.,
Guizzardi, G., Ma, H., and Pastor, O., editors, Inter-
national Conference on Conceptual Modeling, pages
325–339. IEEE/ACM Trans Comput Biol Bioinform.
Borgman, C. L. (2015). Big Data, Little Data, No Data:
Scholarship in the Networked World. MIT Press,
Cambridge, MA.
Brandizi, M., Singh, A., Parsons, J., Rawlings, C., and
Hassani-Pak, K. (2022). Integrative data analysis
and exploratory data mining in biological knowledge
graphs. In Integrative Bioinformatics: History and
Future, pages 147–169. Springer Singapore, Singa-
pore.
Center, D. C. (2020). Data documentation initiative al-
liance.
Chan, L. M. and Zeng, M. L. (2006). Metadata interoper-
ability and standardization - a study of methodology
part i: Achieving interoperability at the schema level.
D-Lib magazine, 12(6).
Cragin, M. H., Palmer, C. L., Carlson, J. R., and Witt, M.
(2010). Data sharing, small science and institutional
repositories. Philosophical Transactions of Royal So-
ciety, 368:4023–4038.
Curty, R. G., Crowston, K., Specht, A., Grant, B. W., and
Dalton, E. D. (2017). Attitudes and norms affecting
scientists’ data reuse. PLOS ONE, 12(12):1059–1078.
Darwin Core Maintenance Interest Group (2023). Darwin
core quick reference guide. Darwin Core TDWG.
DataCite Metadata Working Group (2021). DataCite Meta-
data Schema Documentation for the Publication and
Citation of Research Data and Other Research Out-
puts. DataCite e.V.
Dublin Core (2020). Dublin Core. DCMI.
Formenton, D., Ferreira de Castro, F., de Souza Gracioso,
L., Furnival, A. C. M., and de Melo Sim
˜
oes, M. d. G.
(2017). Os padr
˜
oes de metadados como recursos tec-
nol
´
ogicos para a garantia da preservac¸
˜
ao digital. Bib-
lios, (68):82–95.
Framework, F. D. O. (2020). Fair digital object framework.
Gregory, K., Groth, P., Scharnhorst, A., and Wyatt, S.
(2020). Lost or found? discovering data needed for
research.
He, L. and Nahar, V. (2016). Reuse of scientific data in aca-
demic publications: An investigation of dryad digital
repository. Aslib Joural of Informantion Management,
68:478–494.
Jones, M., O’Brien, M., Mecum, B., Boettiger, C., Schild-
hauer, M., Maier, M., Whiteaker, T., Earl, S., and
Chong, S. (2019). Ecological Metadata Language
version 2.2.0.
Koesten, L. M., Vougiouklis, P., Bontas Simper, E. P., and
Groth, P. T. (2020). Dataset reuse: Toward translating
principles to practice. Patterns, 1.
Loffler, F., Wesp, V., Konig-Ries, B., and Klan, F. (2021).
Dataset search in biodiversity research: Do meta-
data in data repositories reflect scholarly information
needs? PLoS ONE, 16.
Mutschke, P., Le Franc, Y., Klas, C.-P., Magagna, B.,
Scharnhorst, A., and Schiffner, D. (2020). Fair dig-
ital objects for cross-domain data searching, linking
and semantic interoperability.
(NISO), N. I. S. O. (2004). Understanding Metadata. NISO
Press, Bethesda. 16 p.
Park, J.-r. and Tosaka, Y. (2010). Metadata creation prac-
tices in digital repositories and collections: Schemata,
selection criteria, and interoperability. Information
Technology and Libraries, 29:104–116.
Pomerantz, J. P. (2015). Metadata. MIT Press, Cambridge,
MA.
Porto, F. and Spaccapietra, S. (2011). Data model for sci-
entific models and hypotheses. In Kaschek, R. and
Delcambre, L., editors, The Evolution of Conceptual
Modeling, volume 6520 of Lecture Notes in Computer
Science, pages 302–322. Springer, Berlin, Heidelberg.
Silva, F. C. C. d. (2019). Gest
˜
ao de dados cient
´
ıficos. Edi-
tora Interci
ˆ
encia, Rio de Janeiro.
Specka, X., G
¨
artner, P., Hoffmann, C., Svoboda, N.,
Stecker, M., Einspanier, U., Senkler, K., Zoarder,
M. M., and Heinrich, U. (2019). The bonares meta-
data schema for geospatial soil-agricultural research
data – merging inspire and datacite metadata schemes.
Computers & Geosciences, 132:33–41.
St. Pierre, M., Paul, S. K., Simmonds, A., and LaPlante,
W. (1999). We used to call it publishing-issues in
crosswalking content metadata standards. Against the
Grain, 11(4):31.
Thomer, A. K. (2022). Integrative data reuse at scientifically
significant sites: Case studies at yellowstone national
park and the la brea tar pits. Journal of the Association
for Information Science and Technology, 73(8):1155–
1170.
Wallis, J., Rolando, E., and Borgman, C. L. (2011). If we
share data, will anyone use them? data sharing and
reuse in the long tail of science and technology. PLoS
ONE, 8(6).
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Apple-
ton, G., Axton, M., Baak, A., Blomberg, N., Boiten,
J.-W., Santos, L. B. d. S., Bourne, P. E., Bouwman,
J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I.,
Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R.,
Gonzalez-Beltran, A., Gray, A. J., Groth, P., Goble,
C., Grethe, J. S., Heringa, J., ’t Hoen, P. A. C., Hooft,
R., Kuhn, T., Kok, R., Kok, J., Lusher, S. J., Mar-
tone, M. E., Mons, A., Packer, A. L., Persson, B.,
Rocca-Serra, P., Roos, M., van Schaik, R., Sansone,
S.-A., Schultes, E., Sengstag, T., Slater, T., Strawn,
G., Swertz, M. A., Thompson, M., van der Lei, J.,
van Mulligen, E., Velterop, J., Waagmeester, A., Wit-
tenburg, P., Wolstencroft, K., Zhao, J., and Mons, B.
(2016). The fair guiding principles for scientific data
management and stewardship. Scientific Data, 3.
Zimmerman, A. (2003). Data sharing and secondary use of
scientific data: Experiences of ecologists. PhD thesis,
University of Michigan School of Information.
Zimmermann, A. (2008). New knowledge from old data.
Science, Technology, & Human Values, 33:631–652.
Proposal for a Common Conceptual Schema for Metadata Integration and Reuse Across Diverse Scientific Domains
285
