Enhancing NnU-Net for Improved Medical Image Segmentation: A
Comparative Study with TotalSegmentator
Jingyi Wu
a
Sydney Institute of Intelligent Technology, Northeastern University Qinhuangdao, Qinhuangdao, China
Keywords: Medical Image Segmentation, nnU Net, Loss Function Optimization, Data Augmentation, Multimodal Imaging.
Abstract: In this paper, an optimization method is proposed that relies on the no-new-Net (nnU Net) architecture to
improve the performance of medical image segmentation tasks. Medical image segmentation is an important
component of disease diagnosis, treatment planning, and surgical assistance. Since its launch in 2018, nnU
Net has become a fundamental tool in this field by adapting its architecture, preprocessing, and training
strategies. However, current models still have shortcomings in handling data imbalance and multimodal
images. For this purpose, the paper optimized the loss function and data augmentation strategy of nnU Net.
By increasing the Dice loss weight, the model can more effectively handle small structures and imbalanced
data, improving segmentation accuracy. Furthermore, by incorporating higher rotation probability, noise
enhancement, and low-resolution simulation into the improved data augmentation technique, the model's
robustness and capacity for generalization are greatly increased. The experimental results demonstrate that
the upgraded nnU Net performs much better than TotalSegmentor in terms of segmentation accuracy and
complicated boundary handling, especially when compared to metrics like Dice Score, IoU, and Hausdorff
Distance.
1 INTRODUCTION
A basic task in medical image analysis, medical
picture segmentation is essential for many
applications, including disease diagnosis, therapy
planning, and surgical support. One of the approaches
that is most frequently utilized in this field is the U-
Net architecture and its variations. With its self-
configuring framework that automatically adjusts its
architecture, preprocessing, and training algorithms
to each dataset, no-new-Net (nnU-Net), which was
introduced in 2018, revolutionized the domain.
Despite its success, further improvements are
necessary in areas such as data augmentation and loss
function optimization, as specific adjustments could
yield better performance, particularly when handling
diverse datasets.
By utilizing the most recent developments in nnU-
Net, TotalSegmentator expands its capabilities to
multi-class segmentation in Magnetic resonance
imaging (MRI) as well as Computed Tomography
(CT) image modalities, producing impressive
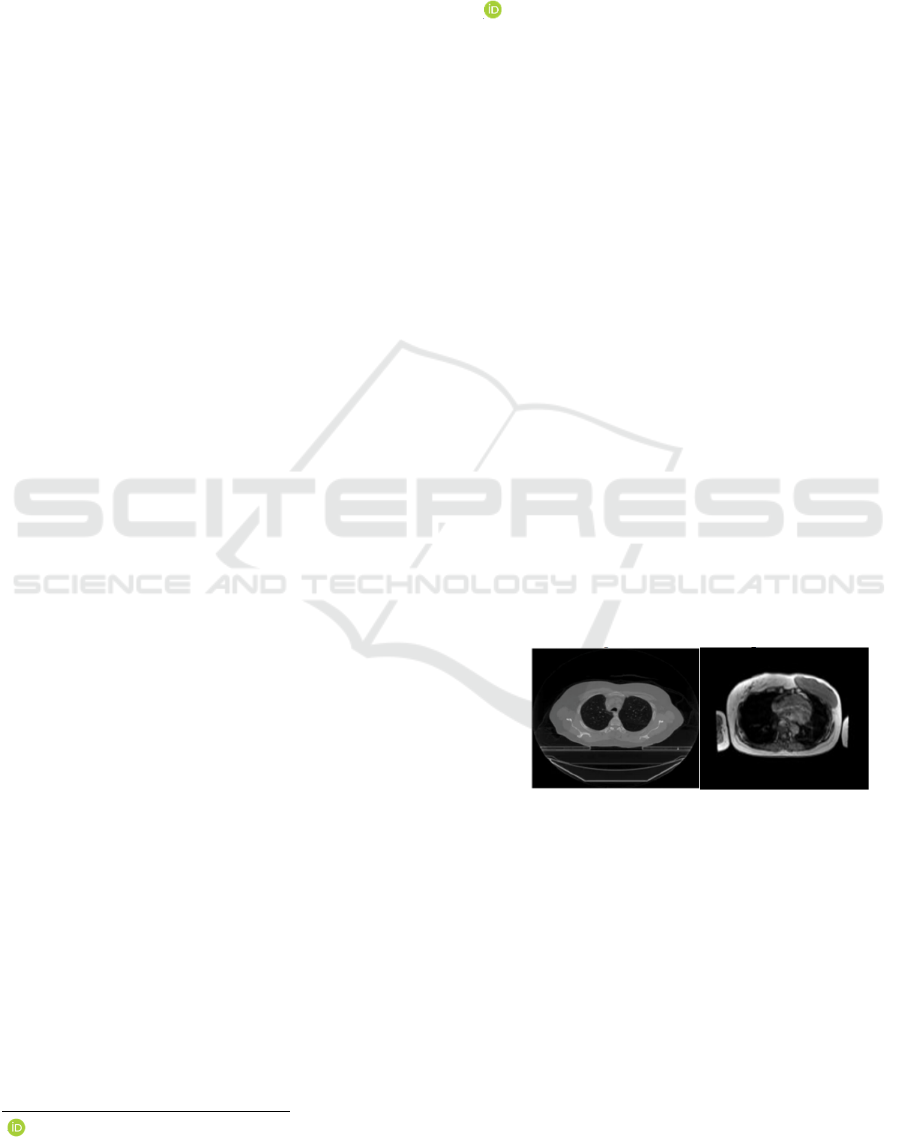
outcomes. Figure 1 illustrates an MRI and CT scan
a
https://orcid.org/0009-0005-3472-5768
example. However, there remains room for
improvement, especially in balancing the loss
function and enhancing training data through more
sophisticated augmentation techniques.
Figure 1: Example of CT and MRI (
Kumar et al, 2021)
In order to overcome the current obstacles in
medical picture segmentation, this research optimizes
two crucial nnU-Net model components: (1)
Adjusting the loss function weights to better balance
segmentation precision across different anatomical
structures and improve performance on imbalanced
datasets; (2) Enhancing the data augmentation
strategy to improve the model's robustness to
variations in medical imaging data, aiming to boost
segmentation accuracy and generalization in real-
246
Wu and J.
Enhancing NnU-Net for Improved Medical Image Segmentation: A Comparative Study with TotalSegmentator.
DOI: 10.5220/0013515100004619
In Proceedings of the 2nd International Conference on Data Analysis and Machine Learning (DAML 2024), pages 246-251
ISBN: 978-989-758-754-2
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
world applications. These improvements strengthen
the model’s resilience and ensure broader
applicability in practical scenarios.
Building upon the nnU-Net framework, the paper
introduced targeted optimizations to further enhance
its performance in segmentation tasks. Through
extensive experimentation, the results show that these
modifications significantly improve both accuracy
and generalization compared to the original nnU-Net
and TotalSegmentator models. The primary
contributions of this paper are as follows:
• The paper proposes a novel adjustment to the
loss function in nnU-Net, optimizing the weight
distribution to better handle class imbalance.
• The model's generalization to new and unseen
data is improved by the paper's use of more varied and
realistic transformations in the data augmentation
technique.
• The experimental results validate the efficacy
of the approach by showing that the upgraded nnU-
Net regularly outperforms TotalSegmentator across
key assessment parameters.
In the following sections, the paper will provide a
detailed description of the methodology,
experimental setup, and the results validating the
proposed improvements.
2 RELATED WORKS
Medical image segmentation, a fundamental task in
medical image analysis, plays an important role in
various applications such as organ localization, lesion
detection, and treatment planning. Early
segmentation methods mainly relied on rule-based or
feature-based techniques such as region growing,
watershed, and level set methods (Fischl et al., 2004).
With the rise of deep learning, convolutional neural
networks (CNNs) emerged as the leading technology
in medical image segmentation, particularly after the
introduction of the U-Net model, which led to
significant advancements (Ronneberger et al., 2015).
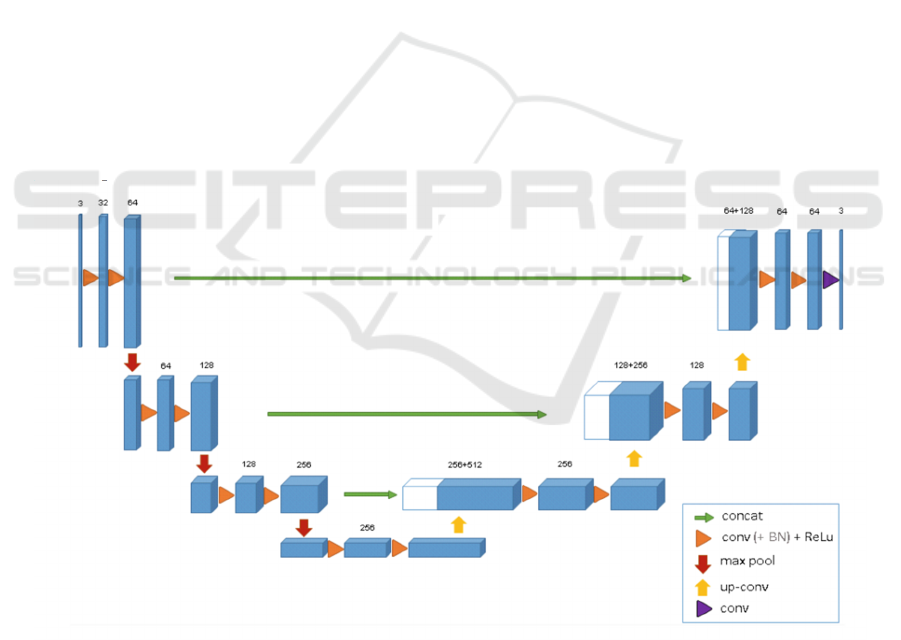
The U-Net architecture, as depicted in Figure 2, is
renowned for its U-shaped design, featuring skip
connections between the encoder and decoder, which
greatly enhance segmentation accuracy (Çiçek et al.,
2016). Introduced in 2018, NnU-Net is a self-
adapting version of U-Net that serves as a general
baseline for medical image segmentation by
automating architecture tweaks, preprocessing, and
training procedures to suit various datasets (Isensee et
al., 2021). This model has excelled in multiple
international segmentation challenges, showcasing
high versatility and adaptability.
Figure 2: The architecture of U-Net (Çiçek et al., 2016)
Despite nnU-Net’s success, recent studies suggest
that its performance on specific tasks can be further
optimized. Research shows that introducing adaptive
weighting in loss functions and improving data
augmentation strategies can enhance both robustness
and precision (Roy et al., 2018). This study builds on
nnU-Net’s framework, with a focus on improving
performance in handling imbalanced data and highly
diverse medical imaging datasets.
Enhancing NnU-Net for Improved Medical Image Segmentation: A Comparative Study with TotalSegmentator
247

TotalSegmentator, an open-source model based
on the nnU-Net framework, was initially developed
for CT image segmentation and later extended to
perform multi-structure segmentation in MRI images
(Wasserthal et al., 2023). TotalSegmentator is a
versatile tool for multi-modality segmentation tasks,
thanks to its sequence-independent nature, enabling it
to segment 59 anatomical structures, including
organs, bones, muscles, and vessels (Akinci
D'Antonoli et al., 2023). By integrating large clinical
datasets, TotalSegmentator demonstrates robustness
in various applications, especially in handling
different MRI sequences. However, its performance
is still challenged in the segmentation of fine
structures, such as those in blurred or low-contrast
regions (Hatamizadeh et al., 2021). This opens an
opportunity to enhance segmentation performance by
optimizing nnU-Net’s loss function and data
augmentation strategies.
In addition to TotalSegmentator, other U-Net-
based segmentation models have emerged in recent
years. For example, 3D U-Net (Çiçek et al., 2016)
extends U-Net to process 3D image data, while
SwinUNETR (Hatamizadeh et al., 2021) combines
Transformer architecture with U-Net to capture long-
range dependencies. However, these models often
come with higher computational costs and fall short
in multi-modality and sequence diversity tasks
compared to TotalSegmentator.
The design of loss functions plays a crucial role in
deep learning-based segmentation tasks, particularly
when dealing with class imbalance and small target
segmentation. Traditional cross-entropy loss often
favors large classes, leading to poor performance in
smaller classes (Sudre et al., 2017). To address this
issue, weighted loss functions such as Dice loss
(Milletari et al., 2016) and Tversky loss (Salehi et al.,
2017) have been introduced to handle imbalanced
data and multi-class segmentation tasks more
effectively. By adjusting the weights of different
classes, these methods improve segmentation
accuracy for small classes and boundary regions.
In terms of data augmentation, traditional
techniques such as rotation, scaling, and translation
are commonly used. However, recent studies have
shown that more advanced augmentation techniques,
such as random cropping, brightness and contrast
adjustment, and elastic deformation, can significantly
improve model robustness (DeVries & Taylor, 2017).
These techniques generate more diverse training data,
enabling models to better generalize to unseen
clinical images. Moreover, adaptive data
augmentation techniques based on deep learning are
continuously evolving, allowing dynamic adjustment
of augmentation strategies based on data
characteristics, further enhancing model performance
(Zhang et al., 2018).
The innovation of this study lies in modifying
nnU-Net’s loss function weights and optimizing its
data augmentation strategy to further improve
performance in medical image segmentation tasks.
These modifications build on previous research
findings and demonstrate superior performance
compared to TotalSegmentator in practical
applications.
3 METHODOLOGIES
3.1 Loss Function Adjustment
The loss function plays a critical role in guiding the
optimization of deep learning models, particularly in
medical image segmentation, where it directly
impacts model performance on complex and
imbalanced datasets. Dice loss emphasizes improving
segmentation accuracy for small structures, while
cross-entropy loss focuses on the overall
segmentation accuracy. Balancing the weights of
these two losses is crucial for achieving optimal
model accuracy.
The paper increased the weight of the Dice loss
from 1 to 1.5 and set the cross-entropy loss weight to
0.5. This adjustment directs the model to focus more
on small structures, prioritizing their segmentation
during optimization while maintaining the overall
accuracy of larger structures and global segmentation.
These adjustments help the model perform better
on imbalanced data, particularly for small targets,
allowing for more precise segmentation. This is
crucial in medical image segmentation tasks, such as
tumor or lesion detection, where increasing the Dice
loss weight reduces the model’s tendency to
overemphasize the background or large structures,
thereby improving the segmentation accuracy of
smaller targets. These changes enhance the model’s
sensitivity to small object recognition, ultimately
improving overall segmentation accuracy and
boundary detail handling.
3.2 Data Augmentation Strategy
Optimization
In order to enhance the generalization ability of the
model and avoid overfitting, data augmentation
requires introducing various random transformations
(such as rotation, scaling, and noise) into the training
set. By exposing the model to more diverse data, it
enhances real-world performance and strengthens its
DAML 2024 - International Conference on Data Analysis and Machine Learning
248

robustness and adaptability in testing or inference
processes. This is particularly important in medical
image segmentation, as data variability arises from
differences in patients, imaging conditions, and noise
levels.
The paper increased the rotation probability to 0.3
to simulate anatomical structures from different
orientations. The paper also extended the variance
range of Gaussian noise to (0, 0.2) and set its
application probability to 0.15 to help the model
handle varying levels of image noise. For low-
resolution simulation, the paper adjusted the scaling
range to (0.7, 1) and increased the application
probability to 0.3, allowing the model to adapt to low-
quality or down-sampled images.
These adjustments significantly improved the
model’s adaptability to data variations. Increasing the
rotation probability allowed the model to handle more
diverse anatomical orientations, while noise
augmentation improved stability in noisy
environments. Low-resolution simulation ensured
that the model could handle varying image
resolutions, maintaining high segmentation accuracy
even with low-quality input. These improvements are
particularly valuable in medical image segmentation,
where models need to be robust and generalizable in
clinical applications.
3.3 Deep Supervision and Multi-scale
Loss
Deep supervision and multi-scale loss help guide the
model at different resolutions, making feature
extraction across various scales more accurate. Deep
supervision enables the model to learn segmentation
information at multiple levels during training, which
is particularly useful for handling complex
anatomical structures with intricate boundaries.
Multi-scale loss weighting ensures that the model
remains efficient during fine-grained segmentation.
In the DeepSupervisionWrapper, the paper
adjusted the multi-scale loss weights by assigning
higher weights to high-resolution outputs, thereby
enhancing the model's focus on fine-grained
segmentation. This adjustment ensures that the model
maintains a balance in feature extraction across
different resolutions while emphasizing high-
resolution outputs.
This modification improves the model's ability to
handle complex boundaries, particularly when
segmenting small or blurred anatomical structures.
By increasing the weight of high-resolution outputs,
the model is better equipped to handle anatomical
detail, significantly reducing Hausdorff distance and
producing more precise segmentation boundaries.
4 EXPERIMENTAL SETUP
4.1 Dataset
This experiment's brain MRI dataset, which includes
samples required for both training and testing, was
obtained from TotalSegmentator. The labels and
photos are included with the data, which is supplied
in nii.gz format. To make sure the model can be
applied to different situations, a five-fold cross-
validation technique is used. This dataset is perfect
for evaluating and verifying the effectiveness of
medical picture segmentation algorithms because to
its intricate anatomical structures and thorough
labeling. The model's capacity to handle complicated
medical pictures, notably in segmenting small
structures and handling multimodal problems, may be
assessed by the study using this dataset.
4.2 Evaluation Metrics
The segmentation performance of the model was
thoroughly evaluated by the article through the
utilization of several metrics. Dice Score is a useful
tool for assessing segmentation accuracy in tiny
regions and handling imbalanced data since it
assesses the overlap between expected outcomes and
ground truth. By determining the ratio between the
intersection and union of the anticipated and actual
regions, Intersection over Union (IoU) offers a more
rigorous evaluation that gauges prediction accuracy.
The model's capacity to identify the target regions and
steer clear of false positives is measured by sensitivity
and specificity, respectively. These two metrics,
which show how well the algorithm detects lesions
while ignoring normal tissue, are crucial for medical
picture segmentation. Last but not least, Hausdorff
Distance assesses segmentation boundary precision
to make sure the model faithfully represents intricate
structural elements. These metrics were selected
because they allow for a thorough evaluation of the
model's performance in a number of areas, from
overall segmentation accuracy to boundary
management and false detection control—a crucial
component of medical picture segmentation model
optimization and assessment.
4.3 Experimental Procedure
The model training was conducted on a high-
performance computing environment equipped with
an NVIDIA RTX 3090 GPU, AMD 5800X CPU,
32GB of RAM, and over 200GB of storage space.
The system operated on Python 3.10.12 and the
Pytorch 2.4.0+cu121 deep learning framework,
Enhancing NnU-Net for Improved Medical Image Segmentation: A Comparative Study with TotalSegmentator
249

ensuring efficient training in an optimized hardware
and software environment. M.2 SSD was utilized for
data storage to maximize data read and write speeds.
The training followed the standard nnU-Net five-fold
cross-validation pipeline. First, preprocessing was
applied to the brain MRI data, including adjusting the
format and resolution. Each fold was trained using
high-resolution 3D data. After completing the
training, five models were generated for performance
evaluation. The training process also incorporated
deep supervision and multi-scale loss strategies,
ensuring the model could learn detailed features at
various scales, thus enhancing segmentation
precision.
The PolyLRScheduler dynamically adjusted the
learning rate during training, with the initial learning
rate for hyperparameter values set to 1e-2. With a
weight decay of 3e-5 and a momentum parameter of
0.99, SGD was the optimizer that was employed. Data
augmentation strategies were adjusted to improve
model generalization by increasing the application
probability of rotation, noise, and low-resolution
simulation. These strategies enabled the model to
better handle real-world complex medical images,
showing robust performance in dealing with noise,
resolution variations, and other challenges.
5 RESULTS AND DISCUSSION
5.1 results
The results show that improved nnU-Net significantly
outperforms TotalSegmentator in terms of
segmentation accuracy, as demonstrated by its higher
Dice Score and IoU. The following table summarizes
the performance comparison:
Table 1: Experimental result
Metric TotalSegmentator Improved
nnU-Net
Dice Score 0.6241 0.99967
IoU 0.4536 0.99935
Sensitivit
y
0.4600 0.99935
S
p
ecificit
y
0.9973 1.0
95% Hausdorff
Distance
26.23 0.0
99.9%
Hausdorff
Distance
34.67 1.0
100% Hausdorff
Distance
55.24 8.31
Based on the comparison in table 1, improved
nnU-Net significantly outperforms TotalSegmentator
across all key performance metrics. improved nnU-
Net achieves a Dice Score of 0.99967, while
TotalSegmentator only reaches 0.6241, indicating
near-perfect alignment of improved nnU-Net’s
segmentation with ground truth labels. Additionally,
improved nnU-Net’s IoU score of 0.99935 is much
higher than TotalSegmentator’s 0.4536, reflecting
greater overlap between predicted segmentation and
actual labels. In terms of sensitivity, improved nnU-
Net excels with a score of 0.99935, far surpassing
TotalSegmentator’s 0.4600, demonstrating its
superior ability to detect relevant foreground regions.
While both models perform well in specificity,
improved nnU-Net achieves a perfect score of 1.0,
indicating its near-flawless ability to avoid false
positives in background regions. In terms of
Hausdorff Distance, improved nnU-Net holds a
significant advantage: its 99.9% Hausdorff Distance
is 1.0, and the 100% Hausdorff Distance is 8.31, far
lower than TotalSegmentator’s 95% Hausdorff
Distance of 26.23 and 100% Hausdorff Distance of
55.24. This shows that improved nnU-Net provides
far more accurate boundary delineations of
anatomical structures. In summary, improved nnU-
Net’s adaptive architecture and finely tuned
configurations offer substantial advantages in
medical image segmentation tasks, particularly where
boundary precision and sensitivity are critical.
5.2 Discussion
The improved nnU-Net significantly outperforms
TotalSegmentator across several metrics due to the
optimizations made to its loss function and data
augmentation strategy. By increasing the weight of
Dice loss, the model more effectively handles small
targets and imbalanced data, resulting in greater
precision when segmenting small regions.
Furthermore, adjustments to the data augmentation
strategy increased the model’s robustness to various
image perturbations such as noise, rotation, and
resolution changes. These improvements have led to
superior performance in metrics like Dice Score and
IoU, while significantly reducing Hausdorff
Distance, indicating more accurate boundary
segmentation.
Nevertheless, enhanced nnU-Net has several
drawbacks. The model's application in resource-
constrained contexts may be limited due to its lengthy
training timeframes and high processing
requirements. Further validation on a range of
datasets is necessary to establish generalizability, as
DAML 2024 - International Conference on Data Analysis and Machine Learning
250

the efficacy of data augmentation procedures may
also depend on the particular features of the dataset.
6 CONCLUSIONS
Through data augmentation techniques and loss
function tuning, this study greatly enhanced the nnU-
Net model's performance in medical picture
segmentation tasks. By increasing the weight of Dice
loss, the model showed enhanced performance in
handling small targets and data imbalance, while the
improvements in data augmentation made the model
more resilient to perturbations like noise and rotation.
These enhancements boosted accuracy, boundary
handling, and robustness, outperforming
TotalSegmentator in metrics like Dice Score, IoU,
and Hausdorff Distance.
Future research will aim to reduce the model's
training time by exploring more efficient
optimization algorithms and ensemble learning
techniques. Additionally, efforts will focus on
validating the model's adaptability and ensuring the
generalizability of its data augmentation strategies
across various types of medical image datasets,
ultimately seeking to enhance performance and
reliability across a broader range of applications.
REFERENCES
Akinci, D., Antonoli, T., Yang, S., & Braren, R. F. (2023).
TotalSegmentator MRI: Sequence-independent
segmentation of 59 anatomical structures in MR
images. arXiv preprint. arXiv:2301.10693.
https://doi.org/10.48550/arXiv.2301.10693
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., ... &
Anandkumar, A. (2021). TransUNet: Transformers
make strong encoders for medical image segmentation.
arXiv preprint. arXiv:2102.04306.
https://doi.org/10.48550/arXiv.2102.04306
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., &
Ronneberger, O. (2016). 3D U-Net: Learning dense
volumetric segmentation from sparse annotation. In
International Conference on Medical Image Computing
and Computer-Assisted Intervention (pp. 424-432).
Springer, Cham. https://doi.org/10.1007/978-3-319-
46723-8_49
DeVries, T., & Taylor, G. W. (2017). Improved
regularization of convolutional neural networks with
cutout. arXiv preprint. arXiv:1708.04552.
https://doi.org/10.48550/arXiv.1708.04552
Fischl, B., Salat, D. H., van der Kouwe, A. J. W., Makris,
N., Ségonne, F., Quinn, B. T., & Dale, A. M. (2004).
Sequence-independent segmentation of magnetic
resonance images. NeuroImage, 23(S1), S69-S84.
https://doi.org/10.1016/j.neuroimage.2004.07.016
Hatamizadeh, A., Yin, Y., Kuo, W.-L., & Myronenko, A.
(2021). Swin UNETR: Swin transformers for semantic
segmentation of brain tumors in MRI images. In
Proceedings of the International Conference on
Medical Image Computing and Computer-Assisted
Intervention (pp. 272-284). Springer, Cham.
https://doi.org/10.1007/978-3-030-87237-3_24
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., &
Maier-Hein, K. H. (2021). nnU-Net: A self-configuring
method for deep learning-based biomedical image
segmentation. Nature Methods, 18(2), 203-211.
https://doi.org/10.1038/s41592-020-01008-z
Kumar, N., Verma, R., & Arora, S. (2021). Three-stage
segmentation of lung region from CT images using
deep neural networks. BMC Medical Imaging, 21(1), 1-
12. https://doi.org/10.1186/s12880-021-00589-8
Milletari, F., Navab, N., & Ahmadi, S.-A. (2016). V-Net:
Fully convolutional neural networks for volumetric
medical image segmentation. In 2016 Fourth
International Conference on 3D Vision (pp. 565-571).
IEEE. https://doi.org/10.1109/3DV.2016.79
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net:
Convolutional networks for biomedical image
segmentation. In Proceedings of the International
Conference on Medical Image Computing and
Computer-Assisted Intervention (pp. 234-241).
Springer, Cham. https://doi.org/10.1007/978-3-319-
24574-4_28
Roy, A. G., Conjeti, S., Navab, N., & Wachinger, C. (2018).
Concurrent spatial and channel 'Squeeze & Excitation'
in fully convolutional networks. In Proceedings of the
International Conference on Medical Image Computing
and Computer-Assisted Intervention (pp. 421-429).
Springer, Cham. https://doi.org/10.1007/978-3-030-
00928-1_47
Salehi, S. S. M., Erdogmus, D., & Gholipour, A. (2017).
Tversky loss function for image segmentation using 3D
fully convolutional deep networks. In Proceedings of
the International Conference on Machine Learning in
Medical Imaging (pp. 379-387). Springer, Cham.
https://doi.org/10.1007/978-3-319-67389-9_44
Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S., &
Cardoso, M. J. (2017). Generalised dice overlap as a
deep learning loss function for highly unbalanced
segmentations. In Deep Learning in Medical Image
Analysis (pp. 240-248). Springer, Cham.
https://doi.org/10.1007/978-3-319-67558-9_28
Wasserthal, J., Meyer, M., Breit, H. C., Cyriac, J., Yang, S.,
& Segeroth, M. (2023). Totalsegmentator: Robust
segmentation of 104 anatomic structures in CT images.
Radiology: Artificial Intelligence, 5(2), e220198.
https://doi.org/10.1148/ryai.220198
Zhang, H., Cisse, M., Dauphin, Y. N., & Lopez-Paz, D.
(2018). Mixup: Beyond empirical risk minimization. In
International Conference on Learning Representations
(ICLR). https://doi.org/10.48550/arXiv.1710.09412
Enhancing NnU-Net for Improved Medical Image Segmentation: A Comparative Study with TotalSegmentator
251
