
Detecting Brain Tumors Through Multimodal Neural Networks
Antonio Curci
a
and Andrea Esposito
b
Department of Computer Science, University of Bari Aldo Moro, Via E. Orabona 4, 70125 Bari, Italy
Keywords:
DenseNet, Brain Tumor, Classification, Multimodal Model.
Abstract:
Tumors can manifest in various forms and in different areas of the human body. Brain tumors are specifically
hard to diagnose and treat because of the complexity of the organ in which they develop. Detecting them
in time can lower the chances of death and facilitate the therapy process for patients. The use of Artificial
Intelligence (AI) and, more specifically, deep learning, has the potential to significantly reduce costs in terms
of time and resources for the discovery and identification of tumors from images obtained through imaging
techniques. This research work aims to assess the performance of a multimodal model for the classification
of Magnetic Resonance Imaging (MRI) scans processed as grayscale images. The results are promising, and
in line with similar works, as the model reaches an accuracy of around 99%. We also highlight the need for
explainability and transparency to ensure human control and safety.
1 INTRODUCTION
Brain tumors refer to a heterogeneous group of tu-
mors arising from cells within the Central Nervous
System (CNS) (WHO Classification of Tumours Ed-
itorial Board, 2022). These tumors can manifest in
various forms, ranging from benign to malignant, and
may originate within the brain tissue or spread from
other parts of the body through metastasis (Lapointe
et al., 2018). In this regard, it is crucial to underline
that tumors that spread in brains are incredibly com-
plex to treat because of the extreme delicacy that the
organ in question is characterized by.
Brain tumors can rise several symptoms in indi-
viduals who suffer from them, such as strong and
recurring headaches, nausea, altered mental status,
papilledema, and seizures; the implications of these
symptoms in individuals can worsen over time if the
tumor is not detected in time, resulting, eventually, in
death (Alentorn et al., 2016). This implies that the
prompt detection, diagnosis, and removal of tumors
must be supported by proper tools and techniques to
assist professionals and increase their efficiency when
performing these tasks. Therefore, there is the need
for ools and instruments featuring the newest tech-
nologies that can support and facilitate this process
for physicians (McFaline-Figueroa and Lee, 2018).
The aid of technology, more specifically Artifi-
a
https://orcid.org/0000-0001-6863-872X
b
https://orcid.org/0000-0002-9536-3087
cial Intelligence (AI), can provide significant advan-
tages concerning the precision, speed, and overall ef-
ficacy of detecting these tumors, thereby improving
therapy outcomes and quality of life (Ranjbarzadeh
et al., 2023). In fact, the landscape of AI models for
the detection of brain tumors is vivid (Anaya-Isaza
et al., 2023; Vermeulen et al., 2023; Huang et al.,
2022; Ranjbarzadeh et al., 2023).
Traditionally, brain tumors are diagnosed by us-
ing imaging techniques, such as Magnetic Reso-
nance Imaging (MRI), Computed Tomography (CT),
or Positron Emission Tomography (PET), which are
incredibly useful and effective. However, the in-
tegration of AI in this context can further improve
and enhance their outputs and maximize efficiency
(Villanueva-Meyer et al., 2017). Recent research has
focused on using machine learning and deep learning
techniques for brain tumor classification, segmenta-
tion, and feature extraction, as well as developing AI
tools to assist neurosurgeons during treatment (Ver-
meulen et al., 2023; Huang et al., 2022).
The current scenario of the application of Neu-
ral Networks employed in the field of medicine and
in brain tumor detection encompasses various mod-
els and techniques, and still represents a very chal-
lenging issue. For instance, Mohesen et al. use Deep
Neural Networks (DNN), combined with Principal
Component Analysis (PCA), and Discrete Wavelet
Transform, achieving a good accuracy, around 97%
(Mohsen et al., 2018). Pei et al., instead, used 3D
Curci, A. and Esposito, A.
Detecting Brain Tumors Through Multimodal Neural Networks.
DOI: 10.5220/0012608600003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 995-1000
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
995

Convolutional Neural Networks (CNN), reaching a
training accuracy of around 81% and a validation ac-
curacy of around 75% (Pei et al., 2020). In addition,
Nayak et al. developed another CNN as a variant of
Efficient DenseNets with dense and drop-out layers,
obtaining an accuracy close to 99% (Nayak et al.,
2022). The employment of these models in classifi-
cation tasks in medicine can be significantly useful.
At the same time, it remains crucial for profession-
als to maintain control and be able to check the out-
put of these instruments to have the final say over
the model’s predictions. The employment of multi-
modal models, instead, is still under development and
research in the literature. It is possible to find cases in
which these models are built with Multi-Layer Per-
ceptrons (MLP) or with DenseNets for 3D images
image classification, in which researchers could not
achieve high-performance rates (Ma and Jia, 2020;
Latif et al., 2017). Different modalities provide dif-
ferent types of information. Images can visual in-
formation about the tumor’s location, size, and ex-
ternal characteristics, while tabular data can include
insights about other aspects and peculiarities either
highlighted by the physician or numerical data ex-
tracted from the images themselves. Combining these
modalities can improve the AI model when it comes
to learning how to discriminate between tumor and
non-tumor cases. Multimodal AI can also provide
a more comprehensive decision support system for
healthcare professionals, leading to better clinical
decision-making and treatment planning (Soenksen
et al., 2022; Yang et al., 2016).
This research work aims at creating and employ-
ing a multi-modal model to classify brain images as
healthy or ill (i.e., containing a tumor) and propos-
ing an approach towards stronger explainability and
transparency to increase physicians’ trust levels when
using AI in medicine. The model in question was built
through a Densely Connected Convolutional Network
(DenseNet) and it was trained over a labeled dataset
composed of tabular data and 2D brain tumor images.
This paper is organized as follows: section 2 en-
compasses all the materials used during this study,
defining the dataset, its provenance, and the distribu-
tion of the classes; section 3 explores the model, its
structure, and the parameters set for the experiment.
In section 4, we describe the tools used to carry out
the experiment and we analyze the results; section 5,
instead, provides an overview of the research work,
its outcomes, and the future directions that we intend
to undertake for this project, highlighting the need for
explainability and control.
2 MATERIALS
This research work was conducted using a dataset de-
rived from the BRATS 2015 challenge (Menze et al.,
2015), freely available on Kaggle.com (Jakesh Bo-
haju, ). The dataset comprises 3762 instances. Each
instance consists in a 240 × 240 three-channel MRI
scans of the brain, and in a set of 13 numeric features
(with an additional feature that allows to identify the
scan associated with the numeric values). The dataset
is fully labeled. The labels are binary and mutually
exclusive: a value of “0” represents the absence of
a tumor (in the following, we will refer to this class
as “healthy”); a value of “1” indicates the presence
of a tumor (in the following, we will refer to this
class as “ill”). The tabular data has 13 features of
first- an second-order; they were extracted by the au-
thors of the dataset from the images, which are the
processed output of MRI scans. The first-order fea-
tures are Mean, Variance,Standard Deviation, Skew-
ness, Kurtosis, while those of second-order are En-
tropy, Contrast, Entropy, Energy, Dissimilarity, Cor-
relation, Coarseness, ASM (Angular second moment),
Homogeneity.
The dataset is slightly unbalanced, with 2079 in-
stances labeled as healthy and 1683 labeled as ill.
To avoid the potential introduction of artifacts or
unrealistic samples using data augmentation (Chlap
et al., 2021), the class-imbalance problem was solved
by dropping randomly selected instances from the
“healthy” class. The numeric features of the dataset
(a) Healthy scan.
(b) Scan presenting a tu-
mor.
(c) Highlighted lesion of
the ill brain.
Figure 1: Examples of MRI scans available in the dataset.
NeroPRAI 2024 - Workshop on Medical Condition Assessment Using Pattern Recognition: Progress in Neurodegenerative Disease and
Beyond
996
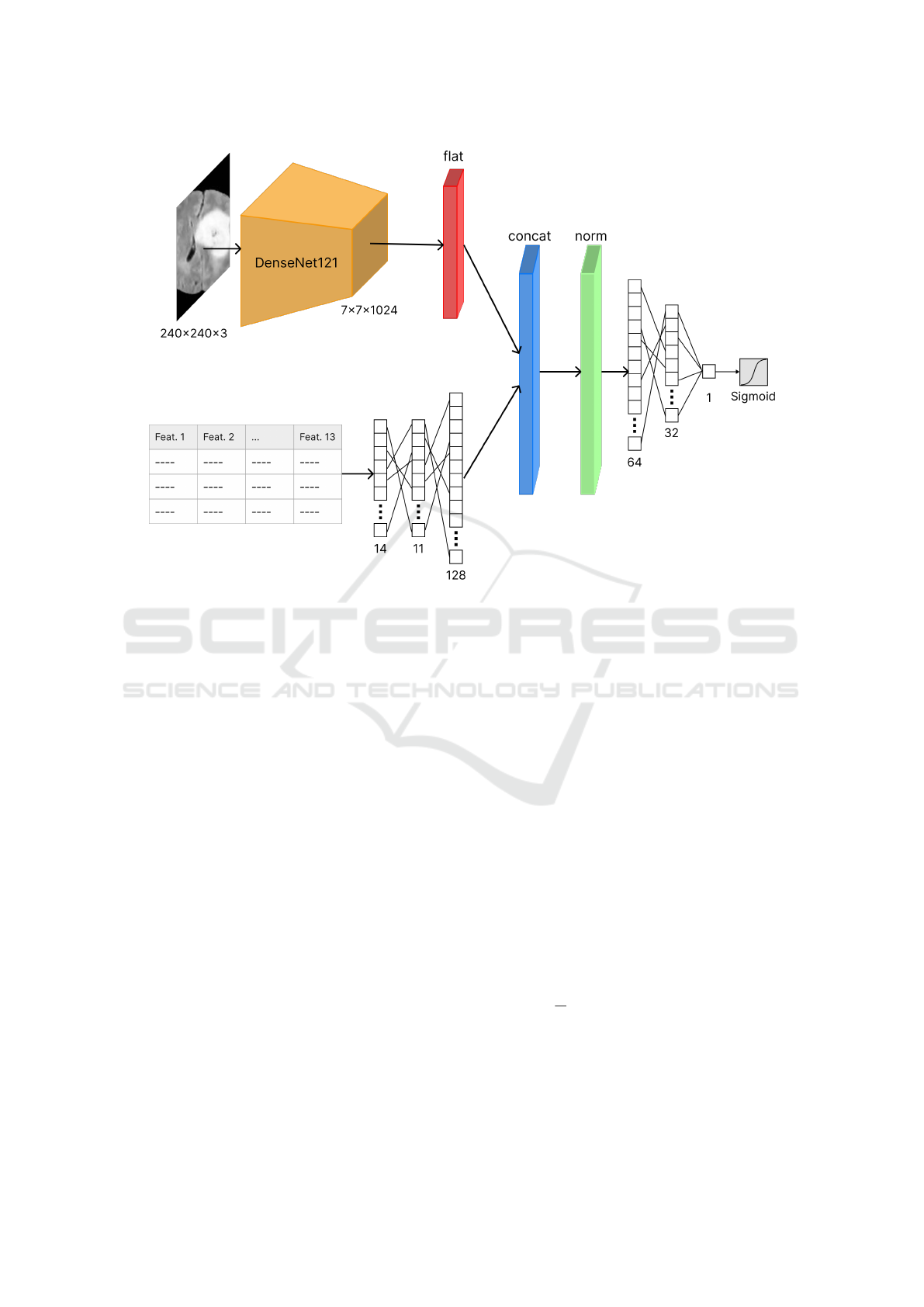
Figure 2: Architecture of the multi-modal deep neural network.
have been standardized in order to have mean µ = 0
and variance σ
2
= 1. The dataset has no missing val-
ues, making it unnecessary to perform any additional
pre-processing.
Figure 1 provides examples of images labeled as
healthy and ill. More specifically, Figure 1b is the im-
age of the scan of a brain containing a tumor, which is
found in its lower-right part as a white area that stands
out from the rest of the organ; the latter is pointed in
Figure 1c in the highlighted red rectangle.
3 METHODS
The model used for this research work is a multi-
modal neural network. The model architecture, de-
picted in Figure 2, is composed of two heads (one
for each type of input data). The first head is respon-
sible for the feature extraction from the MRI scans:
it consists in a DenseNet121 network (Huang et al.,
2018) with input size 240 × 240 × 3 and output size
7 × 7 ×1024, that is then flattened. The second head,
responsible for the encoding of the tabular data, con-
sists in a simple fully-connected neural network, us-
ing the Rectivied Linear Unit (ReLU) activation func-
tion. The outputs of the two heads are then concate-
nated and normalized. The resulting vector is then
provided as input to an additional fully-connected
neural network (also using the ReLu activation func-
tion), which terminates in two SoftMax-activated neu-
rons that provide the final prediction. The model is
shown in Figure 2.
4 RESULTS
The experiment was performed using an Apple Sil-
icon M2 Pro chip with an integrated 16-core GPU,
using the TensorFlow library.
To evaluate the proposed method, a stratified 10-
fold cross-validation was used (i.e., each fold con-
tained roughly the same proportion of the two class
labels). For the training phase, we used binary cross-
entropy as the loss function, defined in Equation 1,
where y
i
is the ground truth label, while p
i
is the
model output for an individual observation.
H (y, p) =
1
N
N
∑
i=1
−(y
i
log(p
i
) + (1 − y
i
)log(1 − p
i
))
(1)
Cross-entropy was minimized using the Adam op-
timizer, with a static learning rate of 10
−3
and a batch
size of 32. The maximum number of epochs was set
to 10
2
, with an early stopping criterion based on the
Detecting Brain Tumors Through Multimodal Neural Networks
997
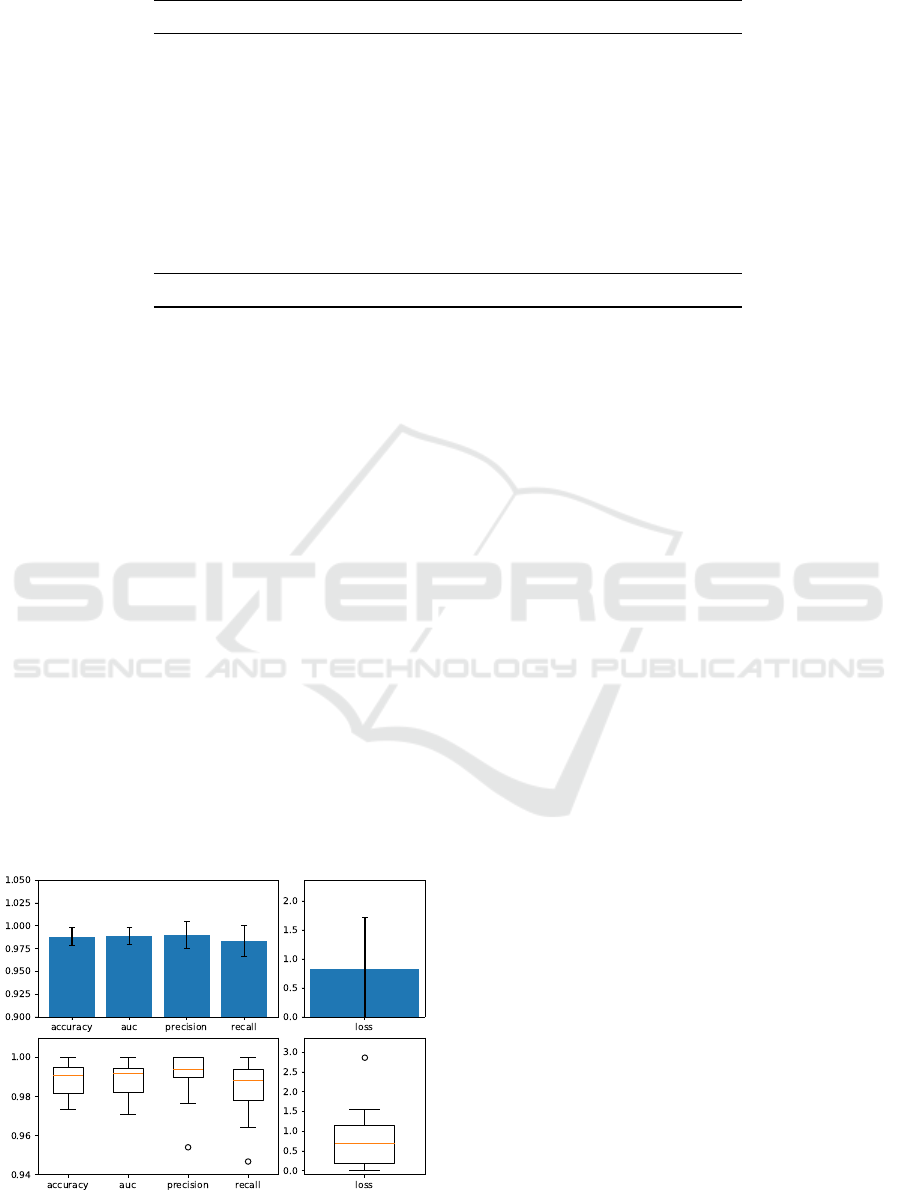
Table 1: Results of the cross validation.
CV Fold Accuracy AUC Loss Precision Recall F1-Score
1 0.99 0.99 0.18 0.99 0.98 0.99
2 0.97 0.97 1.5 0.99 0.95 0.97
3 0.99 0.99 5.6e-05 0.99 0.99 0.99
4 0.98 0.98 1.3 0.98 0.98 0.98
5 0.97 0.98 0.67 0.95 0.99 0.97
6 0.99 0.99 0.84 0.99 0.99 0.99
7 0.99 0.99 0.72 0.99 0.99 0.99
8 0.98 0.98 2.9 0.99 0.96 0.98
9 0.99 0.99 0.22 0.99 0.99 0.99
10 0.99 0.99 0 0.99 0.99 0.99
Avg. 0.99 0.99 0.83 0.99 0.98 0.98
validation loss with a minimum delta of 10
−4
and a
patience of 5 epochs.
As performance metrics, we opted for the most
commonly used metric in classification problems:
• Accuracy: defined as the proportion of the cor-
rectly classified samples (both positives and neg-
atives) in the selected population.
• Recall: which refers to the proportion of diseased
subjects who have been classified as ill;
• Precision: that is the proportion of the correctly
classified samples among all ill-classified sam-
ples;
• F1-Score: that is the harmonic mean between the
precision and recall;
• Area Under ROC-Curve (AUC): that indicates the
probability that, given a healthy and an ill sample,
the classifier is able to correctly distinguish them.
The training phase on the 10 folds exhibited quite
good performances, shown in Table 1; each fold gen-
erated accuracy rates higher than 97%, with an av-
erage of 98.80%. The average values for all metrics
Figure 3: Results of the cross-validation.
are available in Table 1. The loss has values less than
1.5, as shown in Figure 2. The only exception is the
eighth fold, which has a loss value close to 2.9: fur-
ther inspection is needed to uncover the reasons for
this sudden peak.
5 CONCLUSIONS AND FUTURE
WORKS
In this article, we explore the use of multi-modal
DenseNets for brain tumor images classification. The
presented model is useful when dealing with data of
different types with intrinsically different representa-
tions, in this case, tabular data and images. The multi-
modal Deep Neural Network created and exploited in
this case study provides promising results for classi-
fying brain tumor images, achieving an average ac-
curacy of 98%. The results are on par with other
techniques found in the literature (Nayak et al., 2022;
Mohsen et al., 2018).
Although the dataset used in this work was also
used by other researchers in the community, a multi-
modal model was never chosen as the approach to un-
dertake to perform a classification task. Arora et al
created a model consisting of a Convolutional Neural
Network (CNN) that reached a 90% accuracy using
a VGG16 Neural Network (Arora and Sharma, 2021;
Simonyan and Zisserman, 2015). Herm et al, instead,
employed a CNN with 13 layers, obtaining an accu-
racy of around 89% (Herm et al., 2023). Another re-
search work was performed on this dataset by Morris
L., whose model achieved 87% of accuracy by using a
Deep Neural Network with MobileNetV2 (Lee, 2022;
Sandler et al., 2019). It emerges that the work pre-
sented in this article provides a starting ground for fu-
ture research, exploring how the exploitation of differ-
ent types of data can be for the classification of brain
NeroPRAI 2024 - Workshop on Medical Condition Assessment Using Pattern Recognition: Progress in Neurodegenerative Disease and
Beyond
998

tumor images.
An initial aspect that needs further exploring is the
model generalizability: future work may delve in test-
ing the model with additional parameters and/or an-
other dataset with the same structure and belonging
to the same medical domain, to observe its behavior
and efficacy in different settings. Moreover, future
work involves also the comparison of the performance
of this multi-modal model with standard classifiers,
meaning models that are trained merely on tabular
data or on images. The objective is to determine how
the characteristics of the model proposed in this work
can be beneficial to the medical field with respect to a
more traditional approach.
In addition, explainability and transparency are
needed to provide users (i.e., physicians) with more
efficient instruments to understand and comprehend
the outputs it provides. As neural networks’ out-
puts are usually obscure to users without expertise in
computer science and, specifically, in AI, explainabil-
ity has the potential of demystifying the process that
lies behind the final predictions and output of mod-
els. Moreover, it is crucial for physicians to fully
understand the reasons why an AI systems provided
a specific outcome (Combi et al., 2022), as this en-
sure human control. In fact, from an ethical point
of view, the responsibility that physicians undertake
when making decisions about the health state of their
patients cannot depend merely on algorithms that they
do not comprehend properly. Explainability plays
an important role for physicians because it allows to
check and keep track of which features were relevant
for the prediction outputted by the AI model and de-
tecting potential mistakes that can be corrected thanks
to their expertise. The motivation behind this lies in
the fact that AI systems are never perfectly accurate,
thus, the clinical revision process has to be carried
out precisely and meticulously by professionals, im-
plying that having complete and blind trust is not fea-
sible for legal reasons, too (Amann et al., 2020). It
emerges that the goal is to approach a symbiotic re-
lationship between AI and humans. The use of AI in
medicine, especially Neural Networks, can be benefi-
cial both diagnostically and to foster and guide future
research (e.g., through machine teaching (Selvaraju
et al., 2016)).
The multi-modal neural network presented in this
article provides an interesting proving ground, to
evaluate the balance between accuracy, model com-
plexity, and explainability in a challenging high-risk
domain.
ACKNOWLEDGEMENTS
The research of Antonio Curci is supported by the
co-funding of the European Union - Next Genera-
tion EU: NRRP Initiative, Mission 4, Component 2,
Investment 1.3 – Partnerships extended to universi-
ties, research centers, companies, and research D.D.
MUR n. 341 del 15.03.2022 – Next Generation
EU (PE0000013 – “Future Artificial Intelligence Re-
search – FAIR” - CUP: H97G22000210007).
The research of Andrea Esposito is funded by a
Ph.D. fellowship within the framework of the Italian
“D.M. n. 352, April 9, 2022” - under the National Re-
covery and Resilience Plan, Mission 4, Component 2,
Investment 3.3 - Ph.D. Project “Human-Centered Ar-
tificial Intelligence (HCAI) techniques for supporting
end users interacting with AI systems”, co-supported
by “Eusoft S.r.l.” (CUP H91I22000410007).
REFERENCES
Alentorn, A., Hoang-Xuan, K., and Mikkelsen, T. (2016).
Presenting signs and symptoms in brain tumors. In
Handbook of Clinical Neurology, volume 134, pages
19–26. Elsevier.
Amann, J., Blasimme, A., Vayena, E., Frey, D., Madai, V. I.,
and the Precise4Q consortium (2020). Explainability
for artificial intelligence in healthcare: A multidisci-
plinary perspective. BMC Medical Informatics and
Decision Making, 20(1):310.
Anaya-Isaza, A., Mera-Jim
´
enez, L., Verdugo-Alejo, L., and
Sarasti, L. (2023). Optimizing MRI-based brain tumor
classification and detection using AI: A comparative
analysis of neural networks, transfer learning, data
augmentation, and the cross-transformer network. Eu-
ropean Journal of Radiology Open, 10:100484.
Arora, S. and Sharma, M. (2021). Deep Learning for Brain
Tumor Classification from MRI Images. In 2021 Sixth
International Conference on Image Information Pro-
cessing (ICIIP), pages 409–412, Shimla, India. IEEE.
Chlap, P., Min, H., Vandenberg, N., Dowling, J., Holloway,
L., and Haworth, A. (2021). A review of medical im-
age data augmentation techniques for deep learning
applications. Journal of Medical Imaging and Radia-
tion Oncology, 65(5):545–563.
Combi, C., Amico, B., Bellazzi, R., Holzinger, A., Moore,
J. H., Zitnik, M., and Holmes, J. H. (2022). A
Manifesto on Explainability for Artificial Intelligence
in Medicine. Artificial Intelligence in Medicine,
(133):102423.
Herm, L.-V., Heinrich, K., Wanner, J., and Janiesch, C.
(2023). Stop ordering machine learning algorithms by
their explainability! A user-centered investigation of
performance and explainability. International Journal
of Information Management, 69:102538.
Huang, G., Liu, Z., van der Maaten, L., and Weinberger,
Detecting Brain Tumors Through Multimodal Neural Networks
999

K. Q. (2018). Densely Connected Convolutional Net-
works.
Huang, J., Shlobin, N. A., Lam, S. K., and DeCuypere,
M. (2022). Artificial Intelligence Applications in Pe-
diatric Brain Tumor Imaging: A Systematic Review.
World Neurosurgery, 157:99–105.
Jakesh Bohaju. Brain Tumor.
Lapointe, S., Perry, A., and Butowski, N. A. (2018).
Primary brain tumours in adults. The Lancet,
392(10145):432–446.
Latif, G., Mohsin Butt, M., Khan, A. H., Omair Butt,
M., and Al-Asad, J. F. (2017). Automatic Multi-
modal Brain Image Classification Using MLP and 3D
Glioma Tumor Reconstruction. In 2017 9th IEEE-
GCC Conference and Exhibition (GCCCE), pages 1–
9, Manama. IEEE.
Lee, M. (2022). Brain Tumor, Detection from MRI images
[Deep CN]. Accessed 10-01-2024.
Ma, X. and Jia, F. (2020). Brain Tumor Classification with
Multimodal MR and Pathology Images. In Crimi,
A. and Bakas, S., editors, Brainlesion: Glioma, Mul-
tiple Sclerosis, Stroke and Traumatic Brain Injuries,
volume 11993, pages 343–352. Springer International
Publishing, Cham.
McFaline-Figueroa, J. R. and Lee, E. Q. (2018). Brain
Tumors. The American Journal of Medicine,
131(8):874–882.
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J.,
Farahani, K., Kirby, J., Burren, Y., Porz, N., Slot-
boom, J., Wiest, R., Lanczi, L., Gerstner, E., We-
ber, M.-A., Arbel, T., Avants, B. B., Ayache, N.,
Buendia, P., Collins, D. L., Cordier, N., Corso, J. J.,
Criminisi, A., Das, T., Delingette, H., Demiralp, C.,
Durst, C. R., Dojat, M., Doyle, S., Festa, J., Forbes,
F., Geremia, E., Glocker, B., Golland, P., Guo, X.,
Hamamci, A., Iftekharuddin, K. M., Jena, R., John,
N. M., Konukoglu, E., Lashkari, D., Mariz, J. A.,
Meier, R., Pereira, S., Precup, D., Price, S. J., Ra-
viv, T. R., Reza, S. M. S., Ryan, M., Sarikaya, D.,
Schwartz, L., Shin, H.-C., Shotton, J., Silva, C. A.,
Sousa, N., Subbanna, N. K., Szekely, G., Taylor, T. J.,
Thomas, O. M., Tustison, N. J., Unal, G., Vasseur, F.,
Wintermark, M., Ye, D. H., Zhao, L., Zhao, B., Zi-
kic, D., Prastawa, M., Reyes, M., and Van Leemput,
K. (2015). The Multimodal Brain Tumor Image Seg-
mentation Benchmark (BRATS). IEEE Transactions
on Medical Imaging, 34(10):1993–2024.
Mohsen, H., El-Dahshan, E.-S. A., El-Horbaty, E.-S. M.,
and Salem, A.-B. M. (2018). Classification using deep
learning neural networks for brain tumors. Future
Computing and Informatics Journal, 3(1):68–71.
Nayak, D. R., Padhy, N., Mallick, P. K., Zymbler, M., and
Kumar, S. (2022). Brain Tumor Classification Using
Dense Efficient-Net. Axioms, 11(1):34.
Pei, L., Vidyaratne, L., Hsu, W.-W., Rahman, M. M., and
Iftekharuddin, K. M. (2020). Brain Tumor Classifi-
cation Using 3D Convolutional Neural Network. In
Crimi, A. and Bakas, S., editors, Brainlesion: Glioma,
Multiple Sclerosis, Stroke and Traumatic Brain In-
juries, volume 11993, pages 335–342. Springer Inter-
national Publishing, Cham.
Ranjbarzadeh, R., Caputo, A., Tirkolaee, E. B., Ja-
farzadeh Ghoushchi, S., and Bendechache, M. (2023).
Brain tumor segmentation of MRI images: A com-
prehensive review on the application of artificial in-
telligence tools. Computers in Biology and Medicine,
152:106405.
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and
Chen, L.-C. (2019). Mobilenetv2: Inverted residuals
and linear bottlenecks.
Selvaraju, R. R., Das, A., Vedantam, R., Cogswell, M.,
Parikh, D., and Batra, D. (2016). Grad-CAM: Why
did you say that? Visual explanations from deep
networks via gradient-based localization. CoRR,
abs/1610.02391.
Simonyan, K. and Zisserman, A. (2015). Very deep convo-
lutional networks for large-scale image recognition.
Soenksen, L. R., Ma, Y., Zeng, C., Boussioux, L., Villalo-
bos Carballo, K., Na, L., Wiberg, H. M., Li, M. L.,
Fuentes, I., and Bertsimas, D. (2022). Integrated mul-
timodal artificial intelligence framework for health-
care applications. npj Digital Medicine, 5(1):149.
Vermeulen, C., Pag
`
es-Gallego, M., Kester, L., Kranendonk,
M. E. G., Wesseling, P., Verburg, N., de Witt Hamer,
P., Kooi, E. J., Dankmeijer, L., van der Lugt, J., van
Baarsen, K., Hoving, E. W., Tops, B. B. J., and de Rid-
der, J. (2023). Ultra-fast deep-learned CNS tumour
classification during surgery. Nature, 622(7984):842–
849.
Villanueva-Meyer, J. E., Mabray, M. C., and Cha, S. (2017).
Current Clinical Brain Tumor Imaging. Neurosurgery,
81(3):397–415.
WHO Classification of Tumours Editorial Board, editor
(2022). WHO Classification of Tumours: Central Ner-
vous System Tumours. World Health Organization,
Lyon, 5th edition edition.
Yang, C.-H., Chang, P.-H., Lin, K.-L., and Cheng, K.-S.
(2016). Outcomes comparison between smartphone
based self-learning and traditional speech therapy for
naming practice. In 2016 International Conference on
System Science and Engineering (ICSSE), pages 1–4,
Puli, Taiwan. IEEE.
NeroPRAI 2024 - Workshop on Medical Condition Assessment Using Pattern Recognition: Progress in Neurodegenerative Disease and
Beyond
1000
