
Simultaneous Optimization of Edge Bundling and Node Layout Using
Genetic Algorithm
Junsei Meikari
1
and Ryosuke Saga
2a
1
Graduate School of Humanities and Sustainable System Sciences, Osaka Prefecture University, Sakai, Japan
2
Graduate School of Informatics, Osaka Metropolitan University, Sakai, Japan
Keywords: Visualization, Edge Bundling, Node Layout, Genetic Algorithm.
Abstract: This paper describes an evolutionary visualization method that performs edge bundling during the execution
of the genetic algorithm. There are several node layout algorithms and edge bundling, however, there are no
methods considering both algorithms simultaneously. This paper proposes an algorithm to optimize the fitness
function of GABEB, which is genetic algorithm-based edge bundling, and Zhang’s node layout
simultaneously. The experiments for the sample graphs show the improved result from the viewpoints of
several evaluation criteria.
1 INTRODUCTION
The graph, consisting of edges representing
connections between nodes, is used to visually
represent various relationships among data
represented as nodes, such as in aviation charts or
relation diagrams (Saga 2012). However, as the scale
and complexity of the graph increase, it becomes
challenging for users to comprehend the relationships
between nodes, leading to visual clutter problem. To
address such issues, various visualization techniques
have been proposed over time, including force-
directed graph drawing (Kamada, 1989) and
compound graph layout (Sugiyama 1991)(Dogrusoz
2009).
In recent years, edge bundling has been proposed
as a method to solve visual clutter problem. Edge
bundling involves spatially grouping edges according
to certain rules, reducing the visual complexity of the
graph and making it easier to visually grasp the
connection relationships between nodes.
Various edge bundling techniques have been
proposed, and their effectiveness has been
demonstrated (Lhuillier 2017). On the other hand, in
recent years, stochastic optimization approaches such
as Genetic Algorithms (GA) have also been
implemented to address these problems. The
advantages of using such GA approaches include the
potential for obtaining visualization results not
a
https://orcid.org/0000-0003-1528-6534
anticipated by humans and the ability to apply
stochastic optimization methods regardless of the
continuity of the visualization problem space.
The studies like Ferreira et al. (Ferreira 2018) and
Saga et al. (Saga 2020) have proposed edge bundling
using GA, receiving certain evaluations. While
various edge bundling techniques have been proposed,
those that include the process of adjusting node
positions in the edge bundling process have been
rarely suggested. When the process of adjusting node
positions is not included in edge bundling techniques,
even if the method effectively bundles edges before
application, the visualization effects of edge bundling
may not be fully realized due to the inappropriate
positions of nodes in the graph before the application
of the method, especially in cases with a high number
of edge crossings.
Considering the aspects, Meikari et al. (Meikari
2022) proposed an initial approach to simultaneously
optimize node layout and edge bundling using a
genetic algorithm. However, the solution to the
algorithm is the position of the nodes and the
combination of edges for bundles, and actual edge
bundling takes place after the completion of the
genetic algorithm. Therefore, the process of bundling
edges and node layout can be considered separate.
In this paper, our goal is to propose an
evolutionary visualization method where node layout
and edge bundling are optimized simultaneously.
Meikari, J. and Saga, R.
Simultaneous Optimization of Edge Bundling and Node Layout Using Genetic Algorithm.
DOI: 10.5220/0012574200003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 1: GRAPP, HUCAPP
and IVAPP, pages 741-748
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
741

Additionally, we aim to validate the visualization
effects of this method by running the implemented
program and comparing the obtained results with
those of the methods by Meikari’s method.
2 RELATED WORKS
2.1 Edge Bundling
Edge bundling is a graph visualization technique that
involves spatially grouping edges within a graph
according to certain rules. This bundling aims to
reduce the visual clutter of the graph, making it easier
to visually comprehend the connection relationships
between nodes connected by edges. Figure 1
illustrates an example of a graph with edge bundling.
Originating in 1989 with the simplification of
graph edges through spatial concentration (Newbery
1989), edge bundling has evolved into various
methods such as Hierarchical Edge Bundling (Holten
2006) and others, demonstrating its effectiveness
(Telea, 2010) (Ersoy, 2011) (Hurter, 2012) (Cui,
2008) (Lhuillier, 2017). Another well-known method
is Force-Directed Edge Bundling (FDEB), proposed
by Holten et al. (Holten, 2009). FDEB employs a
mechanical approach, modeling edges as springs that
attract each other. It evenly allocates splitting points
along the edges, calculates the magnitude and
direction of forces acting on each splitting point based
on the distance between points and the geometric
similarity between edges, and forms smooth curves
through iterative movements of splitting points and
information updates.
Figure 1: Edge Bundling.
2.2 Edge Bundling Using Genetic
Algorithm
In recent years, probabilistic optimization approaches
such as Genetic Algorithms (GA) have been
implemented. The benefits of using such approaches
include the potential to obtain visualization results not
anticipated by humans and the ability to apply
stochastic optimization methods regardless of
whether the visualization problem space is
continuous. Additionally, there is an implementation
advantage in terms of ease.
Evolutionary Edge Bundling (EEB), proposed by
Ferreira et al. (Ferreira 2018), is an edge bundling
method that utilizes genetic algorithms to determine
bundles (sets of edges) that should be grouped
together. Therefore, the process of bundling edges is
not included in the genetic algorithm's process, and
the actual bundling process uses FDEB after the
genetic algorithm concludes. The graph is evaluated
based on the similarity in angle and length of edges
within bundles that should be maximized and the
number of bundles that should be minimized.
Evolutionary Node Layout and Edge Bundling
(ENLEB), proposed by Meikari et al. (Meikari 2022),
integrates EEB and node layout methods into a single
genetic algorithm. This was the first approach aiming
to simultaneously optimize edge bundling and node
layout in evolutionary visualization. Therefore, it
allows obtaining visualization effects considering
both edge bundling and node layout. The solution
obtained by ENLEB includes the positions of nodes
and bundles of edges, and similar to EEB, the process
of bundling edges using FDEB occurs after the
genetic algorithm concludes. Hence, the processes of
node layout and edge bundling are executed
separately.
Genetic Algorithm Based Edge Bundling
(GABEB), proposed by Saga et al. (Saga 2020), is an
edge bundling method that uses genetic algorithms by
placing control points on edges and optimizing their
positions. Control point movements are treated as
genes, and genetic operations such as crossover and
mutation are applied. The evaluation of individuals
includes four aesthetic criteria related to edge
bundling: Mean Edge Length Difference (MELD),
Mean Occupation Area (MOA), Edge Density
Distribution (EDD) proposed by Saga et al.(Saga
2016), and Path Quality proposed by Cui et al. (Cui
2008).
2.2.1 Mean Edge Length Difference
Mean Edge Length Difference (MELD) is a criterion
to express the difference from the original edges after
edge bundling. A smaller change of edge lengths
indicates superior edge bundling because of over-
bundling, whereas a large change often leads to a loss
of the meaning of the original network. MELD is
calculated as
𝑀𝐸𝐿𝐷
∑ |
𝐿
𝑒
−𝐿
𝑒
|
∈
(1
)
where n is the number of edges, E is the edge set, and
L(e) and L’(e) are the lengths of edge e before and
after edge bundling, respectively. In our approach, we
aim to minimize the MELD.
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
742

2.2.2 Mean of Occupation Area
Mean of Occupation Area (MOA) indicates the
degree among the compressed areas before and after
edge bundling. Based on the idea that better bundling
can compress the area occupied by the edges, MOA
is calculated as
𝑀𝑂𝐴=
1
𝑁
𝑂
𝑒
∈
(2)
where N is the number of total areas, O(e) is the set of
areas occupied by edge e based on an occupation
degree (we use 5% of unit area), and | | indicates the
number of elements contained by a set. Minimising
the MOA is one of our optimization goals.
2.2.3 Edge Density Distribution
Edge Density Distribution (EDD) is rooted in the idea
that a better edge bundling method can gather edges
within a unit area and that the density per unit is high.
EDD is calculated as
𝐸𝐷𝐷=
||
∑
𝐻
𝑝
−𝐻
∈
(3)
where P is a set of pixels, H(p) is the number of edges
pathing pixel p, and H is the average of H(p). We aim
to maximise the EDD.
2.2.4 Path Quality
Path Quality (PQ) expresses the degree of zigzag. The
lower the PQ, the better the edge bundling. PQ is
calculated by the summation of angle differences
between neighbours as
with
∆
=
𝐴
−𝐴
|
𝐴
−𝐴
|
−2𝜋
2𝜋+
|
𝐴
−𝐴
|
i
f
−𝜋<
|
𝐴
−𝐴
|
< 𝜋
i
f
|
𝐴
−𝐴
|
> 𝜋
i
f
|
𝐴
−𝐴
|
< −𝜋
(5)
and
𝛾
=
0
1
i
f
sign
∆
=sign
∆
i
f
sign
∆
≠sign
∆
(6)
, where m is the number of segments divided by
control points+1, and A
i
is the angle between the
original edge and the segment edge. In our GA, we
try to maximize PQ. We use the above four criteria
separately and perform multi-objective optimization.
3 PROPOSED METHODS
In this paper, we address the issue present in EEB,
where the processes of edge bundling and node layout
were executed separately. We propose an
evolutionary visualization method that
simultaneously performs edge bundling and node
layout while optimizing both using a genetic
algorithm. To achieve this, we integrate the genetic
algorithm-based edge bundling method GABEB and
Zhang et al.'s node layout method (Zhang 2005) into
a single genetic algorithm.
The reason for using GABEB is that it operates as
a genetic algorithm solely for manipulating control
points on edges, without considering the impact on
node positions. This makes it less likely to encounter
problems when simultaneously performing node
layout and edge bundling. In the context of our
method aiming to perform edge bundling and node
layout concurrently, GABEB is well-suited for
conducting edge bundling within the genetic
algorithm.
Additionally, we choose Zhang et al.'s node layout
method for two main reasons. First, it is a genetic
algorithm-based node layout method. Second, by
adjusting the weights of the evaluation function in
Zhang et al.'s method, our proposed method can more
easily reflect elements such as the number of edge
crossings that need improvement in the graph
evaluation, especially in the node layout part.
3.1 The Process of Genetic Algorithm
Figure 2 illustrates the flow of the genetic algorithm
for this method. The genetic algorithm of this method
repeats the process of crossover, mutation, joining,
individual evaluation after generating the initial
individuals, and generation update until the
termination condition is satisfied. When a control
point is subject to crossover or mutation, if the control
point is bound to one of the other control points, it is
unbound to prevent an anomaly in the information on
the movement of the control point.
Figure 2: Process of Generic Algorithm.
Generate Initial Population
Evaluate population
Crossover, Mutation
Join Control Points
Update Generation
Unjoin Control Points
𝑃𝑄=
−
𝛾
|
∆
|
∈
(4)
Simultaneous Optimization of Edge Bundling and Node Layout Using Genetic Algorithm
743
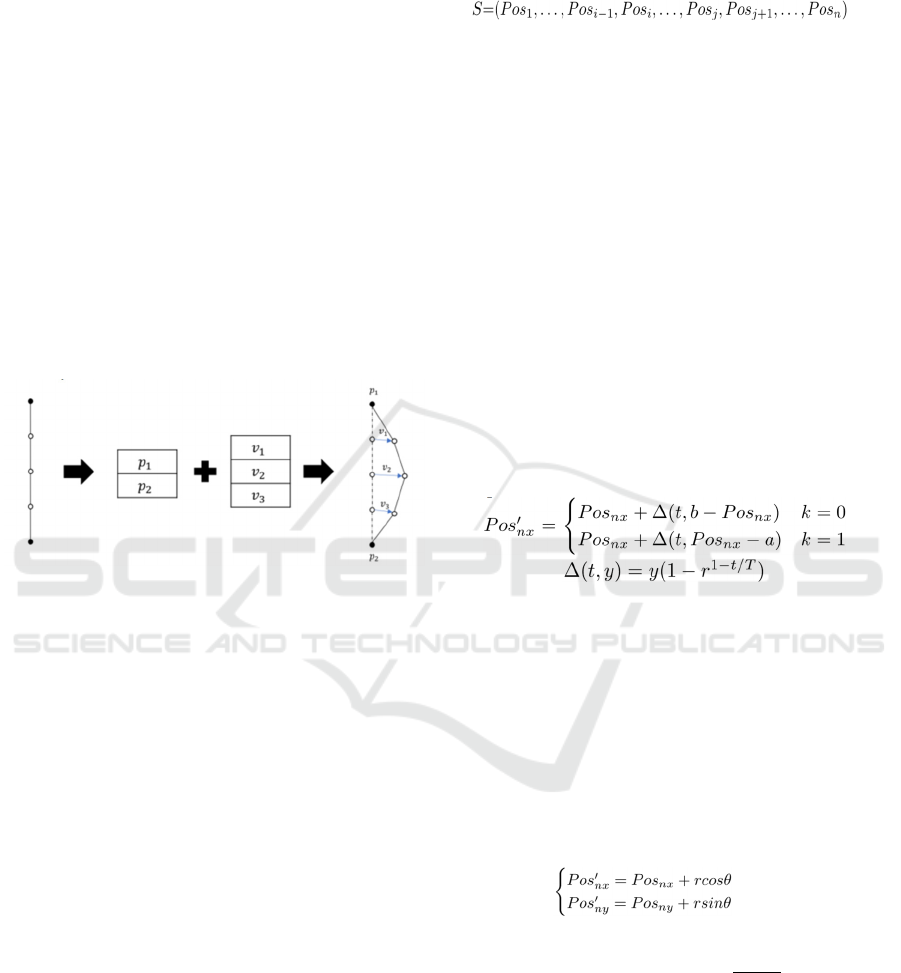
3.2 Chromosomes
Figure 3 shows an image of the individual
representation of the method. Based on GABEB, the
individual representation incorporates the concept of
control points in FDEB. In FDEB, edges are equally
divided by the number of control points, and edge
bundling is performed by moving control points
based on calculations based on mechanical rules such
as the distance between control points. In contrast,
GABEB treats the amount of movement of each
control point as a gene, which represents each
individual. As in FDEB, the original positions of the
control points are assumed to be equally divided by
the number of control points. In addition to the
information on the amount of control point movement
for edge bundling, this method also treats the node
position of each edge as a gene for the purpose of
node layout.
Figure 3: Genetic Representation.
3.3 Crossover
The crossover is performed with respect to the
operation on the position of a node and the amount of
movement of the control point of an edge. For the
former, a simple crossover is performed, in which the
position of one node is exchanged between two parent
individuals, and an inversion, in which the position of
a node of one parent is manipulated. For the latter
operation on the mobility of the control point, we
perform the blended crossover (BLX-α) (α=0.5)
(Eshelman 1993).
The inversion of this method is an operation to
change the position of a node by numbering the
collected positions of the nodes at both ends of an
individual edge, and then inverting the numbers
within a randomly set interval of numbers. Let S be
the list of node positions before applying inversion, S’
be the list of node positions after applying inversion,
i to j be the interval of node positions to be
manipulated, and Pos
i
be the collected node position
information, the operation by inv ersion is as in
quations (7) and (8). After the crossover is applied to
the position of a node, the change in the position of
the node is reflected in the entire gene. Each
crossover is assigned an independent crossover
probability.
(7
)
𝑆ʹ = 𝑃𝑜𝑠
1
,...,𝑃𝑜𝑠
1
, 𝑃𝑜𝑠
, 𝑃𝑜𝑠
1
,..,
𝑃𝑜𝑠
1
, 𝑃𝑜𝑠
, 𝑃𝑜𝑠
1
,...,𝑃𝑜𝑠
(8
)
3.4 Mutation
Mutation involves two genetic manipulations to
change the position of the node as indicated by the
node layout method of Zhang et al. and one genetic
manipulation to change the amount of movement of
the control point.
The mutations shown by Zhang et al. include non-
uniform mutation, in which the shift of node position
becomes smaller with each generation, and single-
vertex-neighbourhood mutation, in which the
position shifts within a circular range from the
original node position.
Let Pos
nx
be the X-coordinate of the nth node of
an individual and Pos
nx
’ be the X-coordinate of the
node after the move, the shift of a node's position due
to non-uniform mutation is calculated by the
equations (9) and (10).
(9)
(10)
In this case, 𝑏 is the maximum value of the X-axis of
the set graph plotting range, and 𝑎 is its minimum
value. 𝑘 is a value of 0 or 1 randomly determined
each time, 𝑇 is the maximum number of generations,
and 𝑡 is the current number of generations. For the
sake of illustration, we have only described the
operation on the X coordinate, but the operation on
the Y coordinate is similar, and they are performed
simultaneously.
The shift of a node's position by single-vertex-
neighborhood mutation is calculated by the equations
(11) and (12).
(11)
𝑟= 𝑑
∗
1 −𝑡/𝑇
(12)
In this case, 𝜃 is randomly determined between 0 and
2π. And d
ideal
is calculated by
𝑠/𝑛
where s is the
area of the drawing area. As in crossover, after the
mutation on the node position is applied, the change
in node position is reflected in the whole gene.
Mutation of a control point is performed by
assigning a new random displacement to the target
control point that is less than the set maximum
displacement. The mutation probability for the
position of the node and the mutation probability for
the displacement of the control point are assigned
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
744
independent mutation probabilities. When it is
decided that a mutation on the node position is to be
made, one mutation is made at random among the two
mutations.
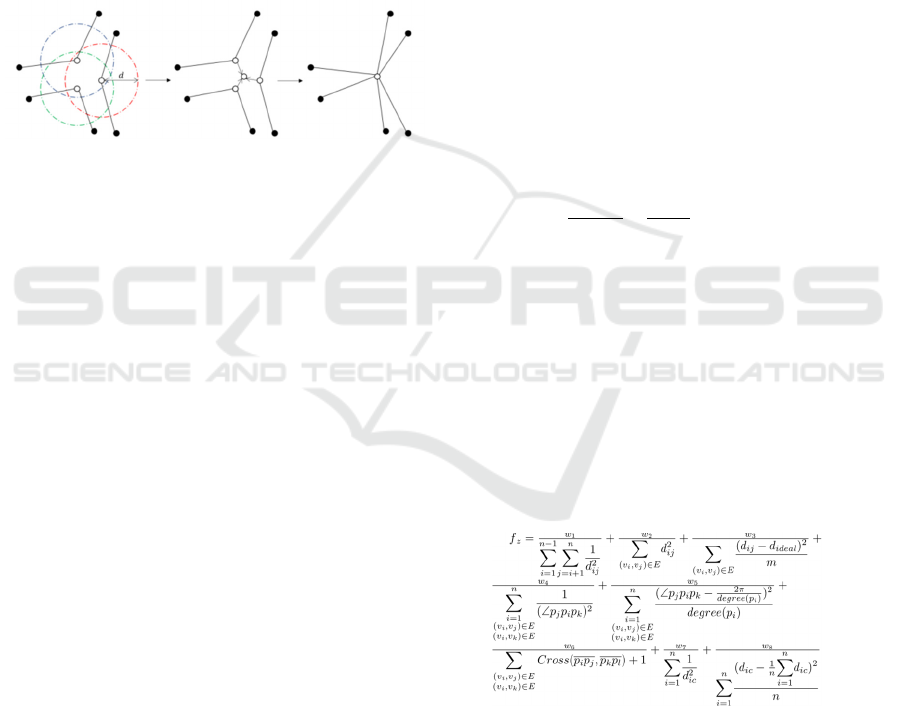
3.5 Control Points Joining
To alleviate the problem of insufficient edge bundling
because control points that existed in GABEB rarely
exist at the same coordinates, the join operation,
which places control points that are close enough
together at exactly the same coordinates(Saga 2023).
The join process of the control points is performed by
the following process (Figure 4).
Figure 4: Join process of control points.
1. Find all control points in each edge for which the
distance between control points is less than or
equal to the set distance (d) that admits a join,
and for which no other control points of the edge
containing the joined control points are joined to
any of the control points.
2. The pair of control points with the shortest
distance between them is first joined. If a pair of
control points that has been joined before has an
edge in common, the pair is not joined.
3. Calculate the average coordinate of each set of
control points for which the coupling has been
determined and assign the amount of movement
from the reference point of the control point to
the average coordinate as the amount of
movement of each control point. In such a case,
the amount of movement before the coupling is
stored for use when the coupling is released.
If a control point that is subject to crossover and
mutation in edge bundling or a control point whose
edge contains a node that is subject to crossover and
mutation in node layout is bound to another control
point, the binding is unbundled. The reason for not
performing the crossover and mutation operation
while the control points are still joined is to prevent
any of the joined control points from moving beyond
an acceptable distance from the edge containing the
control point or node position due to the crossover or
mutation, which would cause visual confusion.
The unjoin of the control points between two
points is done by assigning to each control point the
amount of movement before the coupling was
performed. To unjoin two or more control points,
only the control points that have been subject to
crossover or mutation are assigned the amount of
movement before the join, and a new join is processed
for the remaining control points, starting from the
position of the control points before the join.
3.6 Fitness Function
The fitness function f of this method (equation (13))
is the sum of 𝑓
, which measures the quality of edge
bundling with reference to the individual evaluation
index used in GABEB, and the evaluation function f
Z
shown in the node layout method by Zhang et al.
where 𝑤
G
and 𝑤
Z
are weights for each evaluation
function.
𝑓
= 𝑤
𝑓
+ 𝑤
𝑓
(13)
The fitness function for edge bundling f
G
is the
sum of the values for edge bundling used in GABEB:
EDD and PQ should be maximized, but MELD and
MOA should be minimized, so the inverse is taken for
these two values.
𝑓
=
1
𝑀𝐸𝐿𝐷
+
1
𝑀𝑂𝐴
+ 𝐸𝐷𝐷+ 𝑃𝑄
(14)
The fitness function (equation (15)) presented in
the node layout method of Zhang et al. provides an
aesthetic evaluation of node locations and the edges
affected by them. Here, d
ij
is the distance between
nodes p
i
and p
j
, 𝐸 is the entire set of edges, and m is
the number of edges. Also, ∠𝑝
𝑝
𝑝
is the angle
between the edge with p
j
and the edge with p
k
that
share node p
i
, degree(p
i
) is the degree of node p
i
. And
d
ic
is the distance between the location of the center
of the drawing area and node p
i
. Finally, w
i
is a weight
that the user can set to emphasize any aesthetic
feature.
(15)
Each term with these symbols of the equation (15)
has the following meaning. The first term increases as
the distance between nodes increases, and the second
term increases as the distance between edges
decreases. This prevents the distance between edges
from becoming too large while still evaluating low
node densities. The third term aims to unify the edge
lengths by bringing them closer to d
ideal
. The fourth
and fifth terms are evaluations of the angles between
Simultaneous Optimization of Edge Bundling and Node Layout Using Genetic Algorithm
745
edges and are intended to increase the angles between
edges and unify the angles between edges. The sixth
term, Cross(𝑝
𝑝
, 𝑝
𝑝
), is the value calculated by the
equation (16) and is intended to reduce the number of
edge crossings. Finally, the seventh and eighth terms
evaluate the symmetry of the graph.
(16)
3.7 Update and Termination
From the population that has undergone crossover
and mutation and the population of the current
generation, the top individuals are selected in a
number equal to the number of individuals in the
current generation to form the population of the next
generation. The termination condition is the
completion of the generation update for the set
maximum number of generations, or when the highest
evaluation value is not updated for 100 consecutive
generations.
3.8 Example
The parameters of the genetic algorithm used to
generate the examples are shown in Table1. The
parameters of the weights in 𝑓
are set to (0.02, 10, 1,
20, 200, 100000, 0.001, 1). The parameters are very
different from each other in order to equalize the
magnitude of each item's evaluation value. In
particular, the weight of 𝑤
6
is large, but this is because
the evaluation of crossing is treated low as the graph
scale increases, and because this method places
particular importance on the problem of edge crossing,
which cannot be solved by edge bundling. The
weights 𝑤
for the evaluation function of GABEB
and 𝑤
for the evaluation function of the node layout
method of Zhang et al. are set to 1, respectively.
Table 1: Parameters.
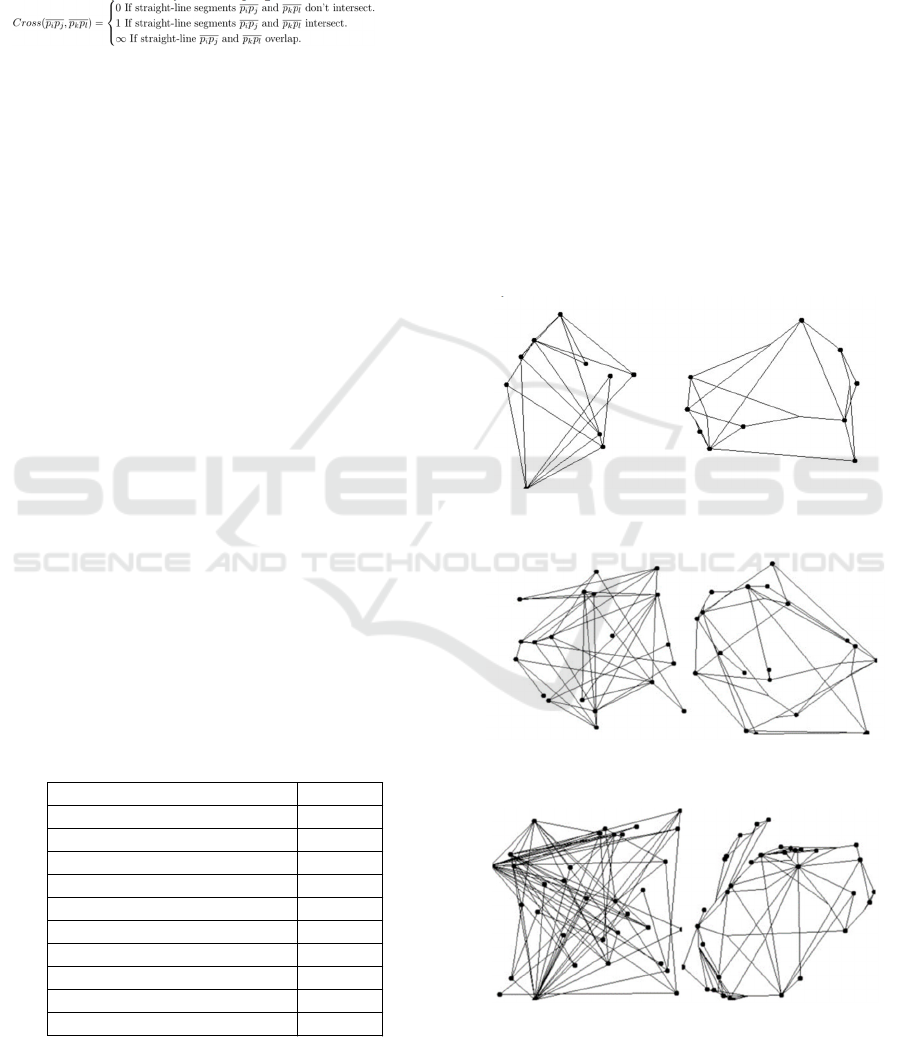
The graphs of Figure 5 to Figure 7 (the left are
examples of graphs before applied, and the graphs on
the right are examples after applied) show that the
effect of the parameter on the number of edge
crossings, which was set particularly high, is applied,
and the number of edge crossings is greatly reduced.
This is because the parameters for the evaluation
value that increases the distance between nodes and
the parameter for the evaluation value that unifies the
distance between nodes are set so large, depending on
the scale of the graph, that the other evaluation values
are not evaluated properly. This is because the
parameter setting prevents other evaluation values
from being evaluated appropriately because they
become very large depending on the scale of the
graph. In addition, the effect of edge bundling is not
so apparent in Figure 5 because the number of edges
is small and the effect of node layout is sufficient, but
in Figure 7, dense edges are bundled together in the
lower left corner and the effect of edge bundling is
fully shown.
Figure 5: An example applied to a graph with 10 nodes and
20 edges.
Figure 6: An example applied to a graph with 20 nodes and
40 edges.
Figure 7: An example applied to a graph with 34 nodes and
78 edges.
Parameters Value
Populations 100
Generations 100000
Crossover probability of Edge Bundlling 0.9
Mutation Probability of Edge Bundling 0.05
Crossover probability of node layout 0.75
Inversion probability of node layout 0.25
Mutation probability of node layout 0.25
Distance to allow coupling (d )10
Number of Control point 3
Length of one side of the MOA unit area 5
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
746
4 EXPERIMENTS
4.1 Environment
In order to compare the visualization effects with
ENLEB, the target graphs for the experiments were
the same as those used in the ENLEB experiments.
That is, randomly generated graphs G1 (10 nodes, 20
edges) and G2 (20 nodes, 40 edges), and three graphs
representing real world graphs G3 well-known as
karate club (34 nodes, 78 edges) (Zachary 1977), G4
as le miserable (62 nodes, 160 edges) (Girvan 2002),
G5 (77 nodes, 254 edges) (Knuth 1993) is the target
of this experiment. The criteria used for the
measurements were the same five aesthetic
evaluation criteria as in ENLEB. The evaluation
criteria are shown in Table 2, and the right column of
the table shows the desired results.
Table 2: Evaluation criteria.
In the experiments, the algorithm of the proposed
method was implemented and run on a PC with an
Intel Core i7-7500 3.4GHz CPU and 8 GB RAM. The
parameters of the genetic algorithm and fitness
function in the experiments are the same as those
shown in the application example of the proposed
method.
4.2 Result and Discussion
The results of the experiment are shown in Table 3.
The first row in each graph is for ENLEB, and the
second row is for this method. The results in Table 3
show that the symmetry of the graphs is consistently
improved compared to ENLEB.
For G1 to G3, where the graph scale is smaller,
there is an improvement in the number of edge
crossings (C1) and the similarity of the angles of the
edges in the bundle (C4). It can be attributed to the
positive effect of the inversion operation, which was
not implemented in ENLEB. On the other hand, the
deterioration in C2 and improvement in C4 can also
be attributed to the fact that the appropriate node
placement results in more opportunities for the
control points in the edges with high similarity to be
spatially close to each other, and less opportunities for
the control points in the edges that are not so close to
each other.
On the other hand, as the graph scale increased
like G4 and G5, the number of edge crossings (C1)
and the similarity of the angles (C3) and lengths of
the edges (C4) in the bundles deteriorated. The
number of bundles (C2) improved. The increase of C1
as the graph scale increases can be attributed to the
fact that the evaluation values other than the number
of edge crossings increased as the number of edges
and nodes increased, and the evaluation value for the
number of edge crossings was treated low, resulting
in insufficient optimization for C1. The change in C2
through C4 can also be attributed to the fact that as
C1 increases, control points in edges have more
opportunities to come closer together, resulting in
more bundling, but has also increased the chances of
bundling edges with low similarity between edges.
Table 3: Experimental results.
Table 4: Calculation Time(s).
The above results indicate that the number of edge
crossings, the similarity of the angles of the edges in
the bundles, and the symmetry of the graphs
improved for small-scale graphs, but as the scale
increased, the number of edge crossings increased,
which had a negative impact on the visualization
effect. Therefore, it is considered necessary to
improve the weighting of evaluation values and the
calculation method to deal with the phenomenon that
evaluation values other than the number of edge
crossings, which is the cause of the increase in the
Desired
outcome
Criteria
DecreaseNumber of edge crossingsC1
DecreaseNumber of bundlesC2
Increase
Average of similarity of edge lengths
within a bundle
C3
Increase
Average of the similarity of the angles of
the edges in the bundle
C4
IncreaseGraph symmetryC5
C5C4 C3 C2 C1 EdgeNode-
336.1
1414.1
0.83
0.86
0.87
0.84
10.2
15.8
21.0
10.5
2010 G1
189.5
563.3
0.83
0.85
0.87
0.79
19.7
25.7
103.1
50.1
4020 G2
44.9
253.2
0.80
0.90
0.85
0.80
31.1
40.8
375.3
94.6
7834 G3
27.2
116.9
0.83
0.77
0.84
0.74
65.8
64.0
1711.9
2394.8
16062 G4
42.5
112.0
0.82
0.79
0.81
0.74
109.2
78.4
4932.0
8298.0
25477 G5
Times(s)EdgeNode-
12.6
2797.7
2010 G1
48.4
3957.1
4020 G2
270.4
7361.0
7834 G3
1496.0
22551.6
16062 G4
4405.1
32437.5
25477 G5
Simultaneous Optimization of Edge Bundling and Node Layout Using Genetic Algorithm
747

number of edge crossings, become larger as the
number of edges and nodes increases.
The obtained computation time (Table 4) shows
that the computation time increased significantly
compared to ENLEB. This is thought to be due to the
extremely large computation time for coupling and
uncoupling, as well as the computation time for the
GABEB evaluation values. Therefore , it is necessary
to improve the search algorithm using kd-tree, etc.
and to reduce the computation time using GPGPU.
5 CONCLUSIONS
In this paper, to solve the problem that the processes
of edge bundling and node layout are actually
executed separately in ENLEB, we proposed an
evolutionary visualization method that performs
simultaneous optimization of edge bundling and node
layout based on GABEB and Zhang’s algorithm. To
examine the effectiveness, the experiment results of
our method were compared with those of ENLEB.
ACKNOWLEDGMENT
This work was supported by KAKENHI(22K12116)
REFERENCES
Barreto, A. D. M. S., Barbosa, H. J. (2000). Graph layout
using a genetic algorithm. In Proceedings Sixth
Brazilian Symposium on Neural Networks, 179-184.
Branke, J., Bucher, F., Schmeck, H. (1996). Using genetic
algorithms for drawing undirected graphs. In The Third
Nordic Workshop on Genetic Algorithms and their
Applications, 193–206.
Cui, W., Zhou, H., Qu, H., Wong, P. C., Li, X. (2008).
Geometry-based edge clustering for graph
visualization. IEEE Transactions on Visualization and
Computer Graphics, 14(6), 1277-1284.
Dogrusoz, U., Giral, E., Cetintas, A., Civril, A., Demir, E.
(2009). A layout algorithm for undirected compound
graphs. Information Sciences, 179(7), 980-994.
Ersoy, O., Hurter, C., Paulovich, F., Cantareiro, G., Telea,
A. (2011). Skeleton-based edge bundling for graph
visualization. IEEE Transactions on Visualization and
computer graphics, 17(12), 2364-2373.
Eshelman, L. J., Schaffer, J. D. (1993). Real-coded genetic
algorithms and interval-schemata. Foundations of
genetic algorithms, 2, 187-202.
Ferreira, J. D. M., Do Nascimento, H. A., Foulds, L. R.
(2018). An evolutionary algorithm for an optimization
model of edge bundling. Information, 9(7), 154.
Girvan, M., Newman, M. E. (2002). Community structure
in social and biological networks. In Proceedings of the
National Academy of Sciences, 99(12), 7821-7826.
Holten, D. (2006). Hierarchical edge bundles: Visualization
of adjacency relations in hierarchical data. IEEE
Transactions on Visualization and Computer graphics,
12(5), 741-748.
Holten, D., Van Wijk, J. J. (2009). Force-directed edge
bundling for graph visualization. Computer Graphics
Forum, 28(3), 983-990.
Hurter, C., Ersoy, O., Telea, A. (2012). Graph bundling by
kernel density estimation. Computer Graphics Forum
31(3), 865-874
Kamada, T., Kawai, S. (1989). An algorithm for drawing
general undirected graphs. Information Processing
Letters, 31(1), 7-15.
Knuth, D. E. (1993). The Stanford GraphBase: a platform for
combinatorial computing (Vol.1). N.York: ACM Press
Sugiyama, K., Misue, K. (1991). Visualization of structural
information: Automatic drawing of compound
digraphs. IEEE Transactions on Systems, Man, and
Cybernetics, 21(4), 876-892.
Lhuillier, A., Hurter, C., Telea, A. (2017). State of the art
in edge and trail bundling techniques. Computer
Graphics Forum, 36(3), 619-645.
Meikari, J., Saga, R. (2022). Evolutionary node layout and
edge bundling. In Proceedings of 2022 IEEE Congress
on Evolutionary Computation (CEC), 1-6.
Newbery, F. J. (1989). Edge concentration: A method for
clustering directed graphs. In Proceedings of the 2nd
International Workshop on Software configuration
management, 76-85.
Saga, R. (2016). Quantitative evaluation for edge bundling
based on structural aesthetics. In Proceedings of the
Eurographics/IEEE VGTC Conference on
Visualization: Posters, 17-19.
Saga, R., Baek, J. (2023). Evolutionary edge bundling with
concatenation process of control points, In Proceeding
of International Conference in Central Europe on
Computer Graphics, Visualization and Computer
Vision, 284-291.
Saga, R., Terachi, M., Tsuji, H. (2012). FACT-Graph: trend
visualization by frequency and co-occurrence.
Electronics and Communications in Japan, 95(2), 50-58.
Saga, R., Yoshikawa, T., Wakita, K., Sakamoto, K.,
Schaefer, G., Nakashima, T. (2020). A genetic algorithm
optimising control point placement for edge bundling. In
VISIGRAPP (3: IVAPP, 217-222.
Telea, A., Ersoy, O. (2010). Image-based edge bundles:
Simplified visualization of large graphs. Computer
Graphics Forum 29(3), 843-852.
Tamassia, R., Di Battista, G., Batini, C. (1988). Automatic
graph drawing and readability of diagrams. IEEE Tran-
sactions on Systems, Man, and Cybernetics, 18(1), 61-79.
Zachary, W. W. (1977). An information flow model for
conflict and fission in small groups. Journal of
anthropological research, 33(4), 452-473.
Zhang, Q. G., Liu, H. Y., Zhang, W., Guo, Y. J. (2005).
Drawing undirected graphs with genetic algorithms. In
Advances in Natural Computation: First International
Conference, ICNC 2005, 28-36
IVAPP 2024 - 15th International Conference on Information Visualization Theory and Applications
748
