
Selection of Backbone for Feature Extraction with U-Net in Pancreas
Segmentation
Alexandre de Carvalho Ara
´
ujo
a
, Joao Dallyson Sousa de Almeida
b
,
Anselmo Cardoso de Paiva
c
and Geraldo Braz Junior
d
Applied Computing Group (NCA), Federal University of Maranh
˜
ao (UFMA), S
˜
ao Lu
´
ıs, MA, Brazil
Keywords:
Image Segmentation, Pancreas Segmentation, U-Net.
Abstract:
The survival rate for pancreatic cancer is among the worst, with a mortality rate of 98%. Diagnosis in the
early stage of the disease is the main factor that defines the prognosis. Imaging scans, such as Computerized
Tomography scans, are the primary tools for early diagnosis. Computer Assisted Diagnosis tools that use these
scans usually include in their pipeline the segmentation of the pancreas as one of the initial steps for diagnosis.
This paper presents a comparative study of the use of different backbones in combination with the U-Net. This
study aims to demonstrate that using pre-trained backbones is a valuable tool for pancreas segmentation and
to provide a comparative benchmark for this task. The best result obtained was 85.96% of Dice in the MSD
dataset for the pancreas segmentation using backbone efficientnetb7.
1 INTRODUCTION
Compared to other cancers, pancreatic cancer is rela-
tively rare. Some symptoms associated with this can-
cer are weight loss, jaundice, pain, anemia, and oth-
ers. In Brazil alone, this type of cancer accounts for
about 1% of all cancer diagnoses, but it represents 5%
of all cancer deaths in the country. The prognosis for
pancreatic cancer is unfavorable. Among all tumor
forms, pancreatic cancer has one of the lowest sur-
vival rates, with a mortality rate of 98%, and diagno-
sis in the early stages of the disease is the main factor
that defines the prognosis (Cheng, 2018).
Small lesions on imaging exams, such as abdomi-
nal ultrasound, computer tomography (CT), and mag-
netic resonance imaging (MRI), which are the main
tools used for early diagnosis, make it challenging for
the specialist to identify the early stages of pancre-
atic cancer. This is one of the main challenges in the
early diagnosis of this type of cancer (Dietrich and
Jenssen, 2020). These tools also come with a few
more challenges. A professional is required to ana-
lyze the numerous images produced by a CT scan, for
instance. This analysis is highly complex, thus requir-
a
https://orcid.org/0000-0002-0250-6211
b
https://orcid.org/0000-0001-7013-9700
c
https://orcid.org/0000-0003-4921-0626
d
https://orcid.org/0000-0003-3731-6431
ing the attention and experience of the specialist. By
its nature, this analysis is a repetitive process, which
may lead to physical and mental fatigue which can
cause distraction of the specialist. This makes it pos-
sible for injuries to go unnoticed, which could result
in consequences from the cancer and the medical pro-
cedures required to treat it. For this reason, technolo-
gies that supplement these image-based examinations
are needed.
Many studies in the biomedical field focus on
Computer-Aided Diagnosis (CAD) as a tool to facil-
itate disease detection, decrease errors in diagnosis,
aid and reduce invasive procedures, and save time and
costs related to analysis. Automatic segmentation of
the pancreas is an essential topic in this area, since it
is an initial step often used in cancer analysis, lesion
detection and three-dimensional visualization of the
pancreas (Gong et al., 2019). In CT scans, this step
is hampered by the fact that the pancreas occupies a
minimal part of the scan, besides having shape, size,
and location in the abdomen with drastic variance be-
tween patients (Zheng et al., 2020).
Although there are examples of pre-trained back-
bones being used for feature extraction for pancreas
segmentation (Yu et al., 2019; Liu et al., 2019; Hu
et al., 2020), as well as the original U-Net architecture
being used as a coarse segmentation step, we have
not found a comparison between different backbones
for pancreas segmentation. That being the case, in
822
Araújo, A., Sousa de Almeida, J., Cardoso de Paiva, A. and Braz Junior, G.
Selection of Backbone for Feature Extraction with U-Net in Pancreas Segmentation.
DOI: 10.5220/0012573900003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
822-829
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

this paper, a comparative study is done between dif-
ferent networks as encoders or feature extractors in a
U-Net model for pancreas segmentation. This study
benchmarks U-Net architectures in pancreas segmen-
tation, providing a useful baseline for comparison for
architectural modifications of the U-Net model. In
our pipeline, we apply Hounsfield value windowing
and Histogram Matching as preprocessing to increase
the contrast between the pancreas and neighboring
structures and decrease the variation in the contrast
between scans from different CT scanners. As CT
scan slices are single-channel images, we also apply
an initial convolutional layer in our models with the
purpose of generating 2 maps with the same dimen-
sions as the original slice and those maps will serve as
the second and third channel instead of simply repeat-
ing the single channel slice for the second and third
channel. This is a requirement for the ImageNet pre-
trained weights used in our backbone networks.
The rest of this paper is organized as follows: The
section 2 describes works related to the problem ad-
dressed. The sections 3 and 4 describe the method
used for this work, as well as the results obtained, dis-
cussions and comparisons with other works. Finally,
the section 5 presents the final considerations about
the results and proposals for future work.
2 RELATED WORK
For pancreas segmentation and other segmentation
tasks in medical images, U-Net (Ronneberger et al.,
2015) is one of the most widely used architectures in
the literature. This is due to the quality of segmenta-
tion provided by this network. Due to its simple yet
effective structure, composed of an encoder and a de-
coder, many proposals in the literature aim to modify
the U-Net to improve its ability to segment the pan-
creas.
One example is changes to the original network
blocks with the addition of layers within the blocks
(Fan and Tai, 2019). Another possibility is to modify,
in addition to the network blocks, the skip connections
of the network (Oktay et al., 2018; Ma et al., 2021;
Dai et al., 2023). Another possible improvement is
in the bottleneck layer, such as using dilated convo-
lutions to increase the receptive field of convolutions
(Giddwani et al., 2020).
It can also be observed the use of more than one
U-Net to perform the segmentation of the pancreas,
such as the combination of two 3D U-Net proposed on
(Zhao et al., 2019), two Deformable U-Net on (Huang
et al., 2019) and one 2.5D U-Net with Multi-View 2D
U-Net (Li et al., 2021a). In these approaches, the out-
put of the first network is a coarse segmentation that
serves as input to the second network that refines the
segmentation to obtain the final result. Other possibil-
ities are to use a U-Net in combination with another
network, sequentially (Yang et al., 2019), in a par-
allel manner (Cai et al., 2019) or with an ensemble
of U-Net’s (Liu et al., 2019). In these approaches,
one combines the U-Net with FCN’s or more com-
plex networks in a sequential manner to improve the
segmentation provided by the U-Net.
However, one point that needs to be tackled is us-
ing different backbones as a U-net encoder for pan-
creas segmentation. Using pre-trained backbones can
provide good results, with different backbones having
different applications where they stand out (Ahmed
et al., 2022). Hence, a study of the use of different
backbones for U-Net is performed in this paper. Ta-
ble 1 compares the related works.
Table 1: Researches with U-Net to segment the pancreas.
Authors Dataset Dice
(Fan and Tai, 2019) NIH 68.45%
(Cai et al., 2019) MSD 74.30%
(Boers et al., 2020) NIH 78.10%
(Giddwani et al., 2020) NIH 83.30%
(Liu et al., 2019) NIH 84.10%
(Zhao et al., 2019) NIH 85.99%
(Huang et al., 2019) NIH 87.25%
(Yang et al., 2019) NIH 87.82%
(Ma et al., 2021) NIH 88.48%
(Li et al., 2021a) MSD 88.52%
(Dai et al., 2023) MSD 91.22%
3 METHOD
This section details the preprocessing techniques, as
well as the backbone choice method used in combi-
nation with U-net.
We use the well-known U-Net architecture (Ron-
neberger et al., 2015) to address our pancreas segmen-
tation task. The U-Net architecture is a fully convolu-
tional network in the encoder-decoder neural network
family. The spatial information is downsampled in
the encoding stage using convolution and pooling op-
erations. This section serves as a feature extraction
stage. The spatial information is upsampled back to
the original size in the decoding section using convo-
lution transpose. High-resolution features from the
encoder are concatenated to the corresponding fea-
tures from the decoder via skip connections, infusing
high-resolution information into the decoder. The ad-
dition of skip connections provides the network with
Selection of Backbone for Feature Extraction with U-Net in Pancreas Segmentation
823
its “U” shape.
In our work, we experiment with different network
architectures pre-trained on the ImageNet dataset as
the feature extractor, replacing the standard encoder
of U-Net.
3.1 Preprocessing
The preprocessing step aims to increase the contrast
of the pancreas relative to the other organs present
in the scan, providing better features for the follow-
ing steps of the method. To achieve this, we per-
form Hounsfield (HU) values windowing and apply
Histogram Matching.
HU value windowing is a process in which the
grayscale values of the voxel values of a CT are
truncated to highlight specific structures (Seeram,
2015). Two thresholds define this process. From
these thresholds, the truncation of HU values is per-
formed. Any HU value greater than the upper thresh-
old is truncated to the upper threshold value, and any
HU value less than the lower threshold is truncated to
the lower threshold value. The thresholds used were
[-150, 250]. This range highlights soft tissue in the
abdomen, a category to which the pancreas belongs
(Mo et al., 2020).
After windowing the HU values, the Histogram
Matching (HM) algorithm is applied. Given two his-
tograms, this algorithm finds a color mapping that ap-
proximates one histogram to the other (Castleman,
1996). However, a reference histogram must be de-
fined, which will be the approximated histogram. The
reference histogram was defined empirically. First,
we trained a 3D U-Net where the only preprocessing
applied was the windowing of HU values. Then, the
volume of the training set where the network obtained
the best Dice for segmentation of the pancreas was
chosen. Using this volume as a basis, the HM was ap-
plied to all volumes in the set, with the histogram of
the chosen volume as reference. Finally, the volumes
were transformed into slices. We employ 3D U-Net
because using one histogram for the entire scan leads
to superior contrast normalization.
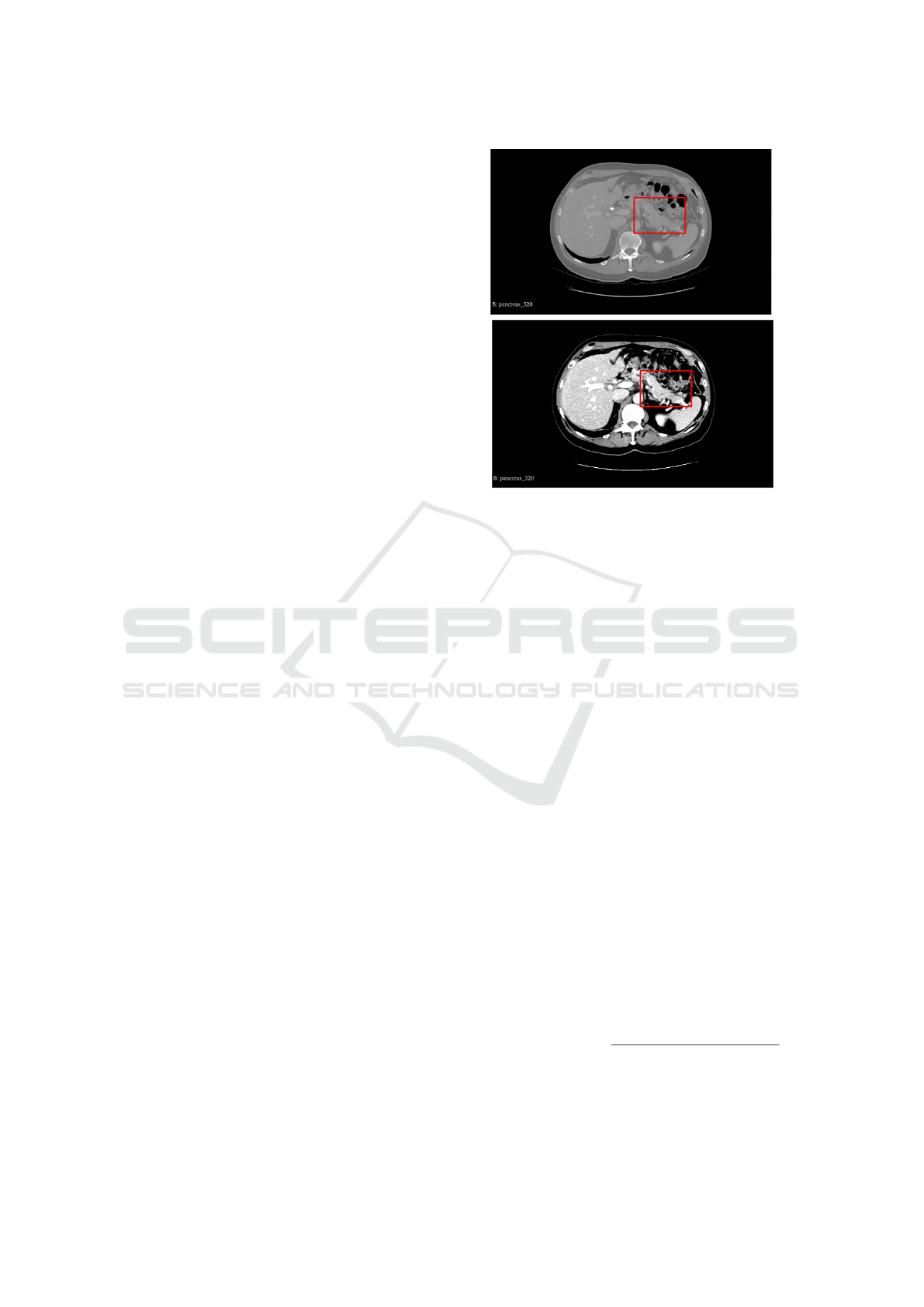
Figure 1 illustrates the application of preprocess-
ing. The red square represents the pancreas and the
area around it. It is possible to see that after pre-
processing, the pancreas becomes more evident when
comparing with surrounding structures.
3.2 Networks for Feature Extraction
For training, the U-Net architecture is used as a basis
and the network encoder is exchanged for different ar-
chitectures pre-trained on the ImageNet dataset such
(A)
(B)
Figure 1: Preprocessing step: (A) original cut and (B) cut
after preprocessing.
as VGG (Simonyan and Zisserman, 2014), ResNet
(He et al., 2016), EfficientNet, (Koonce and Koonce,
2021), MobileNet (Howard et al., 2017), Inceptionv3
(Szegedy et al., 2016) and DenseNet (Iandola et al.,
2014). Two versions of each of these architectures,
the version with the most parameters and the version
with the fewest parameters, were used. Specifically
for EfficientNet three models are used, as this network
has several versions. The smallest (b0) the median
(b3) and the largest (b7) were chosen. The chosen
versions for each architecture can be seen in Table 2.
Top-1 Accuracy refers to the accuracy obtained on the
ImageNet validation set, Number of Parameters refers
to the number of trainable parameters in the architec-
ture, and depth refers to the number of layers with
parameters, such as convolution layers and batch nor-
malization layers.
3.3 Models Training
For the training of the models, we use the same pre-
processing and the same hyperparameters, such as op-
timizer and learning rate for all models. The loss
function used for training is a combination of the Dice
Loss (Sudre et al., 2017) and the Focal Loss (Lin
et al., 2017b). Dice Loss is calculated as:
L
dice
(t p, f p, f n) =
(1 + β
2
) ·t p
(1 + β
2
) · f p + β
2
· f n + f p
(1)
where t p are true positives, f p are false positives and
f n are false negatives.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
824

Table 2: Summary of the backbones used in this study.
Number of parameters in millions.
Method
Top-1
Accuracy
Params Depth
VGG16 71.3% 138.4M 16
VGG19 71.3% 143.7M 19
ResNet50 74.9% 25.6M 107
ResNet152 76.6% 60.4M 311
InceptionV3 77.9% 23.9M 189
InceptionResNetV2 80.3% 55.9M 449
MobileNet 70.4% 4.3M 55
MobileNetV2
71.3% 3.5M 105
DenseNet121 75.0% 8.1M 242
DenseNet201 77.3% 20.2M 402
EfficientNetB0 77.1% 5.3M 132
EfficientNetB3 81.6% 12.3M 210
EfficientNetB7 84.3% 66.7M 438
Meanwhile, Focal Loss is calculated as:
L
f ocal
(gt, pr) = −gtα(1 − pr)
γ
log(pr)
−(1 − gt)αpr
γ
log(1 − pr)
(2)
where gt is ground truth, pr is the model prediction, α
is the weighting factor and γ is the focusing parameter
for modulating factor (1 − p).
These two loss functions address class unbalance,
and their combination aims to take advantage of the
positives of each while minimizing their disadvan-
tages. As the values of Focal Loss function are gener-
ally smaller than the values of Dice Loss function by
a high prediction confidence cases, we multiply the
Focal Loss by a factor of 10 to balance the two met-
rics, as this was the factor observed empirically to best
balance the both functions. The final loss function is
defined in Equation 3.
total loss = Dice loss + 10 ∗ Focal loss (3)
The pre-trained weights are unfrozen. This is nec-
essary as ImageNet is a dataset made of images from
diverse domains, but our targets are CT scan images,
which are not present on the ImageNet dataset. So,
unfreezing the backbone weights enables specialized
learning for CT scan images features extraction based
on already learned feature extraction.
4 RESULTS
4.1 Datasets
We evaluate the performance of the different models
on two publicly available datasets: The first dataset
contains 281 contrast-enhanced CT scans with la-
beled pancreas and pancreatic tumor from the Med-
ical Segmentation Decathlon (MSD) challenge pan-
creas segmentation dataset (Simpson et al., 2019),
where each CT volume has dimensions 512 x 512 x
Z, and Z ∈ [37, 751]. Following similar studies (Chen
et al., 2022), pancreas and pancreatic tumor labels
were combined into a single target label. The sec-
ond dataset contains 82 abdominal contrast-enhanced
CT scans from the National Institutes of Health (NIH)
Clinical Center pancreas segmentation dataset (Roth
et al., 2015). Each volume has dimensions 512 x 512
x Z, where Z ∈ [181, 466].
4.2 Evaluation Metric
To evaluate segmentation performance, we use Dice
similarity coefficient (Dice). Dice is the most com-
mon metric used for evaluating segmentation results
in medical image segmentation (Dai et al., 2023).
This metric represents the harmonic mean of the pre-
cision and sensitivity and is calculated as in Equation
4.
Dice =
2t p
2t p + f p + f n
(4)
Where t p are the true positives, f p are the false
positives, and f n are the false negatives. In our work,
these translate to t p being equivalent to pancreas pix-
els that the model correctly classified as pancreas,
background pixels that the model mistakenly classi-
fied as pancreas as f p and pancreas pixels that the
model wrongly classified as background pixels as f n.
This metric ranges from 0 to 1, with higher values
representing better segmentation.
4.3 Implementation Details
The neural network models were implemented us-
ing the Python 3.8, along with the libraries Ten-
sorflow 2.6.0, Keras 2.9.0 and Segmentation models
(Iakubovskii, 2019). To manipulate the NIFTI vol-
umes, the Nibabel 3.2.1 library was used, and to
manipulate the slices extracted from the volumes,
the Opencvpython 4.5.3.56 library was used. An
NVIDIA RTX 3060 graphics card with 12 GB of
memory was used for all experiments. Due to mem-
ory limitations, the images that entered the network
were resized from 512 x 512 to 256 x 256.
For all experiments, training parameters were kept
the same. The models were trained for 100 epochs
with initial learning rate (lr) set to 0.0001 with the
Adam optimizer and lr decay by a factor of 0.1 if val-
idation loss has not decreased in the last 10 epochs.
For all models, the data was preprocessed and split
identically for all training and test experiments.
Selection of Backbone for Feature Extraction with U-Net in Pancreas Segmentation
825
4.4 Segmentation Result on MSD
Dataset
To evaluate the performance of the trained models, we
compare them with networks that perform well on the
MSD dataset. The results obtained can be seen in Ta-
ble 3, where we compare the different backbones used
with works in the literature for pancreas segmentation
on the MSD dataset. The average Dice for all patient
slices is used to represent the segmentation quality for
the patient. It can be seen that the use of U-Net with a
backbone already trained on ImageNet obtains good
results, being superior to some works found in the lit-
erature. The best result obtained was with the efficien-
tenetb7 architecture as a backbone, obtaining a Dice
2.56% lower than the second best result and 5.26%
lower than the best result found in the literature for
this dataset.
Table 3: Pancreas segmentation on the MSD dataset.
Method
Pacient (Dice)
(Cai et al., 2019) 74.30%
resnet50 76.12%
(Boers et al., 2020) 78.10%
inceptionv3 79.71%
(Zhu et al., 2019)
79.94%
vgg16 81.54%
efficientnetb0 81.88%
vgg19 82.24%
(Zhang et al., 2021b) 82.74%
resnet152 83.10%
inceptionresnetv2 83.78%
mobilenet 84.33%
densenet121 84.52%
mobilenetv2 84.61%
(Fang et al., 2019) 84.71%
efficientnetb3 85.08%
densenet201 85.33%
(Zhang et al., 2021a) 85.56%
efficientnetb7 85.96%
(Li et al., 2021a) 88.52%
(Dai et al., 2023) 91.22%
In Table 3 it can been seen that the results found
vary between [76.12%; 85.96%]. Despite this varia-
tion of 9.84% between the worst and best result ob-
tained, the top 5 backbones vary between [84.52%;
85.96%]. This shows that the choice of backbone has
a high impact and also that backbones such as Mo-
bileNetv2, EfficientNet b3 and b7 and densenet121
and densenet201 have such close results that the
choice of backbone will depend on the application,
where GPU memory limitations or inference time
may be the determining factors for the best backbone.
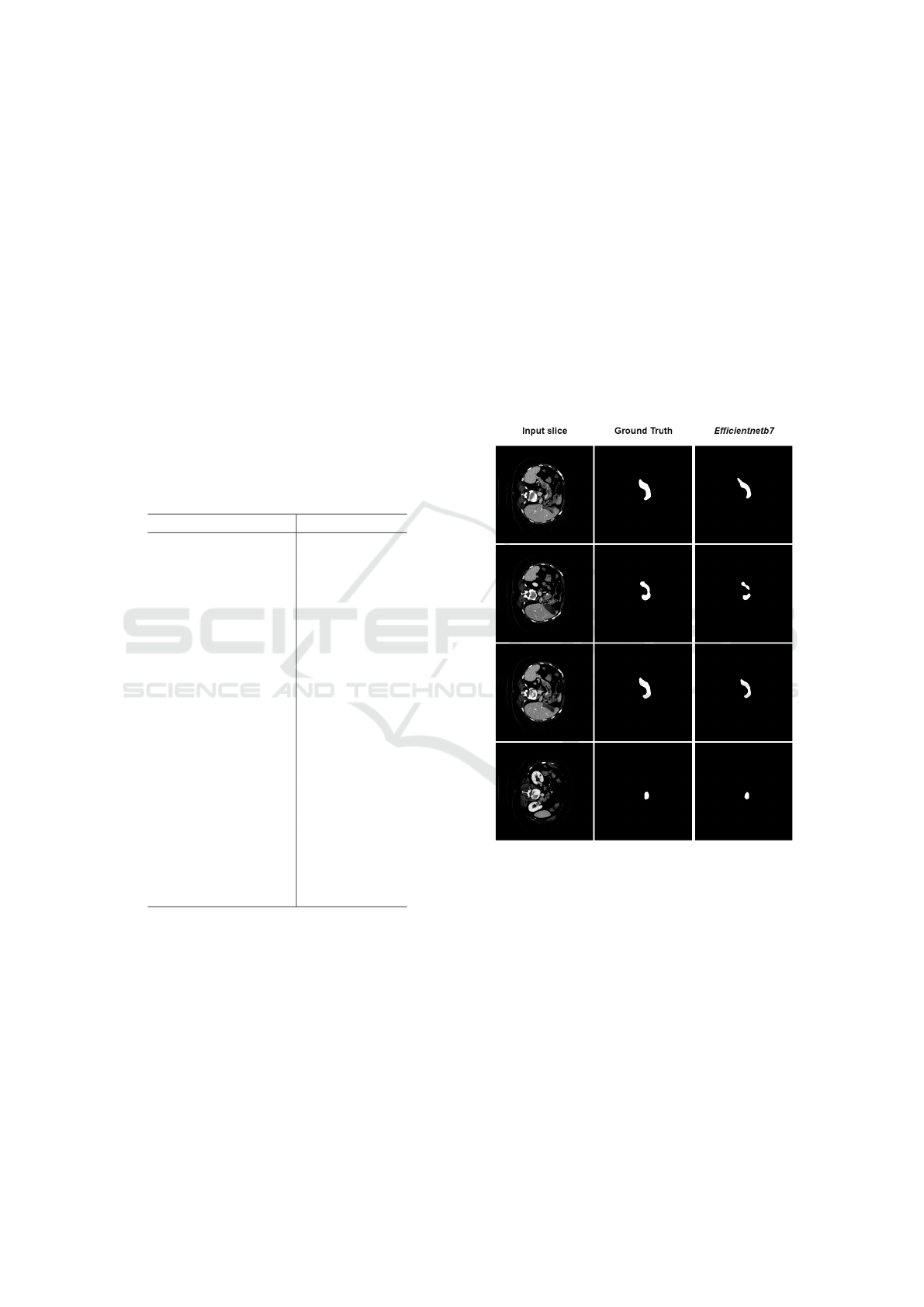
Figure 2 shows examples of the pancreas segmen-
tation performed by the U-Net with efficientnetb7 as
the backbone on multiple slices of the same patient.
For this patient, the Dice obtained was 93.39%, indi-
cating a segmentation very close to the ground truth.
As you can see, in three of the four examples shown,
the segmentation result is quite similar with the label.
In the second slice, an error case is presented, where
the network did not identify the connected body of the
pancreas. One of the reasons that may have caused
this error is the texture change that exists in this slice.
It is possible to notice that in the location where the
failure occurs, the scan presents a change in the inten-
sity of the pancreas pixels, making it difficult for the
network to recognize it.
Figure 2: Examples of pancreas segmentation with the U-
Net using backbone efficientnetb7. From left to right, input
slice, expert labelling (ground truth) and segmentation per-
formed by the model.
Figure 3 shows three examples of comparison be-
tween segmentation with different backbones. In
these examples, the three networks followed the same
pattern observed in Table 3, where ResNet50 obtained
the worst result among the backbones tested, VGG19
obtained a better result than ResNet50, but lower than
EfficientNetb7, which obtained the best average result
for the dataset MSD. ResNet50 had difficulty detect-
ing the pancreas at two separate points in row A, a be-
havior that was repeated in VGG19, while Efficient-
Netb7 was able to capture this non-connectivity at this
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
826
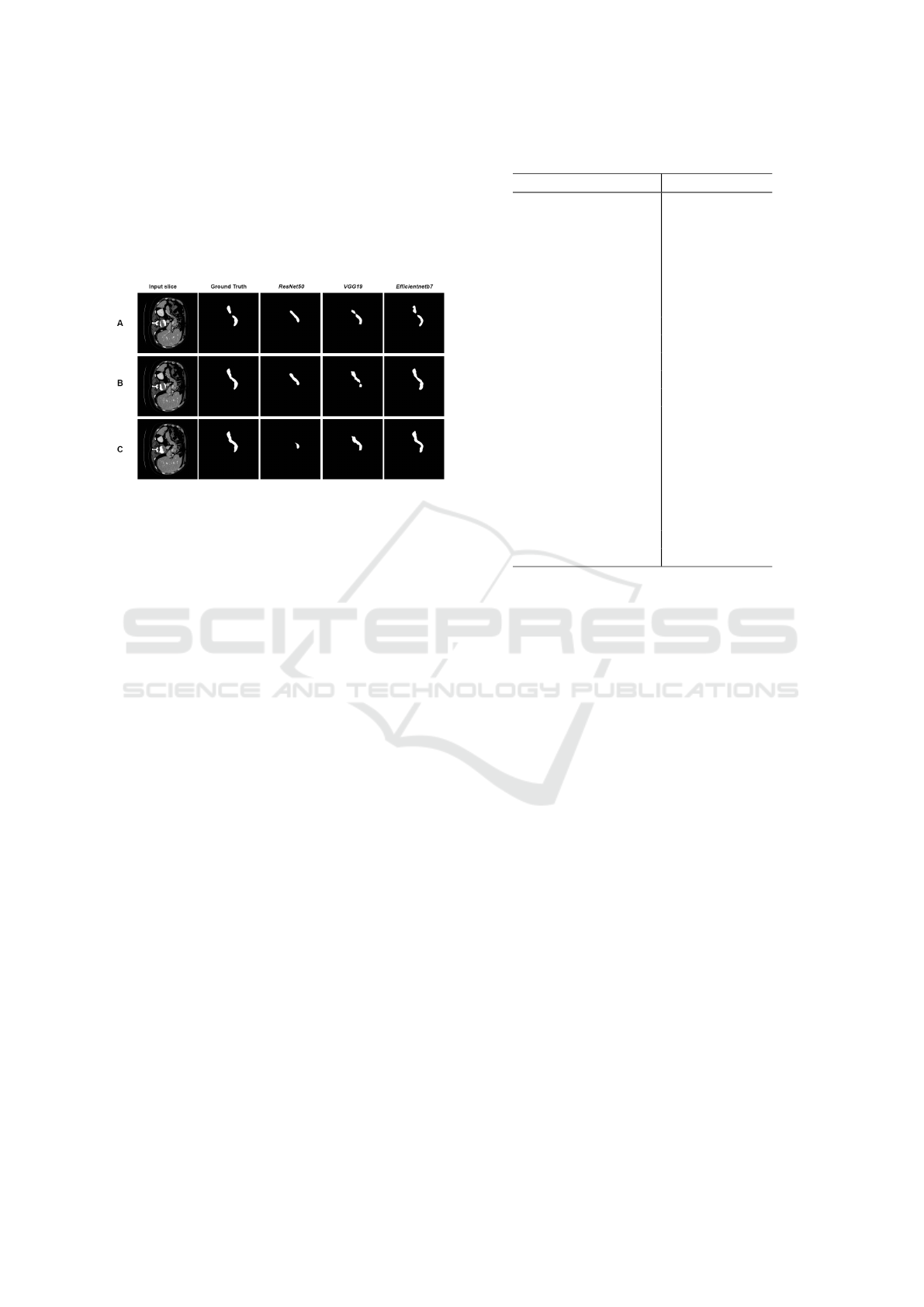
cut. In row C, it is observed that ResNet50 had great
difficulty in segmenting the pancreas. In that same
slice, VGG19 can capture more information from the
pancreas, but there is still noticeable loss. Efficient-
Netb7 achieved a segmentation very close to the ex-
pert’s marking, reflecting the average result obtained
for the dataset as a whole.
Figure 3: Segmentation with 3 different backbones,
ResNet50 (79.02% patient dice), VGG19 (88.36% patient
dice) and EfficientNetb7 (91.11% patient dice).
4.5 Segmentation Result on NIH
Dataset
We also evaluate the performance of the trained mod-
els with networks that perform well on the NIH
dataset. The results obtained can be seen in Table 4.
The average Dice for all patient slices is used to repre-
sent the segmentation quality for the patient. As with
the MSD dataset, the use of U-Net with a backbone al-
ready trained on ImageNet obtains good results, with
being superior to works already found in the litera-
ture. The best result obtained was also with the ef-
ficientenetb7 architecture as a backbone, obtaining a
Dice 2.11% lower than the third best result, 2.18%
lower than the second best result and 4.5% lower than
the best result in the literature for this dataset.
In Table 4 it can be seen that the results found vary
between [63.08%, 85.39%]. This is a much larger gap
than the one found on the MSD dataset. The VGG
family models had difficulty learning relevant features
for segmentation. Such variations can be attributed to
the difference in datasets, as the NIH dataset generally
presents greater difficulty for segmentation (Dai et al.,
2023). The choice of backbone shows an even bigger
impact in this dataset, but The top 5 backbones, even
though with a bigger range in performance, vary be-
tween [83.94%; 85.39%], which is a relatively small
gap in performance. As such, the choice of backbone
will mainly depend on the application, where GPU
memory limitations or inference time may be the de-
termining factors for the best backbone.
Table 4: Pancreas segmentation on the NIH dataset.
Method
Pacient (Dice)
vgg16 63.08%
vgg19 63.09%
densenet201 69.64%
resnet152 73.09%
resnet50 80.53%
efficientnetb0 80.99%
efficientnetb3 82.68%
(Li et al., 2021b) 83.21%
mobilenet 83.44%
inceptionresnetv2 83.94%
densenet121 84.02%
inceptionv3 84.24%
mobilenetv2 84.35%
(Zhang et al., 2021b) 84.47%
(Zhang et al., 2021a) 84.90%
(Chen et al., 2022) 85.19%
(Li et al., 2021a) 85.35%
efficientnetb7 85.39%
(Li et al., 2020b) 87.50%
(Li et al., 2020a) 87.57%
(Dai et al., 2023) 89.89%
5 CONCLUSION
The present study aimed to evaluate the use of differ-
ent backbones as encoder for the U-Net architecture
for pancreas segmentation. Several models pretrained
on the ImageNet dataset were compared. The best
combination found was U-net with EfficientNetb7,
which showed positive results that are competitive
with the literature in the two most commonly used
datasets. One advantage of using backbones as an en-
coder is the good segmentation results with ease of
implementation, which can be useful in initial seg-
mentation steps for more complex models, such as
coarse-to-fine models. This study can also serve as
a benchmark for comparison for pancreas segmenta-
tion, providing an useful baseline for comparison of
architectural modifications for the U-Net model. As
a future work, it is suggested to investigate new ar-
chitectures such as Feature Pyramid Network (FPN)
(Lin et al., 2017a), LinkNet (Chaurasia and Culur-
ciello, 2017) and Pyramid Scene Parsing Network
(PSPN) (Zhao et al., 2017) in place of U-Net as the
main architecture to further improve the baseline for
architectural modifications in those models for pan-
creas segmentation. Another line of research could
be the combination of models in an ensemble, as seen
in (Georgescu et al., 2023).
Selection of Backbone for Feature Extraction with U-Net in Pancreas Segmentation
827

ACKNOWLEDGMENTS
The authors acknowledge the Coordenac¸
˜
ao de
Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
(CAPES),Finance Code 001, Conselho Nacional
de Desenvolvimento Cient
´
ıfico e Tecnol
´
ogico
(CNPq), and Fundac¸
˜
ao de Amparo
`
a Pesquisa
Desenvolvimento Cient
´
ıfico e Tecnol
´
ogico do
Maranh
˜
ao (FAPEMA) (Brazil), Empresa Brasileira
de Servic¸os Hospitalares (Ebserh) Brazil (Grant
number 409593/2021-4) for the financial support.
REFERENCES
Ahmed, I., Carni, D. L., Balestrieri, E., and Lamonaca, F.
(2022). Comparison of u-net backbones for morpho-
metric measurements of white blood cell. In 2022
IEEE International Symposium on Medical Measure-
ments and Applications (MeMeA), pages 1–6. IEEE.
Boers, T., Hu, Y., Gibson, E., Barratt, D., Bonmati,
E., Krdzalic, J., van der Heijden, F., Hermans, J.,
and Huisman, H. (2020). Interactive 3d u-net for
the segmentation of the pancreas in computed to-
mography scans. Physics in Medicine and Biology,
65(6):065002.
Cai, J., Xia, Y., Yang, D., Xu, D., Yang, L., and Roth, H.
(2019). End-to-end adversarial shape learning for ab-
domen organ deep segmentation. In Machine Learn-
ing in Medical Imaging: 10th International Workshop,
MLMI 2019, Held in Conjunction with MICCAI 2019,
Shenzhen, China, October 13, 2019, Proceedings 10,
pages 124–132. Springer.
Castleman, K. R. (1996). Digital image processing. Pren-
tice Hall Press.
Chaurasia, A. and Culurciello, E. (2017). Linknet: Exploit-
ing encoder representations for efficient semantic seg-
mentation. In 2017 IEEE visual communications and
image processing (VCIP), pages 1–4. IEEE.
Chen, H., Liu, Y., Shi, Z., and Lyu, Y. (2022). Pancreas
segmentation by two-view feature learning and multi-
scale supervision. Biomedical Signal Processing and
Control, 74:103519.
Cheng, S. (2018). Punc¸
˜
ao ecoendosc
´
opica de massas
s
´
olidas pancre
´
aticas por t
´
ecnica de press
˜
ao negativa
versus capilaridade: estudo prospectivo e random-
izado. PhD thesis, Universidade de S
˜
ao Paulo.
Dai, S., Zhu, Y., Jiang, X., Yu, F., Lin, J., and Yang,
D. (2023). Td-net: Trans-deformer network for
automatic pancreas segmentation. Neurocomputing,
517:279–293.
Dietrich, C. F. and Jenssen, C. (2020). Modern ultra-
sound imaging of pancreatic tumors. Ultrasonogra-
phy, 39(2):105.
Fan, J. and Tai, X.-c. (2019). Regularized unet for auto-
mated pancreas segmentation. In Proceedings of the
Third International Symposium on Image Computing
and Digital Medicine, pages 113–117.
Fang, C., Li, G., Pan, C., Li, Y., and Yu, Y. (2019). Glob-
ally guided progressive fusion network for 3d pan-
creas segmentation. In Medical Image Computing and
Computer Assisted Intervention–MICCAI 2019: 22nd
International Conference, Shenzhen, China, October
13–17, 2019, Proceedings, Part II 22, pages 210–218.
Springer.
Georgescu, M.-I., Ionescu, R. T., and Miron, A. I. (2023).
Diversity-promoting ensemble for medical image seg-
mentation. In Proceedings of the 38th ACM/SIGAPP
Symposium on Applied Computing, pages 599–606.
Giddwani, B., Tekchandani, H., and Verma, S. (2020). Deep
dilated v-net for 3d volume segmentation of pancreas
in ct images. In 2020 7th International Conference on
Signal Processing and Integrated Networks (SPIN),
pages 591–596. IEEE.
Gong, Z., Zhu, Z., Zhang, G., Zhao, D., and Guo, W.
(2019). Convolutional neural networks based level set
framework for pancreas segmentation from ct images.
In Proceedings of the Third International Symposium
on Image Computing and Digital Medicine, pages 27–
30.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., Andreetto, M., and Adam,
H. (2017). Mobilenets: Efficient convolutional neu-
ral networks for mobile vision applications. arXiv
preprint arXiv:1704.04861.
Hu, P., Li, X., Tian, Y., Tang, T., Zhou, T., Bai, X., Zhu, S.,
Liang, T., and Li, J. (2020). Automatic pancreas seg-
mentation in ct images with distance-based saliency-
aware denseaspp network. IEEE journal of biomedi-
cal and health informatics, 25(5):1601–1611.
Huang, M., Huang, C., Yuan, J., and Kong, D. (2019).
Fixed-point deformable u-net for pancreas ct segmen-
tation. In Proceedings of the Third International Sym-
posium on Image Computing and Digital Medicine,
pages 283–287.
Iakubovskii, P. (2019). Segmentation models. https://gith
ub.com/qubvel/segmentation\ models.
Iandola, F., Moskewicz, M., Karayev, S., Girshick, R., Dar-
rell, T., and Keutzer, K. (2014). Densenet: Imple-
menting efficient convnet descriptor pyramids. arXiv
preprint arXiv:1404.1869.
Koonce, B. and Koonce, B. (2021). Efficientnet. Convo-
lutional Neural Networks with Swift for Tensorflow:
Image Recognition and Dataset Categorization, pages
109–123.
Li, F., Li, W., Shu, Y., Qin, S., Xiao, B., and Zhan, Z.
(2020a). Multiscale receptive field based on resid-
ual network for pancreas segmentation in ct im-
ages. Biomedical Signal Processing and Control,
57:101828.
Li, H., Li, J., Lin, X., and Qian, X. (2020b). A model-driven
stack-based fully convolutional network for pancreas
segmentation. In 2020 5th International Confer-
ence on Communication, Image and Signal Process-
ing (CCISP), pages 288–293. IEEE.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
828

Li, J., Lin, X., Che, H., Li, H., and Qian, X. (2021a). Pan-
creas segmentation with probabilistic map guided bi-
directional recurrent unet. Physics in Medicine and
Biology, 66(11):115010.
Li, M., Lian, F., and Guo, S. (2021b). Automatic pancreas
segmentation using double adversarial networks with
pyramidal pooling module. IEEE Access, 9:140965–
140974.
Lin, T.-Y., Doll
´
ar, P., Girshick, R., He, K., Hariharan, B.,
and Belongie, S. (2017a). Feature pyramid networks
for object detection. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2117–2125.
Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Doll
´
ar, P.
(2017b). Focal loss for dense object detection. In
Proceedings of the IEEE international conference on
computer vision, pages 2980–2988.
Liu, S., Yuan, X., Hu, R., Liang, S., Feng, S., Ai, Y., and
Zhang, Y. (2019). Automatic pancreas segmentation
via coarse location and ensemble learning. IEEE Ac-
cess, 8:2906–2914.
Ma, H., Zou, Y., and Liu, P. X. (2021). Mhsu-net:
A more versatile neural network for medical image
segmentation. Computer Methods and Programs in
Biomedicine, 208:106230.
Mo, J., Zhang, L., Wang, Y., and Huang, H. (2020). It-
erative 3d feature enhancement network for pancreas
segmentation from ct images. Neural Computing and
Applications, 32:12535–12546.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich,
M., Misawa, K., Mori, K., McDonagh, S., Hammerla,
N. Y., Kainz, B., et al. (2018). Attention u-net: Learn-
ing where to look for the pancreas. arXiv preprint
arXiv:1804.03999.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18, pages 234–241.
Springer.
Roth, H. R., Lu, L., Farag, A., Shin, H.-C., Liu, J., Turkbey,
E. B., and Summers, R. M. (2015). Deeporgan: Multi-
level deep convolutional networks for automated pan-
creas segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part I 18, pages 556–564.
Springer.
Seeram, E. (2015). Computed Tomography-E-Book: Phys-
ical Principles, Clinical Applications, and Quality
Control. Elsevier Health Sciences.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Simpson, A. L., Antonelli, M., Bakas, S., Bilello, M.,
Farahani, K., Van Ginneken, B., Kopp-Schneider, A.,
Landman, B. A., Litjens, G., Menze, B., et al. (2019).
A large annotated medical image dataset for the de-
velopment and evaluation of segmentation algorithms.
arXiv.
Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S., and
Jorge Cardoso, M. (2017). Generalised dice over-
lap as a deep learning loss function for highly unbal-
anced segmentations. In Deep Learning in Medical
Image Analysis and Multimodal Learning for Clini-
cal Decision Support: Third International Workshop,
DLMIA 2017, and 7th International Workshop, ML-
CDS 2017, Held in Conjunction with MICCAI 2017,
Qu
´
ebec City, QC, Canada, September 14, Proceed-
ings 3, pages 240–248. Springer.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2016). Rethinking the inception architecture
for computer vision. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2818–2826.
Yang, Z., Zhang, L., Zhang, M., Feng, J., Wu, Z., Ren, F.,
and Lv, Y. (2019). Pancreas segmentation in abdomi-
nal ct scans using inter-/intra-slice contextual informa-
tion with a cascade neural network. In 2019 41st An-
nual International Conference of the IEEE Engineer-
ing in Medicine and Biology Society (EMBC), pages
5937–5940. IEEE.
Yu, W., Chen, H., and Wang, L. (2019). Dense attentional
network for pancreas segmentation in abdominal ct
scans. In Proceedings of the 2nd International Con-
ference on Artificial Intelligence and Pattern Recogni-
tion, pages 83–87.
Zhang, D., Zhang, J., Zhang, Q., Han, J., Zhang, S., and
Han, J. (2021a). Automatic pancreas segmentation
based on lightweight dcnn modules and spatial prior
propagation. Pattern Recognition, 114:107762.
Zhang, Y., Wu, J., Liu, Y., Chen, Y., Chen, W., Wu, E. X.,
Li, C., and Tang, X. (2021b). A deep learning frame-
work for pancreas segmentation with multi-atlas reg-
istration and 3d level-set. Medical Image Analysis,
68:101884.
Zhao, H., Shi, J., Qi, X., Wang, X., and Jia, J. (2017).
Pyramid scene parsing network. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 2881–2890.
Zhao, N., Tong, N., Ruan, D., and Sheng, K. (2019). Fully
automated pancreas segmentation with two-stage 3d
convolutional neural networks. In Medical Im-
age Computing and Computer Assisted Intervention–
MICCAI 2019: 22nd International Conference, Shen-
zhen, China, October 13–17, 2019, Proceedings, Part
II 22, pages 201–209. Springer.
Zheng, H., Chen, Y., Yue, X., Ma, C., Liu, X., Yang, P.,
and Lu, J. (2020). Deep pancreas segmentation with
uncertain regions of shadowed sets. Magnetic Reso-
nance Imaging, 68:45–52.
Zhu, Z., Liu, C., Yang, D., Yuille, A., and Xu, D. (2019). V-
nas: Neural architecture search for volumetric medical
image segmentation. In 2019 International conference
on 3d vision (3DV), pages 240–248. IEEE.
Selection of Backbone for Feature Extraction with U-Net in Pancreas Segmentation
829
