
Numerical Modelling and Simulation of a Lab-on-a-Chip for Blood
Cells’ Optical Analysis
Ahmed Fadlelmoula
1,2
, Vítor Carvalho
3,4
, Susana O. Catarino
1,2
and Graça Minas
1,2
1
Center for MicroElectromechanical Systems (CMEMS-UMinho), University of Minho, 4800-058 Guimaraes, Portugal
2
LABBELS–Associate Laboratory, Braga/Guimaraes, Portugal
3
2Ai, School of Technology, IPCA, 4750-810 Barcelos, Portugal
4
Algoritmi Research Center, University of Minho, 4800-058 Guimaraes, Portugal
Keywords: Lab-on-a-Chip, Microfluidics, Numerical Simulation, COMSOL Multiphysics.
Abstract: Blood is a treasure of information about the functioning of the whole body. Thus, there is a continuous need
for new, accurate, fast, and precise techniques to analyse blood samples. The goal of this work is to design
and numerically simulate a low-cost lab-on-a-chip device, which, in the future, can be used to quickly
diagnose diseases by using a tiny drop of a blood sample from the patient. The designed microdevice includes
two fluid inlets, a serpentine area for achieving a continuous and fully developed flow, as well as a detection
chamber able for optical measurements. The numerical model of the designed microdevice was computed
using COMSOL Multiphysics software, taking into account the flow and tracking of microparticles,
mimicking blood cells. In order to reach the best lab-on-a-chip geometry, i.e., achieving a high and stable
number of particles in the detection chamber during the entire microfluidic assay, the inlet velocity, the
channel width, and the diameter of the detection chamber were individually optimized. A mesh study was
also performed to achieve the best results’ accuracy, with lowest computational effort. From the achieved
results, it was observed that a lab-on-a-chip geometry with a 0.5 mm channel width and a 2- or 3-mm detection
chamber radius, with a fluid inlet velocity of 3 mm/s, was the one with the most interesting results for the
intended application, with a constant number of particles flowing through the detection chamber (142 in
average, for the selected inlet conditions).
1 INTRODUCTION
Blood is a treasure of information about the
functioning of the whole body. Every minute, the
entire blood volume circulates throughout the body,
delivering oxygen and nutrients to every cell and
transporting products from and toward all different
tissues. As a result, blood harbors a massive amount
of information about the functioning of all tissues and
organs in the body Kouzehkanan et al., 2022).
Consequently, blood sampling and analysis are of
prime interest for medical and science applications
and hold a central role in diagnosing several
physiologic and pathologic conditions, localized or
systemic. However, for clinical and scientific
applications, it is necessary to understand, not only
the biology, but also the technologies involved
(Balogh, 2016). The knowledge about blood has
always evolved in parallel with the general
knowledge of biology, and several breakthroughs
were facilitated by technological advances. More
specifically, numerous devices are used to analyze
blood cells with good sensitivity. Various techniques
have been used for detecting platelets, white and red
blood cells (Rohde, 2015), with a continuous need
for fast, and precise techniques for the blood samples
analysis. Microfluidics has demonstrated an
enormous potential in this field. However, designing
a customized microfluidic platform, and gaining a
better understanding of its operation and the
underlying physics and mechanics still pose
significant technical challenges. On one hand,
experimental approaches, although expensive and
laborious, have been commonly used for the
development of microfluidic devices since they are
accurate and evidence-based methods (Fadlelmoula,
2022). Numerical approaches, on the other hand, are
now recognized as a reliable complementary method
aiming a reduction of cost, time, and effort, while
being relatively accurate (Nagarajan, 2017). So, this
Fadlelmoula, A., Carvalho, V., Catarino, S. and Minas, G.
Numerical Modelling and Simulation of a Lab-on-a-Chip for Blood Cells’ Optical Analysis.
DOI: 10.5220/0012571900003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 185-190
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
185

work aims to design, simulate and optimize a low-
cost lab-on-a-chip (LOC) device, which can be used
to quickly evaluate and analyze blood cells using
optical methods. It presents the numerical modelling
and the simulation study of an optimized
microchannel geometry for the intended application.
Therefore, it simulates the flow of a buffer fluid
(representing plasma) filled with microparticles,
mimicking flowing blood cells. The microdevice will
comprise a serpentine region, for achieving a fully
developed flow (Catarino, 2019), as well as a
detection chamber, where the optical measurements
will occur, where the number of particles/cells should
be as high and steady as possible during the entire
duration of the assays.
This paper is organized into 5 sections: Section 2
presents the numerical methods; Section 3 shows the
obtained results; Section 4 presents the discussion;
and Section 5 enunciates the conclusions and further
developments.
2 NUMERICAL METHODS
This section describes the numerical simulation
methods applied for simulating the designed
microdevice.
2.1 2D Geometry Model
Figure 1 shows the initial lab-on-a-chip design
geometry. According to the computational results
(section 3), it will be optimized.
Figure 1: LOC initial design.
As can be observed in Figure 1, the total chip
dimension, 2D top view, is 4 cm length and 1.6 cm
width, there are 2 inlets and an outlet with a 0.5 mm
radius. The serpentine turn width is 0.4 mm, and the
circular detection chamber has a 1.3 mm diameter.
Relatively to the domains’ materials and properties,
the microchannel walls are constituted by
polydimethylsiloxane (PDMS) while, in the interior
of the device, water (1000 kg/m³ density and 0.001
Pa·s viscosity) will be flowing (Norouzi, 2017).
2.2 Governing Equations
The COMSOL Multiphysics Laminar Flow interface
(COMSOL, 2017) is used to compute the velocity and
pressure fields for the flow of a single-phase fluid in
the laminar flow regime. Equations (1) and (2)
present the fluid flow governing equations:
𝜌
𝑢∗∇𝑢
∇𝑃∇.𝜏 (1)
𝜏𝜂
𝛻𝑢 𝛻𝑢
2𝜂 𝐷 (2)
Where ρ represents the density,
∇
represents the
gradient operator, u is the velocity vector, t is time, P
is pressure, τ represents the Newtonian extra stress
tensor, η is the dynamic viscosity, T is the matrix
transpose, and D is the Symmetric rate of strain
tensor.
Besides the laminar flow, particle tracing for the
fluid flow was used as a numerical method for
computing the paths and migration of individual
particles by solving their equations of motion over
time. The particle traceability will be examined under
different conditions, to reach the maximum number
of particles that will pass through the detection
chamber. Equation (3) presents the particle tracing
governing equation:
𝐹𝑡 𝑑
𝑚𝑝𝑣
/𝑑𝑡
(3)
where Ft, mp, and v are, respectively, the total
force, the particle mass, and the particle velocity.
Particles moving through a fluid are subjected to a
force, known as drag force, which acts in the direction
of the fluid’s motion relative to the object. Equation
(4) presents the Stokes’ law equation, that allows to
determine the drag force (FD).
𝐹𝐷 6π ∗ 𝜇 ∗ 𝑟𝑝 ∗ 𝑢𝑠 (4)
where rp is the radius of the sphere and us is the
velocity of the fluid relative to the sphere, also called
slip velocity.
2.3 Boundary and Initial Conditions
The following boundary and initial conditions were
considered:
Laminar Flow: The default boundary condition in
laminar flow is a non-slip wall, which means that the
fluid velocity at the wall is zero.
Two different inlets were considered in the
microchannel, one for the particle’s inlet and other for
the buffer solution (Inlet 1 and Inlet 2). Both of them
are independently described by fluid velocities, that
range from 1 to 8 mm/s. At the outlet, a zero pressure
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
186
boundary is set to assure the outflow. Regarding the
numerical initial conditions, both initial velocity and
pressure were set to zero.
Particle Tracing for Fluid Flow: The boundary
condition in particle tracing was assumed as a
slipping particle wall, which means particles reflect
from the wall, such that the particle momentum is
conserved.
Regarding the inlet of particles, these were
released on both inlets, since the beginning of the
assay (time = 0 s), and at time steps of 0.1 seconds,
for a total duration of 15 seconds. At each 0.1 second
release, 200 rigid, non-charged particles (as an
approximation to flowing cells), entered the
microchannel. The number of particles was selected,
aiming to represent the number of blood cells in a
diluted whole blood sample, flowing in the channel.
In these simulations, the particles have a 5 μm
diameter and a 1050 kg/m³ density, representing the
size and density of red blood cells.
2.4 Mesh
After defining the numerical model, the geometry was
meshed, aiming its computational solution. To reach
the best type of mesh, aiming the most accurate
results with lowest computational cost, a mesh study
was performed. For that, simulations of the fluid flow
were performed in three different regions of the
microdevice, and the maximum fluid velocity
was evaluated in each of those regions, as shown in
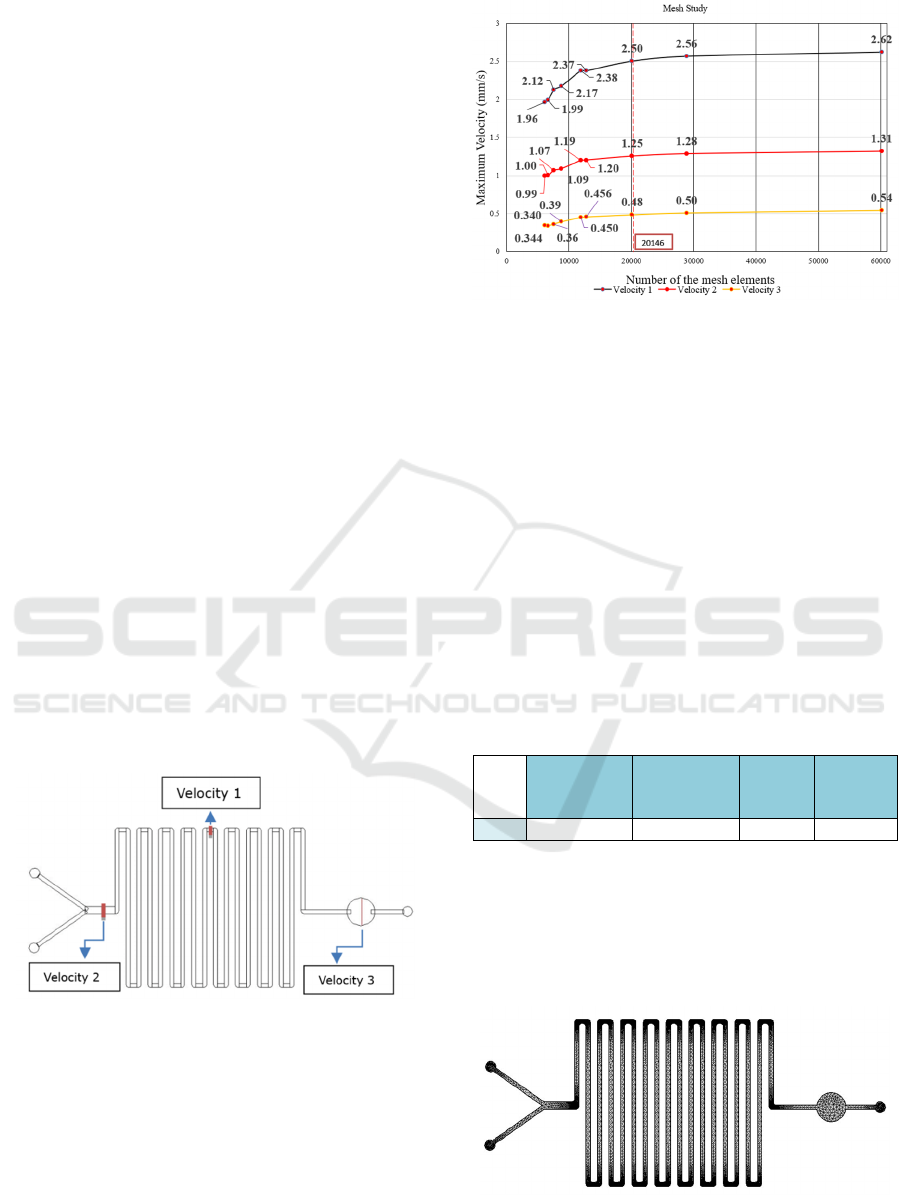
Figure 2.
Figure 2: Schematic of the three regions where the
maximum velocity was evaluated during the mesh study
simulations.
After computing the model for 9 different meshes
(predefined at COMSOL Multiphysics), the
maximum velocity was evaluated in each of the 3
sections. Figure 3 shows the maximum velocity, at
each of the three regions (Velocity 1, Velocity 2 and
Velocity 3), for all the simulated meshes.
Figure 3: Maximum velocity (mm/s) in the microchannel as
a function of the number of mesh elements.
From the presented plot, it can be observed that,
above the 20146 elements mesh (“Finer” mesh, in
COMSOL), even if the number of elements is
increased (obviously with higher computational
efforts, since the number of calculus points’ increases
significantly), there is no significant variation or
improvement in the maximum velocity in any of the
considered regions. As it reaches a plateau, it will be
considered as the ideal mesh for this model, as it
allowed to achieve accurate results, without an
excessive computational cost (both regarding time
and memory). Thus, the statistics of the selected
“Finer” mesh, as predefined in COMSOL
Multiphysics, are shown in table 1.
Table 1: Mesh Statistics.
Minimum
Element
Qualit
y
Average
Element
Qualit
y
Mesh
Vertices
Triangles
Fine
r
0.5016 0.8256 9351 20146
This mesh, with 20146 triangular elements (9351 of
them mesh vertices), achieved the best results in
terms of velocity stability and computational time.
This mesh has a minimum element quality of 0.5016
(0-1 scale) and an average element quality of 0.8256,
and it is represented in figure 4.
Figure 4: Representation of the 2D finer mesh.
Numerical Modelling and Simulation of a Lab-on-a-Chip for Blood Cells’ Optical Analysis
187
2.5 Solver
The laminar flow, as a steady-state condition, was
simulated using a stationary solver. The particle
tracing was simulated considering a time-dependent
solver, for a total duration of 15 seconds, with 0.1 s
time steps.
3 RESULTS AND DISCUSSION
The simulated results, using COMSOL Multiphysics,
were achieved considering the effect of the fluid flow
(velocity and pressure profile along the lab-on-a-chip)
and of the particle’s migration in the detection
chamber. For achieving a high and stable number of
particles in the detection chamber during the entire
microfluidic assay, the inlet velocity, the channel
width, and the diameter of the detection chamber
were individually optimized, outputting the best LOC
geometry.
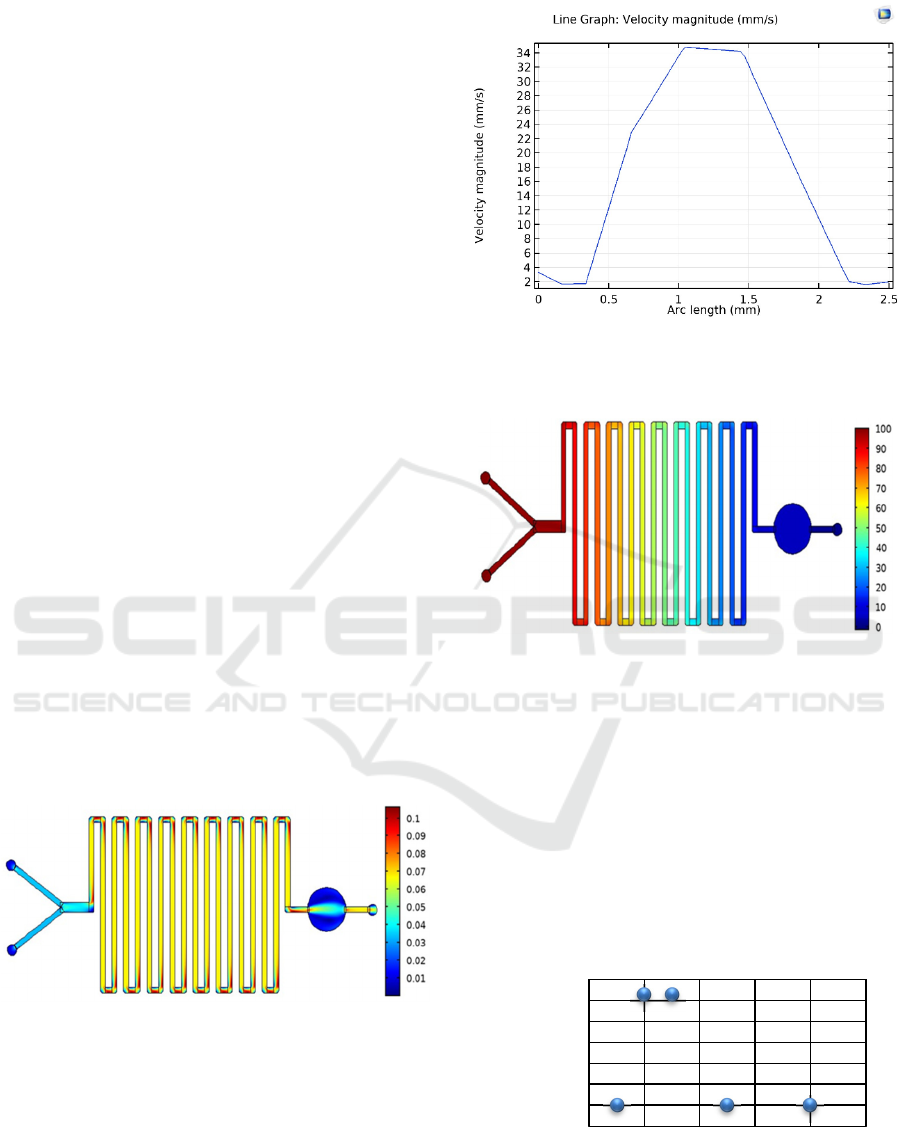
With the layout geometry described in Figure 1,
Figure 5 shows the plot of the fluids’ velocity
magnitude during the migration along the
microdevice. Analyzing Figure 6, that represents the
velocity profile through a half-width cut of one of the
simulated detection chambers, it can be concluded
that the velocity in the near wall region decreases,
being 0 at the wall, and being maximum at the center
of the detection chamber. Moreover, figure 7 shows
that the obtained pressure varies along the LOC
channels from the highest at the beginning of the
microdevice, decreasing gradually until the end of the
microdevice.
Figure 5: Stationary velocity magnitude (mm/s) in the
microdevice.
The first layout geometry optimization was
performed fixing the channel width and the detection
chamber radius, and changing the inlet velocity. The
channels' width was kept at 0.4 mm and the detection
chamber radius as 1 mm. The velocity was changed
being 1, 2, 3, 5 and 8 mm/s. Figures 8 show the
average of the number of particles that passed through
Figure 6: Example of a cross-section plot of the stationary
velocity magnitude (mm/s), at the detection chamber half-
width.
Figure 7: Pressure distribution along the microdevice.
the detection chamber, respectively, during the entire
duration of the experiment (15 seconds). The results
show that modifying the inlet velocity, from 1 to 8
mm/s, has no significant effect in the number of
particles crossing the detection chamber (it was
always around 25 particles at any time). As the 2
mm/s and 3 mm/s inlet velocities reached the exact
same average results, a 3 mm/s inlet velocity was
selected for the next optimization steps due to their
slightly higher particles number passing through the
detection chamber.
Figure 8: Average number of particles that passed through
the detection chamber, for different inlet velocities.
25
25.05
25.05
25 25
24,99
25
25,01
25,02
25,03
25,04
25,05
25,06
0246810
Average number of
particles (a.u.)
Particles velocity at inlet (mm/s)
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
188
In the second experiment, the channel width (0.4 mm)
and the inlet velocity (3 mm/s) were fixed, and the
detection chamber radius was changed from 1 to 3
mm in steps of 0.5 mm. Figures 9 and 10 show the
average and standard deviation of the number of
particles that passed through the detection chamber,
respectively, during the duration of the simulation (15
seconds). The results show that, increasing the radius
of the detection chamber leads to an increase of the
average number of particles in that area. Thus, a 3 mm
radius (with an average number of particles around
35) was selected for the next optimization steps.
Figure 9: Average number of particles that passed through
the detection chamber, for different detection chamber
radius.
Figure 10: Standard deviation of the number of particles
passing through the detection chamber, for different
detection chamber radius.
In the third experiment, the inlet velocity (3 mm/s)
and the detection chamber radius (3 mm) were fixed
and the width of the channels was varied from 0.2 to
1 mm in steps of 0.1 mm. Figures 11 and 12 show the
average and standard deviation of the number of
particles that passed through the detection chamber
during the duration of the assay (15 seconds),
respectively. From the average and the standard
deviation obtained results, an adequate channel width,
combining a high and stable number of particles in the
chamber is 0.5 mm.
Figure 13 shows the number of particles that passed
through the detection chamber when changing the inlet
2 velocity and maintaining the previously optimized
detection chamber radius (3 mm), channel width
Figure 11: Average number of particles that passed the
detection chamber, for different channels’ width.
Figure 12: Standard deviation of the number of particles
passing through the detection chamber, for different
detection channels width.
(0.5mm) and inlet 1 velocity (3 mm/s). It was verified
that no significant changes occurred regarding the
particle number that passed in the detection chamber.
This means that, for the studied geometries and inlet
conditions, the change in the inlet 2 velocity (buffer)
will not change the particle number.
Figure 13: Average number of particles that passed the
detection chamber, for different inlet 2 velocities.
From the results presented in the previous plots,
Figure 14 shows the instantaneous number of
particles passing through the detection chamber when
a 0.5 mm channel width, a detection chamber radius
25.05
26.42
26.10
33.73
35.05
0
10
20
30
40
01234
Average number of
particles (a.u.)
Detection chamber radius (mm)
0.22
0.50
0.31
0.45
0.40
0
0,1
0,2
0,3
0,4
0,5
0,6
01234
Standard deviation
(a.u.)
Detection chamber radius (mm)
69.28
56
73.05
141.58
159
141
395.58
33.33
0
100
200
300
400
500
0 0,2 0,4 0,6 0,8 1
Average number of
particles (a.u.)
Channels width (mm)
3.24
0
0.22
1.37
1.52
1.19
1.85
18.38
0
4
8
12
16
20
0 0,2 0,4 0,6 0,8 1
Standard deviation (a.u.)
Channels width (mm)
94.84
94.69
94.76
94.92
94.69
94,65
94,7
94,75
94,8
94,85
94,9
94,95
0 5 10 15
Average number of
particles (a.u.)
Inlet 2 velocity
Numerical Modelling and Simulation of a Lab-on-a-Chip for Blood Cells’ Optical Analysis
189
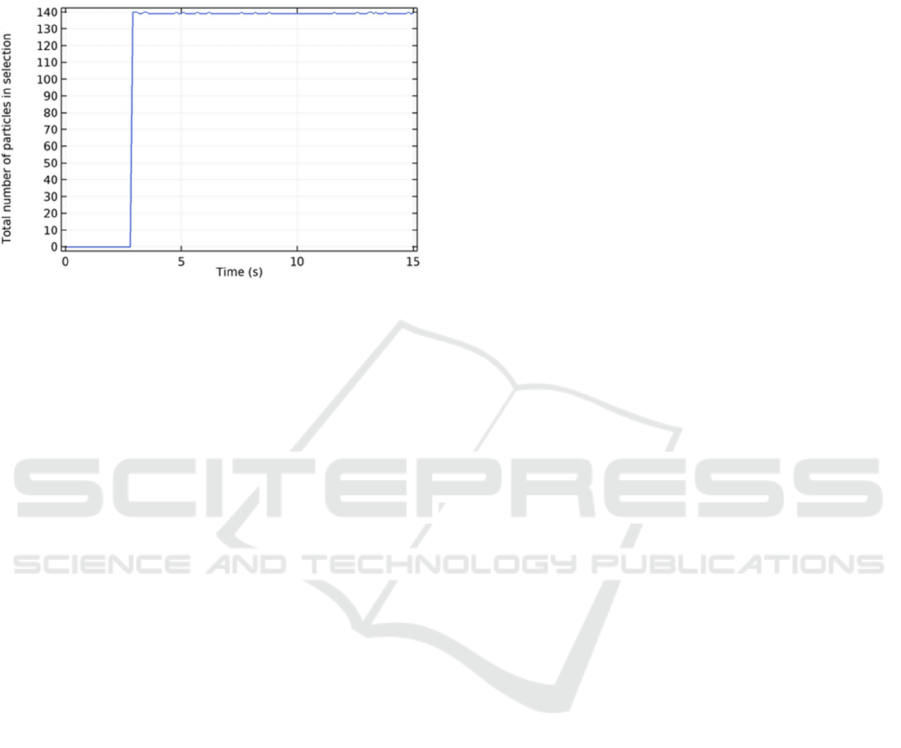
of 3 mm and an inlet velocity of 3 mm/s was
considered. It can be observed that, after the particles
first reach the detection chamber (shortly before 6
seconds), the number of particles keeps stable and
almost constant during the assays.
Figure 14: Total number of particles passing in the detection
chamber over each time instant, considering the optimized
design.
4 CONCLUSION AND FUTURE
WORK
This works presented the design and numerically
simulation of a LOC device with optimized
dimensions, regarding inlets velocities, channel
width, and diameter of the detection chamber for
achieving a high and stable number of particles in the
detection chamber. The numerical model was
computed, using COMSOL Multiphysics software,
taking into account the flow and microparticles
tracking, mimicking the blood cells.
The obtained results showed the ideal design, a 0.5
mm channel width, a detection chamber radius of 2
mm or 3 mm, and an inlet velocity of 3 mm/s,
achieving a total number of 142 particles flowing in
the detection chamber (see figure 11). The change of
the channels width made the major difference, when
compared with the others changed parameters, in the
number of particles passing through the detection
chamber. Regarding the flow, the pressure along the
LOC reached the maximum value at the inlet and
decreased gradually until reached the minimum in the
outlet. The stationary velocity reached the maximum
value in the serpentine channels and at the center of
the detection chamber.
Further work will consolidate the physical
implementation of the simulated LOC model and
their testing, examining the velocity, pressure, and
particle flow inside the chip, and performing design
updates if required.
ACKNOWLEDGMENTS
This work was supported by the R&D Unit Project
Scope: UIDB/04436/2020, UIDB/05549/2020 and
UIDP/05549/2020 funded by the Foundation for
Science and Technology, I.P. (FCT). A.F. thanks the
FCT for his 2023.03312.BD PhD grant. S.O.C. thanks
the FCT for her 2020. 00215.CEECIND contract
funding (DOI: 10.54499/2020.00215.CEECIND/
CP1600/CT0009).
REFERENCES
Balogh, E. P., Miller, B. T., & Ball, J. R. (2016). Improving
diagnosis in health care. In Improving Diagnosis in
Health Care. National Academies Press. https://doi.org/
10.17226/21794
Catarino, S. O., Rodrigues, R. O., Pinho, D., Miranda, J. M.,
Minas, G., & Lima, R. (2019). Blood cells separation
and sorting techniques of passive microfluidic devices:
From fabrication to applications. In Micromachines
(Vol. 10, Issue 9). MDPI AG. https://doi.org/10.3390/
mi10090593
COMSOL Multiphysics User’s Guide. 2017. Available
online: doc.comsol.com/5.4/doc/com.comsol.help.com
sol/COMSOL_ReferenceManual.pdf (accessed on 1
September 2023).
Fadlelmoula, A., Pinho, D., Carvalho, V. H., Catarino, S. O.,
& Minas, G. (2022). Fourier Transform Infrared (FTIR)
Spectroscopy to Analyse Human Blood over the Last 20
Years: A Review towards Lab-on-a-Chip Devices. In
Micromachines (Vol. 13, Issue 2). MDPI.
https://doi.org/10.3390/mi13020187.
Kouzehkanan, Z. M., Saghari, S., Tavakoli, S., Rostami, P.,
Abaszadeh, M., Mirzadeh, F., Satlsar, E. S.,
Gheidishahran, M., Gorgi, F., Mohammadi, S., &
Hosseini, R. (2022). A large dataset of white blood cells
containing cell locations and types, along with
segmented nuclei and cytoplasm. Scientific Reports,
12(1). https://doi.org/10.1038/s41598-021-04426-x
Nagarajan, S., Stella, L., Lawton, L. A., Irvine, J. T. S., &
Robertson, P. K. J. (2017). Mixing regime simulation and
cellulose particle tracing in a stacked frame
photocatalytic reactor. Chemical Engineering Journal,
313, 301–308. https://doi.org/10.1016/j.cej.2016.12. 016.
Norouzi, N., Bhakta, H. C., & Grover, W. H. (2017). Sorting
(Multiphysics®)cells by their density. PLoS ONE,
12(7). https://doi.org/10.1371/journal.pone.0180520
Rohde, T., & Martinez, R. (2015). Equipment and Energy
Usage in a Large Teaching Hospital in Norway. In
Journal of Healthcare Engineering · (Vol. 6, Issue 3).
BIODEVICES 2024 - 17th International Conference on Biomedical Electronics and Devices
190
