
Automatic Computation of the Posterior Nipple Line from
Mammographies
Quynh Nhu Jennifer Tran
1
, Tina Santner
2 a
and Antonio Rodr
´
ıguez-S
´
anchez
1 b
1
Department of Computer Science, University of Innsbruck, Innsbruck, Austria
2
Department of Radiology, Medical University of Innsbruck, Innsbruck, Austria
Keywords:
Mammogram, Posterior Nipple Line Detection, Computer Vision.
Abstract:
Breast cancer is the most commonly diagnosed cancer in female patients. Detecting early signs of malignity
by undergoing breast screening is therefore of great importance. For a reliable diagnosis, high-quality exami-
nated mammograms are essential since poor breast positioning can cause cancers to be missed, which is why
mammograms are subject to strict evaluation criteria. One such criterion is the posterior (or pectoralis) nip-
ple line (PNL). We present a method for computing the PNL length, which consisted of the following steps:
Pectoral Muscle Detection, Nipple Detection, and final PNL Computation. A multidirectional Gabor filter
allowed us to detect the pectoral muscle. For detecting the nipple we made use of the geometric properties
of the breast, applied watershed segmentation and Hough Circle Transform. Using both landmarks (pectoral
muscle and nipple), the PNL length could be computed. We evaluated 100 mammogram images provided
by the Medical University of Innsbruck. The computed PNL length was compared with the real PNL length,
which was measured by an expert. Our methodology achieved an absolute mean error of just 6.39 mm.
1 INTRODUCTION AND
BACKGROUND
Breast cancer is the most commonly diagnosed can-
cer in women, accounting for 11.7% of all cancer
cases in 2020 with approximately 2.3 million new
cases worldwide. According to GLOBOCAN 2020,
a database on cancer statistics, breast cancer is also
the fifth leading cause of cancer mortality with about
685,000 new deaths (Sung et al., 2021).
Early signs of malignity can be detected by exam-
ining screening mammograms, which are X-ray im-
ages of the breast. The ability to make reliable di-
agnoses strongly depends on the quality of the mam-
mograms, where correct breast positioning is particu-
larly important, as poor positioning of the breast can
contribute to breast cancers being missed. Several
tools are available to check and monitor this diagnos-
tic image quality. These are checklists or classifica-
tion systems, where several image quality statements
are assessed and the images are classified based on the
overall score (Waade et al., 2021). Well-known and
internationally (NHSBSP, 1989) in use is the PGMI
a
https://orcid.org/0000-0002-2224-1089
b
https://orcid.org/0000-0002-3264-5060
(perfect-good-moderate-inadequate) (Klabunde et al.,
2001). One positioning criterion of this system is
the posterior (or pectoralis) nipple line (PNL). The
PNL is a line drawn posteriorly and perpendicularly
from the nipple towards the pectoral muscle. Ide-
ally, the PNL in the craniocaudal (CC) view should
be the same length as the PNL in the mediolateral
oblique (MLO) view to ensure that sufficient breast
tissue is included, and a reliable diagnosis can be
made (Sweeney et al., 2017). However, the problem
is that measuring the PNL for both views manually is
time-consuming. In addition, the results are subjec-
tive and inhomogeneous and can vary from person to
person.
Recent approaches have been proposed for auto-
mated assessment of the breast positioning quality in
mammograms based on deep learning. (Gupta et al.,
2020) used transfer learning to predict the two points
representing the PNL in the MLO view. He used
Inception-V3 (Szegedy et al., 2015) as the base net-
work and replaced the last layer with a single out-
put node. The network was initialized with the pre-
trained weights from the ImageNet dataset (Deng
et al., 2009). For detecting the PNL in the CC view, an
algorithm was developed for detecting the radiopaque
marker, which was placed over the nipple during the
Tran, Q., Santner, T. and Rodríguez-Sánchez, A.
Automatic Computation of the Posterior Nipple Line from Mammographies.
DOI: 10.5220/0012570500003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 4: VISAPP, pages
629-636
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
629

data labeling step, using Hough Circle Transform.
The final PNL was a line drawn horizontally from
the radiopaque marker to the image border. Brahim
et al. (2022) trained a convolutional neural network
for binary classification. The output was either 0
(good breast positioning) or 1 (poor breast position-
ing). Hejduk et al. (2023) trained eight deep convo-
lutional neural networks for detecting the presence of
anatomical landmarks and localizing features. Three
of the features are the nipple, pectoralis cranial, and
the pectoralis caudal. Based on them the PNL length
could be calculated.
Even though providing very good results, deep
learning is limited when the quantity of images is lim-
ited or the intermediate steps - i.e. image landmarks -
are not provided, but just the final PNL values.
In this paper, we present a method to automati-
cally compute the length of the PNL in mammograms
without the need for a large labelled dataset contain-
ing image landmarks. The proposed method can help
in assessing the image quality of mammograms by au-
tomatically measuring the PNL length and thus saving
time. This is done by first detecting the pectoralis ma-
jor (pectoral) muscle and the location of the nipple us-
ing computer vision-based methods, which once suc-
cessfully detected, the PNL length can be accurately
calculated. All images used in this paper are from the
dataset provided by the Medical University of Inns-
bruck.
2 METHODS
Our method for Pectoral Muscle Detection follows a
similar approach to the multidirectional Gabor filter
(MDGF)-based approach for pectoral muscle bound-
ary (PMB) detection by (Rahman and Jha, 2022) and
is explained next.
2.1 Region of Interest (ROI) Extraction
In the MLO view, the pectoral muscle usually lies in
the upper left corner, and therefore a triangular ROI
containing the PMB was computed. For this, the
image was converted into a binary image by apply-
ing simple thresholding with a threshold value of 2.
Afterwards, the border following algorithm (Suzuki
and Keiichi, 1985) was applied on the binary image
to retrieve contours, and the contour with the largest
area was selected to be the one of the breast bound-
ary. Next, a mask was created by filling the area
bounded by the contour. The Harris Corner Detec-
tor algorithm (Harris and Stephens, 1988) was then
applied to the mask, where only significant corners
were selected. Only pixels where the Harris corner
response was greater than 20% of the maximum re-
sponse value were considered. The corner with the
smallest Euclidean distance to the top-right corner of
the image was then selected. Finally, the triangular
ROI was defined by the three corner points: top-left
corner, bottom-left corner, and the computed corner
marking the starting point of the breast boundary.
2.1.1 ROI Preprocessing
To increase the image contrast, Contrast Limited
Adaptive Histogram Equalization (CLAHE) (Pizer
et al., 1987) was applied to the ROI with a con-
trast limiting threshold of 1 and 8 ×8 tile size. To
further enhance the ROI, Non-Local Means Denois-
ing (Buades et al., 2011) was used to reduce noise.
The filter strength was set to 10 since a higher value
would remove important details. Besides low con-
trast and noise, breast tissue can pose a challenge in
cases where it overlaps with the PMB. To address this
problem, we applied image inpainting which can re-
store selected regions in an image by filling them with
the information surrounding them (Bertalmio et al.,
2000). Since breast tissue usually has higher pixel
intensities than the muscle region, top-hat filtering
(computes the opening of an image by removing small
objects from the foreground, and subtracts it from the
original image) was applied to the denoised ROI to
enhance small bright objects. Then simple thresh-
olding was performed on the transformed ROI with
a threshold value of 9. The resulting thresholded im-
age indicates which regions needed to be restored and
was passed as an inpainting mask to the inpainting
function.
The ROI image might contain regions of 0-pixels
like the background in mammograms. A strong inten-
sity edge between the 0-pixels region and the nearby
breast region will be formed. This edge might be
falsely detected as the pectoral muscle boundary later
on (Rahman and Jha, 2022). Therefore the Zero
Background Pixel Correction algorithm by (Rahman
and Jha, 2022) was performed on the inpainted ROI,
where each row is scanned and if a 0-pixel is detected,
the mean intensity value of 12 nearby nonzero pixels
along the same row will be assigned to all 0-pixels in
that row.
2.1.2 Multidirectional Gabor Filter (MGF)
After the ROI Preprocessing step, a set of three high-
frequency Gabor filters tuned at different orientations
was created and applied to the processed ROI for cap-
turing the PMB. The parameter values were selected
as follows:
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
630

(a)
(b) (c) (d)
(e)
Figure 1: (a) Source image. (b, c, d) Phase response (left) and Laplacian (right) for orientations 180, 170 and 160 respectively.
(e) Detected pectoral muscle boundary (left) marked in blue and fitted line marked in green (right).
1. The three Gabor filters were each tuned to an ori-
entation (α) of 180, 170 and 160, respectively, to
cover all orientations in which the muscle can ap-
pear.
2. The spatial aspect ratio was set to the value 0.2617
obtained by using the formula
3∗π
2∗M
stated in (Rah-
man and Jha, 2022) with M = 18, where M de-
notes the total number of filters in the interval
[0, π].
3. The spatial frequency bandwidth was set to 1.2
octaves allowing a moderate range of frequencies
around the central frequency in which the filter
can respond well.
4. The wavelength (λ) was set in a similar manner
as in (Rahman and Jha, 2022) and was computed
using the formula b
w
2
∗
√
2
4
c where w denotes the
width of the ROI image to capture high- and mid-
frequency features.
2.1.3 Phase Response of MFG for Pectoral
Muscle Detection
After applying the three Gabor filters to the ROI im-
age, the phase response of each Gabor response im-
age was computed. To detect and extract the textu-
ral edge information the Laplacian of each phase re-
sponse was computed and combined (addition) (Rah-
man and Jha, 2022). The combined Laplacian image
(Fig. 1b,c,d) was converted into a binary image by ap-
plying simple thresholding with a threshold value of
8. In some cases, a strong intensity edge might be re-
flected in two or three phase responses, and because
of this two or three different intensity edges very close
to each other might appear in the combined Laplacian
(Rahman and Jha, 2022). Morphological closing (fill-
ing small holes) was applied to the resulting image
in order to merge nearby edge lines. The PMB was
obtained by selecting the largest connecting compo-
nent with 8-connectivity. Once the pectoral muscle
had successfully been detected, a line was fitted to the
points on the detected boundary using the linear least
squares method. This is necessary because the actual
PMB is curved, but for computing the PNL length a
straight line is needed (Fig. 1e).
3 NIPPLE DETECTION
3.1 ROI Extraction
For detecting the nipple only a small region is of in-
terest and therefore a small ROI containing the nipple
was extracted. This was done similarly as in (Jiang
et al., 2019). First, the topmost corner marking the
starting point of the breast boundary (x
1
, y
1
) com-
puted in figure 2.1 and the bottom-left corner (x
2
, y
2
)
were used as ending points for forming a line. The
angle θ required for rotating this line so that it aligns
with the left image border was calculated
line angle =
tan
−1
(
y
1
−y
2
x
1
−x
2
)
∗
180
π
(1)
θ = | line angle −90 |. (2)
Then the breast mask computed in figure 2.1 was
rotated (only necessary for the MLO view) by θ and
passed to the border following algorithm (Suzuki and
Keiichi, 1985) to get the breast contour, the rightmost
point on the contour was identified and shifted by 35
pixels to the left along the x-axis. A new image with
the same shape as the breast mask filled with 0-pixels
was then created. Pixel values of pixels lying inside
the rectangular region bounded by the corner points
(s
x
, 0), (w, 0), (w, h) and (s
x
, 0) were set to 255 where
(s
x
, s
y
) are the coordinates of the shifted point, w is the
width and h is the height of the image. Afterwards,
Automatic Computation of the Posterior Nipple Line from Mammographies
631
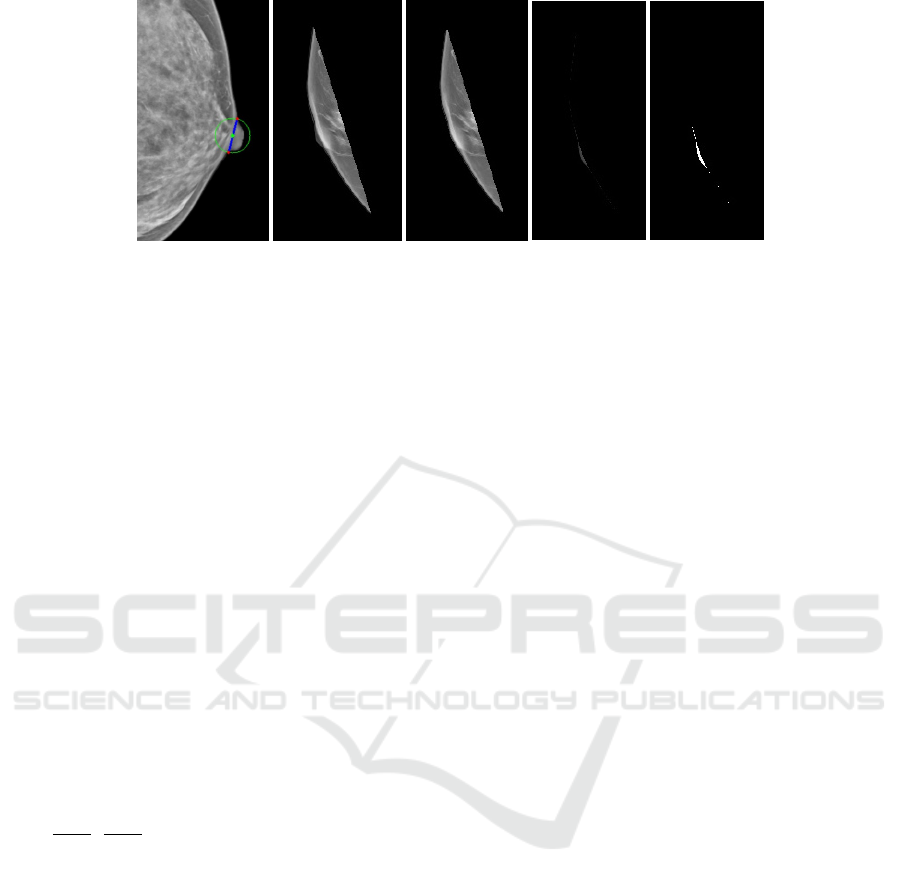
(a)
(b)
(c)
(d) (e)
Figure 2: (a) Starting points are marked with red circles and the location of the nipple base is marked with a green circle. (b)
Breast with nipple. (c) Breast without nipple. (d) Output of intersection between (b) and (c). Thresholded image of (d).
bitwise conjunction between the rotated breast mask
and the new image was performed. The new mask
was then rotated back to its original position (only for
the MLO view). To get the ROI containing only the
nipple region bitwise conjunction between the mask
and the source image was performed.
3.2 Detection of the Nipple
First, large nipples that are in profile are located.
Here, the ROI image was converted into a binary
image, and the contour of the nipple region was
computed following the border (Suzuki and Keiichi,
1985). Then the convex hull of the nipple region was
computed which is the minimum boundary that en-
closes this region. Afterwards, the convexity defects
(deviations of the contour from the convex hull), are
calculated. Large deviations with a depth greater than
2.5 usually indicate the starting points of the nipple.
The two points with the largest deviation (x
1
, y
1
) and
(x
2
, y
2
) were used to find the location of the nipple
base (
x
1
+x
2
2
,
y
1
+y
2
2
) (Fig. 2a).
If no deviation with a depth greater than 2.5 was
found, either a small nipple in profile or a subtle nip-
ple was present, a problem that was overcome as ex-
plained next. Usually, small nipples that are in pro-
file have a distinct nipple base boundary edge and
this characteristic was used to detect them. This was
done by segmenting the nipple from the rest of the
breast using watershed segmentation (Meyer, 1992).
A marker was created by determining which region
belongs to the breast region without the nipple (sure
foreground) and which is the background (sure back-
ground). To obtain the sure background, dilation
(adds pixels to the object boundaries) with a 2 ×2
kernel consisting of ones was applied to the binary
image and for the sure foreground, we applied ero-
sion (removes pixels from boundaries) with a 12 ×12
kernel. By subtracting the sure foreground from the
sure background, a region (unknown) was derived for
which there is no information on whether it belongs
to the foreground or background. This unknown re-
gion encloses the nipple base boundary edge. Next,
the regions in the marker are labelled. The unknown
region was labelled with 0, the background with 1 and
the foreground with 2. The marker was then passed to
the watershed segmentation algorithm to get the con-
tour of the breast region without the nipple. Next, the
intersection between the breast region and the breast
region without the nipple was computed followed by
thresholding. The largest connecting component with
an area greater than 25 was selected as the nipple (Fig.
2).
If no connecting component with an area greater
than 25 was found, a subtle nipple (nipple not in pro-
file) was present. For detecting subtle nipples the
source image was first restricted to the region along
the breast boundary. Next, Non-Local Means Denois-
ing was applied to the ROI image to reduce noise, fol-
lowed by opening (removing small objects from the
foreground) with circular structuring element of size
10 ×10. Afterwards, Hough Circle Transform was
applied to find circles with a circle radius between 10
and 25, and only those with a mean intensity greater
than 65 were selected. The final subtle nipple is the
circle furthest to the right.
3.3 Posterior Nipple Line Computation
Once the pectoral muscle and nipple have success-
fully been detected, the posterior nipple line and its
length can be computed. Here, the pectoral muscle is
represented as a tuple (m
pec
, c
pec
) where m
pec
is the
slope and c
pec
is the y-intercept in the slope-intercept
form y = m ∗x + c of the pectoral muscle line. If the
muscle line cannot be represented in slope-intercept
form, that is, it is a vertical line, the pectoral muscle
is represented by the variable x
pec
instead, which is
set to the value of the x-coordinate of any point on
that line. The nipple is represented by its coordinates
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
632
(a)
(b)
(c)
(d)
(e)
(f)
Figure 3: Examples of correctly (a) and falsely (b) located nipples that are in profile and large. Examples of correctly (c) and
falsely (d) located nipples that are in profile and small. Examples of correctly (e) and falsely (f) located nipples that are not in
profile.
(x
nip
, y
nip
).
In the case where the pectoral muscle is not a ver-
tical line, the slope m
pnl
and the y-intercept c
pnl
of the
linear equation of the PNL were calculated as follows:
m
pnl
=
−1
m
pec
(3)
c
pnl
= (y
nip
−(m
pnl
∗x
nip
)) (4)
Next, the intersection point (x
inter
, y
inter
) of the
pectoral muscle line and the PNL line was calculated
as follows:
x
inter
y
inter
=
−m
pnl
1
−m
pec
1
−1
c
pnl
c
pec
(5)
In the case where the pectoral muscle is a vertical
line, the PNL is a horizontal line with linear equation
y = y
nip
and the intersection point is (x
pec
, y
nip
).
To calculate the length of the PNL, the Euclidean
distance between the intersection point and the nipple
was calculated.
4 EXPERIMENTAL EVALUATION
A dataset of DICOM images provided by the Med-
ical University of Innsbruck was used in our exper-
imental evaluation, which contains mammograms of
both MLO and CC views. For each patient there is an
MLO and a CC view of each breast (left and right),
for some patients, there are only images of one side
of the breast.
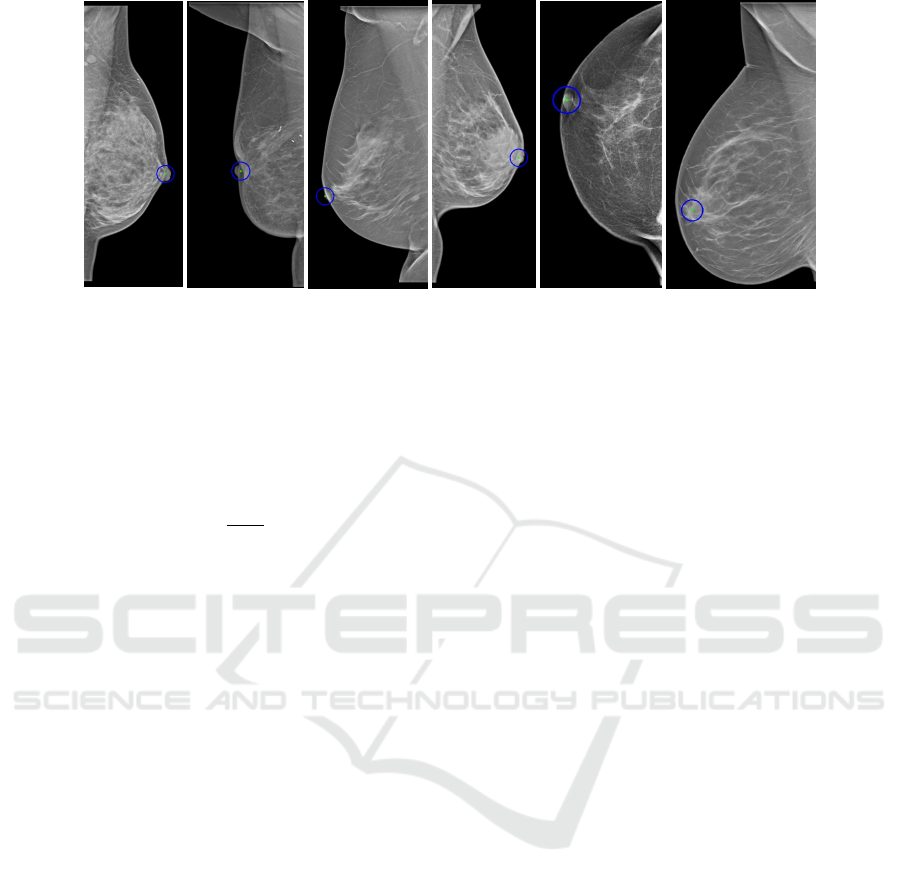
4.1 Qualitative Results on Nipple
Detection
Nipples that were large and had well-defined edges at
the two starting points of the nipple were accurately
identified in most of the cases as shown in figure 3a.
In cases with no sharp edges, the center could be
slightly off since the points with the largest deviation
can not be accurately defined. Our approach failed
to detect the nipple in rare cases as the one shown in
figure 3b, which was due to the breast having an in-
dentation at the location of the nipple.
Our approach was also successful at detecting
small nipples that are in profile. This is because the
edge of the nipple base is usually well-defined making
proper segmentation possible (Fig. 3c). If a small nip-
ple in profile was present but the nipple base edge was
not well-defined, our method failed to correctly locate
the nipple (Fig. 3d), but this case rarely occurred.
The detection of nipples that are not in profile
(subtle nipples) is a challenging task even for experts
since in most cases the nipple is not clearly visible or
not visible at all. Therefore the performance of the
proposed method for detecting nipples for this case
was not good. We obtained good results in cases
where the nipple had a round shape and well-defined
edges (Fig. 3e). For cases where the shape of the nip-
ple was not round or its edges were not well-defined,
the Hough Circle Transform failed to accurately iden-
tify the nipple. The center could be slightly off. There
were also some cases where the nipple was not visible
at all (Fig. 3f).
4.2 Qualitative Results on Pectoral
Muscle Detection
The proposed method for pectoral muscle detection
was very accurate for PMBs that have well-defined
edges as in figure 4a. If dense breast tissue is present
around the border but the edge is still defined, the bor-
der can be still accurately identified (Fig. 4b). The
Automatic Computation of the Posterior Nipple Line from Mammographies
633
(a)
(b)
(c)
Figure 4: Examples of correctly located PMB. The detected edge is marked in blue and the fitted line to the detected edge is
marked in green.
(a)
(b)
(c)
Figure 5: Examples of falsely located PMB. The detected edge is marked in blue and the fitted line to the detected edge is
marked in green.
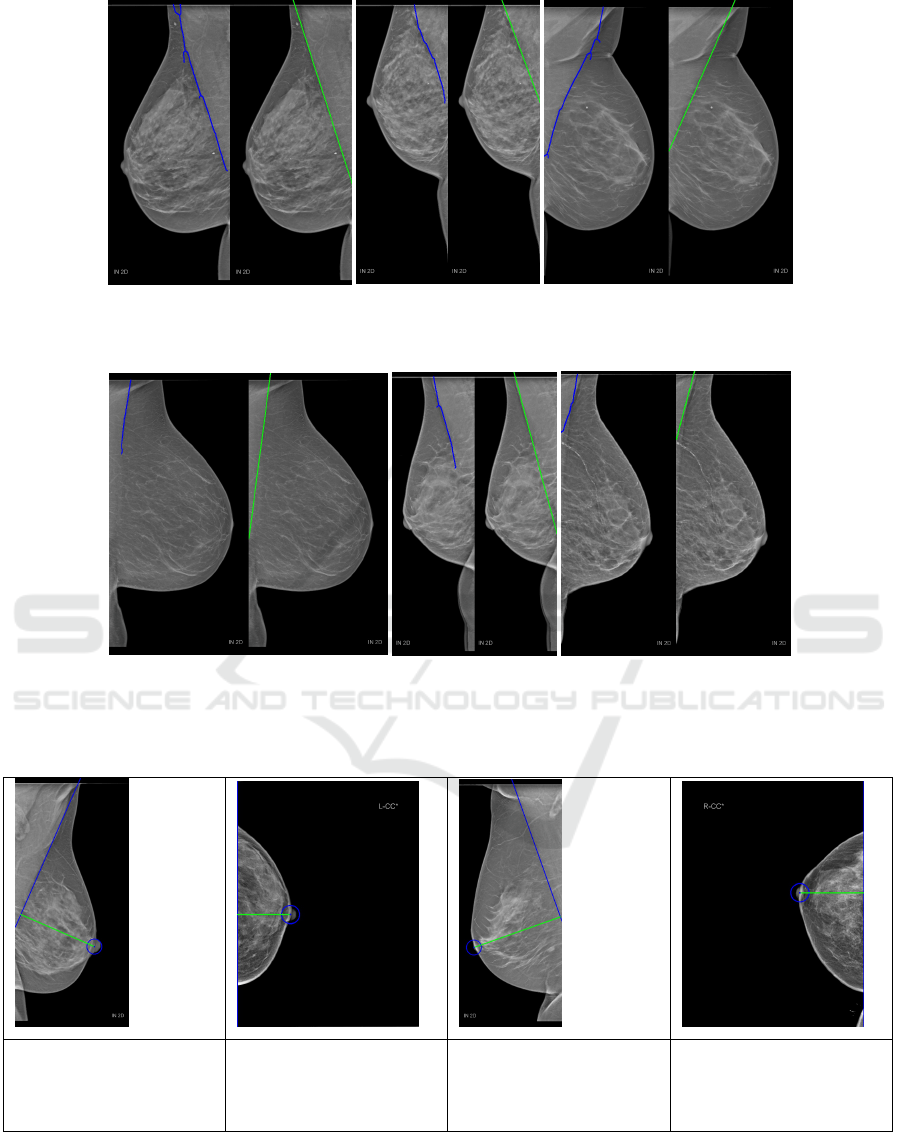
Table 1: Examples for computed PNL that were close to the expert values. PNL is marked in green. Values are in mm.
True PNL: 98.90 mm
Computed PNL: 99.72
mm
Difference: 0.82 mm
True PNL: 50.80 mm
Computed PNL: 54.11
mm
Difference: 3.31 mm
True PNL: 114.70 mm
Computed PNL: 114.30
mm
Difference: 0.40 mm
True PNL: 61.40 mm
Computed PNL: 65.15
mm
Difference: 3.75 mm
breast tissue here does not affect our method since it
will be removed during the inpainting step. In figure
4c the PMB was accurately located even with a occur-
ring fold close to the border. Since the fold has high-
intensity pixels it was removed during the inpainting
step and therefore was not falsely detected as an edge.
There were a few cases where our method failed
to correctly locate the PMB. An example is shown
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
634
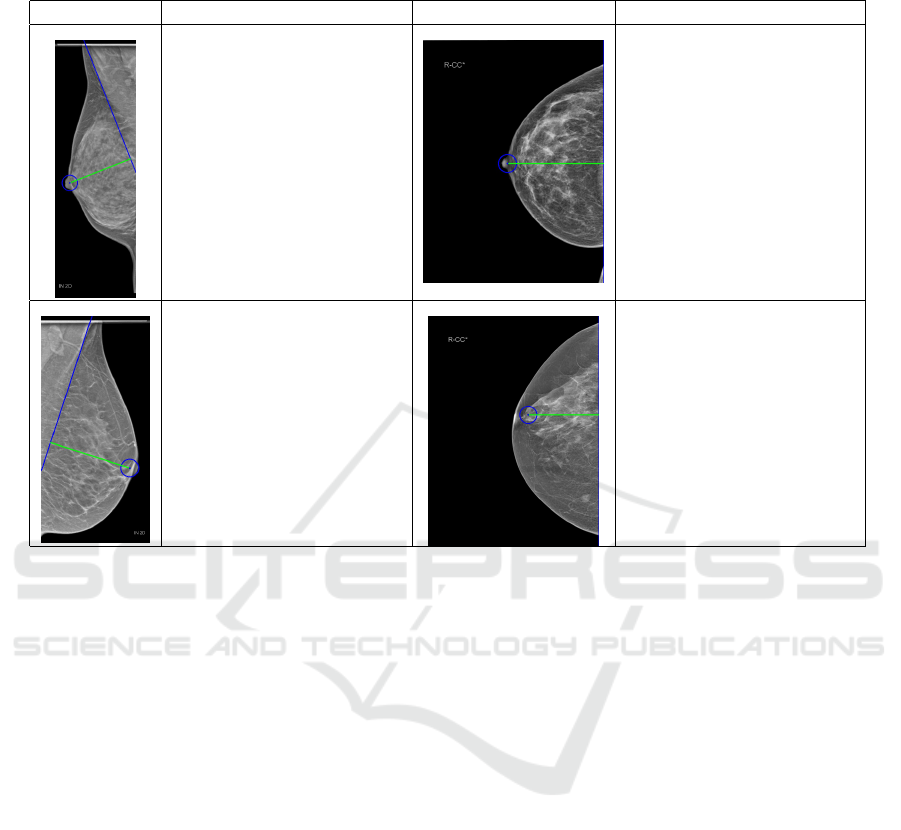
Table 2: Examples for computed PNL that were off when compared to the expert’s. PNL is marked in green. Values are in
mm.
Examples Analysis Examples Analysis
True PNL: 92.00 mm
Computed PNL: 78.41 mm
Difference: 13.59 mm
Here the computed PNL
length is shorter than the
true length. The pectoral
muscle is strongly curved
and therefore the located
boundary is off.
True PNL: 85.90 mm
Computed PNL: 98.61 mm
Difference: 12.71 mm
Here the computed PNL
length is longer than the
true length. The PNL was
measured from the nipple to
the right image border but
should have been measured
from the nipple to the pec-
toral muscle (not visible).
True PNL: 88.80 mm
Computed PNL: 86.56 mm
Difference: 2.24 mm
Here there is a small dif-
ference because the nipple
(in profile and large) was
not located accurately and is
slightly off.
True PNL: 84.80 mm
Computed PNL: 76.54 mm
Difference: 8.26 mm
Here the nipple (not in pro-
file) was falsely detected,
which led to a large differ-
ence between the actual and
computed PNL length.
in figure 5a, here the upper part of the PMB could
be accurately identified since a strong intensity edge
is present, whereas the lower part is strongly blurred
and could not be captured by the Gabor filters. In this
case, using only the upper part to determine the real
PMB did not suffice. The second example is similar
to the previous one except that the problem lies in the
fact that there was dense breast tissue. Therefore the
detected border was slightly off (Fig. 5b). Another
example of a failure can be seen in figure 5c, where
multiple edges that are close to each other are present
making it difficult to identify the real PMB. Even if
the PMB can be accurately detected, fitting the line to
the detected edge can still result in a falsely marked
PMB. This is only the case if the actual pectoral mus-
cle is strongly curved.
4.3 PNL Quantitative Results
For evaluating the performance of the proposed
method 100 images from the dataset were used. Im-
ages with breast implants, not visible nipples, or miss-
ing real PNL length (manually measured by an expert
from the Medical University of Innsbruck) were not
considered. The computed PNL length was compared
with the real PNL length. When using those 100 im-
ages, we obtained an absolute mean error of just 6.39
mm (stdev.= 4.62 mm). Since the location of both
the PMB and the nipple are needed for calculating
the PNL length, both of them need to be accurately
located, It would be enough for just one component
to be incorrectly detected to have a negative impact
on the overall performance. In table 1 four examples
(two for each view) with their results can be seen. The
nipples were accurately located in all of them, and the
boundaries of the pectoral muscle were also correctly
identified in the MLO views. Table 2 shows four ex-
amples of a few cases were results were worse than
expected along with an explanation on the causes.
5 CONCLUSIONS
This thesis presents a method to automatically com-
pute the length of the PNL in mammograms, which
can be divided into three parts: Pectoral Muscle De-
tection, Nipple Detection, and PNL Computation. For
the detection of the PMB, a set of three phase re-
sponses was computed by using three high-frequency
Gabor filters tuned at three different orientations. The
Laplacian was then computed for each phase response
and combined to detect the muscle boundary. For
the nipple detection method, the convex hull of the
nipple region was used to calculate the convexity de-
Automatic Computation of the Posterior Nipple Line from Mammographies
635

fects. Based on this, a distinction could be made be-
tween large and small nipples. Large nipples can be
located by using the two points with the largest devia-
tion (depth greater than 2.5 each) and small nipples
were located with the help of watershed segmenta-
tion. If no nipple could be found, it means that the
nipple does not lie on the breast boundary and for this
case, Hough Circle Transform was used. Once both
the PMB and nipple were detected, the PNL length
was calculated.
By applying the proposed method on 100 images
from the dataset provided by the Medical Univer-
sity of Innsbruck an absolute mean error of 6.39 mm
could be achieved. In cases where the nipple does not
lie along the breast boundary, the method performs
poorly, but otherwise, the accuracy is high, except for
some rare cases. The overall performance of the de-
tection of the PMB is very accurate, although there is
still room for improvement for some cases.
ETHICS APPROVAL
This study was approved by the ethics commission of
the Medical University of Innsbruck (reference num-
ber 1321/2021).
REFERENCES
Bertalmio, M., Sapiro, G., Caselles, V., and Ballester, C.
(2000). Image inpainting. In Proceedings of the 27th
Annual Conference on Computer Graphics and Interac-
tive Techniques, SIGGRAPH ’00, page 417424, USA.
Brahim, M., Westerkamp, K., Hempel, L., Lehmann, R.,
Hempel, D., and Philipp, P. (2022). Automated assess-
ment of breast positioning quality in screening mammog-
raphy. Cancers, 14(19).
Buades, A., Coll, B., and Morel, J. (2011). Non-Local
Means Denoising. IPOL, 1:208–212.
Deng, J., Dong, W., Socher, R., Li, L., Kai, L., and Li,
F. (2009). Imagenet: A large-scale hierarchical image
database. In CVPR 2009, pages 248–255.
Gupta, V., Taylor, C., Bonnet, S., Prevedello, L., Hawley,
J., White, R., Flores, M., and Erdal, B. (2020). Deep
learning-based automatic detection of poorly positioned
mammograms to minimize patient return visits for repeat
imaging: A real-world application.
Harris, C. and Stephens, M. (1988). A combined corner
and edge detector. In Proceedings of the Alvey Vision
Conference, pages 23.1–23.6. doi:10.5244/C.2.23.
Hejduk, P., Sexauer, R., Ruppert, C., Borkowski, K., Un-
kelbach, J., and Schmidt, N. (2023). Automatic and stan-
dardized quality assurance of digital mammography and
tomosynthesis with deep convolutional neural networks.
Insights into Imaging, 14(1):90.
Jiang, J., Zhang, Y., Lu, Y., Guo, Y., and Chen, H. (2019).
A radiomic-feature based nipple detection algorithm on
digital mammography. Medical Physics, 46.
Klabunde, C., Bouchard, F., Taplin, S., Scharpantgen, A.,
and Ballard-Barbash, R. (2001). Quality assurance for
screening mammography: an international comparison.
JECH, 55(3):204–212.
Meyer, F. (1992). Color image segmentation. In 1992 In-
ternational Conference on Image Processing and its Ap-
plications, pages 303–306.
Pizer, S., Amburn, E., Austin, J., Cromartie, R., Geselowitz,
A., Greer, T., ter Haar Romeny, B., Zimmerman, J., and
Zuiderveld, K. (1987). Adaptive histogram equalization
and its variations. CVGIP, 39(3):355–368.
Rahman, M. and Jha, R. (2022). Multidirectional gabor
filter-based approach for pectoral muscle boundary de-
tection. IEEE TRPMS, 6(4):433–445.
Sung, H., Ferlay, J., Siegel, R., Laversanne, M., Soerjo-
mataram, I., Jemal, A., and Bray, F. (2021). Global can-
cer statistics 2020: Globocan estimates of incidence and
mortality worldwide for 36 cancers in 185 countries. CA,
71(3):209–249.
Suzuki, S. and Keiichi, A. (1985). Topological structural
analysis of digitized binary images by border following.
CVGIP, 30(1):32–46.
Sweeney, R., Lewis, S., Hogg, P., and McEntee, M. (2017).
A review of mammographic positioning image quality
criteria for the craniocaudal projection. The British jour-
nal of radiology, 91 1082:20170611.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2015). Rethinking the inception architecture for
computer vision.
Waade, G., Skyrud, D., Holen, A., Larsen, M., Hanestad,
B., Hopland, N., Kalcheva, V., and Hofvind, S. (2021).
Assessment of breast positioning criteria in mammo-
graphic screening: Agreement between artificial intelli-
gence software and radiographers. Journal of Medical
Screening, 28:096914132199871.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
636
