
Utilizing Radiomic Features for Automated MRI Keypoint Detection:
Enhancing Graph Applications
Sahar Almahfouz Nasser
a
, Shashwat Pathak
b
, Keshav Singhal
c
, Mohit Meena
d
,
Nihar Gupte
e
, Ananya Chinmaya
f
, Prateek Garg
g
and Amit Sethi
h
Department of Electrical Engineering, Indian Institute of Technology Bombay, Mumbai, India
Keywords:
Image Matching, Image Registration, Kepoint Detection, Radiomic Features, Brain MRI, GNN.
Abstract:
Graph neural networks (GNNs) present a promising alternative to CNNs and transformers for certain image
processing applications due to their parameter-efficiency in modeling spatial relationships. Currently, an active
area of research is to convert image data into graph data as input for GNN-based models. A natural choice for
graph vertices, for instance, are keypoints in images. SuperRetina is a promising semi-supervised technique
for detecting keypoints in retinal images. However, its limitations lie in the dependency on a small initial
set of ground truth keypoints, which is progressively expanded to detect more keypoints. We encountered
difficulties in detecting a consistent set of initial keypoints in brain images using traditional keypoint detection
techniques, such as SIFT and LoFTR. Therefore, we propose a new approach for detecting the initial keypoints
for SuperRetina, which is based on radiomic features. We demonstrate the anatomical significance of the
detected keypoints by showcasing their efficacy in improving image registration guided by these keypoints.
We also employed these keypoints as ground truth for a modified keypoint detection method known as LK-
SuperRetina for improved image matching in terms of both the number of matches and their confidence scores.
1 INTRODUCTION
Graph neural networks (GNNs) have shown promis-
ing results for reducing the computational require-
ments for certain image processing tasks as shown
in (He et al., 2023). However, converting images into
graphs is an active area of research. One approach is
to break down an image into patches and treat each
patch as a node in a graph while another is to segment
an image into super-pixels and use each super-pixel
as a graph vertex (He et al., 2023). Our method relies
on detecting important keypoints in the images along
with their features and making graphs.
Detecting important keypoints in certain types of
images, such as magnetic resonance images of the
brain is not straightforward due to the lack of well-
a
https://orcid.org/0000-0002-5063-9211
b
https://orcid.org/0000-0002-4508-8531
c
https://orcid.org/0009-0003-2290-4924
d
https://orcid.org/0009-0008-1641-2727
e
https://orcid.org/0009-0004-5972-2088
f
https://orcid.org/0009-0008-5844-5498
g
https://orcid.org/0009-0005-5348-8905
h
https://orcid.org/0000-0002-8634-1804
defined landmarks. In various registration competi-
tions, such as the BraTSReg challenge (Baheti et al.,
2021), experts have had to mark specific landmark
points on 3D brain images to help the participants
align images and evaluate how well their registration
algorithms worked. But this marking is neither easy
nor fast. Also, only a few points – between 6 and 50
per set of images – get marked, which can be inade-
quate for good registration.
Traditional keypoint detection algorithms such as
SIFT (Lindeberg, 2012) fall behind deep learning-
based keypoint detection algorithms which have dif-
ferent types include: supervised, unsupervised, and
semi-supervised methods. Some examples are Un-
superPoint (Christiansen et al., 2019), SuperPoint
(DeTone et al., 2018), GLAMpoints (Truong et al.,
2019), and SuperRetina (Liu et al., 2022).
After trying and failing to find reliable keypoints
in brain images using methods such as SIFT (Linde-
berg, 2012) and LoFTR (Sun et al., 2021), we pro-
posed a new algorithm. Our proposed method finds
keypoints and extract their descriptors using radiomic
features in brain MR images. We assessed the util-
ity of these keypoints in brain images by using them
for image registration. Then we used a dataset that
Nasser, S., Pathak, S., Singhal, K., Meena, M., Gupte, N., Chinmaya, A., Garg, P. and Sethi, A.
Utilizing Radiomic Features for Automated MRI Keypoint Detection: Enhancing Graph Applications.
DOI: 10.5220/0012568800003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 319-325
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
319

we prepared, which consists of MRI images from
OASIS dataset (Marcus et al., 2007) and the key-
points we detected, to train the LK-SuperRetina al-
gorithm (Almahfouz Nasser et al., 2023) to detect
new keypoints. We showed that using a GNN, such
as SuperGlue (Sarlin et al., 2020), for image match-
ing improves the features of detected keypoints. This
method of detecting keypoints opens doors for using
graph-based neural networks for different tasks, in-
cluding brain classification, segmentation, and regis-
tration.
Our contribution can be outlined as follows:
• Introduction of an innovative, fully automated ap-
proach for keypoint detection in brain MRI im-
ages.
• Development of a two-stage method: the first
stage involves detecting initial keypoints by uti-
lizing segmentations of regions of interest in the
brain, employing radiomic features as keypoint
descriptors, and establishing keypoints at the cen-
ters of mass of the segmentations. The sec-
ond stage employs our previously developed algo-
rithm, LK-SuperRetina, gradually increasing the
number of detected keypoints, commencing from
the initial set.
• Enhancement of various tasks, including im-
age registration and image matching, achieved
through the incorporation of detected keypoints.
2 RELATED WORK
Traditional approaches to keypoint detection have
long been prominent in the field of computer vision,
with a focus on identifying keypoints in images that
remain consistent across scaling, rotation, and light-
ing variations. These techniques involve character-
izing the local image patch around these keypoints
using a set of features, allowing for the matching of
keypoints between different images and object recog-
nition. However, these methods come with their limi-
tations, including high computational complexity, re-
duced accuracy in the presence of extreme lighting
and viewpoint changes, and challenges in handling
occlusions and cluttered backgrounds.
In recent years, deep learning-based keypoint de-
tection algorithms have emerged as a promising al-
ternative. These algorithms have the ability to au-
tonomously learn robust and discriminative features
directly from data, making them better suited to han-
dle complex and diverse image variations. As a result,
they have found application in various domains, in-
cluding object detection, semantic segmentation, and
image retrieval.
Within the realm of deep learning, keypoint de-
tection algorithms come in various forms, including
supervised, semi-supervised, self-supervised, and un-
supervised techniques. Supervised techniques require
annotated data, where keypoints are manually labeled
in training images, proving advantageous in scenar-
ios with a substantial volume of labeled data, such
as facial recognition or object detection. Conversely,
unsupervised techniques operate independently of la-
beled data, with the network learning to identify key-
points by maximizing specific objectives, like infor-
mation preservation during feature extraction. These
methods are particularly valuable in contexts where
obtaining labeled data is challenging or expensive,
such as in medical imaging or remote sensing.
Prominent deep learning-based keypoint detection
methods include UnsuperPoint, SuperPoint, GLAM-
points, and SuperRetina. UnsuperPoint (Christiansen
et al., 2019) introduces an innovative unsupervised
training approach, utilizing a combination of differ-
entiable soft nearest neighbor loss and unsupervised
clustering loss. SuperPoint (DeTone et al., 2018),
on the other hand, represents a self-supervised deep
learning-based algorithm for keypoint detection and
description, relying on a novel loss function for train-
ing on unlabeled images, which makes it adaptable
and scalable across various applications.
The primary distinction between UnsuperPoint
and SuperPoint lies in their training methodologies.
While SuperPoint follows a self-supervised approach,
UnsuperPoint takes an unsupervised path. Addition-
ally, UnsuperPoint achieves state-of-the-art perfor-
mance across various benchmarks, even outperform-
ing SuperPoint in challenging scenarios characterized
by significant viewpoint alterations and illumination
shifts. GLAMpoints (Truong et al., 2019) emerges as
a semi-supervised deep learning-based algorithm for
interest point detection and description, employing a
unique greedy training strategy for end-to-end learn-
ing of keypoint detection and description. This strat-
egy involves learning to select the most precise key-
points and their descriptors, resulting in higher accu-
racy and efficiency, especially in demanding scenarios
involving significant viewpoint changes, scaling, ro-
tation, and benchmarks like HPatches (Balntas et al.,
2017). Furthermore, GLAMpoints is well-suited for
accommodating multiple object instances within the
same image, making it suitable for multi-object track-
ing and matching. SuperRetina (Liu et al., 2022) rep-
resents a semi-supervised approach for keypoint de-
tection and description in retinal images, leveraging
both labeled and unlabeled data to enhance the per-
formance of the keypoint detector and descriptor.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
320

In the following section, we introduce our pro-
posed approach for detecting keypoints in brain MRI
images. As we will demonstrate, this task proves to be
highly challenging, and traditional keypoint detection
methods like SIFT, as well as well-known deep learn-
ing techniques such as LoFTR, struggle to identify ro-
bust and repeatable keypoints. Our solution involves
leveraging radiomic features for initial keypoint de-
tection in the brain. Subsequently, we employ LK-
SuperRetina to increase the number of identified key-
points in the images. Finally, we showcase the effec-
tiveness of these keypoints in applications like image
matching and registration, highlighting their positive
impact on the overall performance of these tasks.
3 PROPOSED METHOD
We now introduce our approach for keypoint detec-
tion using radiomic features. Given an image along-
side its segmentation labels, our method identifies ra-
diomic keypoints as the centers of radiomic segmen-
tation labels within the image. These radiomic fea-
tures encompass a range of intensity characteristics
(such as mean intensity, standard deviation, skew-
ness and kurtosis), shape characteristics (such as vol-
ume, surface area, and compactness), texture char-
acteristics (such as gray level co-occurrence matrix
(GLCM), gray level run length matrix (GLRLM), and
gray level run length matrix)), wavelet-Based charac-
teristics, spatial and statistical characteristics (such as
centroid position, eccentricity, and entropy), and frac-
tal characteristics. These radiomic features are spe-
cific to the regions defined by the segmentation map.
To compute these features, we utilized the Pyradiomic
library (Van Griethuysen et al., 2017).
Radiomic keypoints are closely tied to segmenta-
tion regions predicted by neural networks. We trained
Swin UNetR (Hatamizadeh et al., 2021) to predict the
segmentation maps of brain images and used them as
masks to extract the keypoints. Each segmentation
mask yields a keypoint location, accompanied by 53
descriptive radiomic features, see figures 1 and 2 for
more details. Radiomic keypoints exhibit repeatabil-
ity across various brain samples even in the presence
of varying intensity and non-rigid deformations which
make our proposed method a more robust alternative
to methods, such as SIFT (Lindeberg, 2012). These
detected keypoints can serve as a initial keypoints for
detecting additional keypoints through deep learning
techniques as we will show in our results, in which
we trained LK-SuperRetina (Almahfouz Nasser et al.,
2023) to automatically detect these ground truth key-
points and extra keypoints. This underscores the sig-
nificance of these keypoints as essential landmarks
within brain images.
4 EXPERIMENTS AND RESULTS
This section begins with an introduction to the dataset
used in all our experiments. Following that, we ex-
plore various approaches applied for keypoint detec-
tion, explaining how the detected keypoints were uti-
lized to improve registration and identify additional
keypoints. Finally, we will conclude with our results
on GNN-based image matching.
4.1 OASIS Dataset
The OASIS dataset, as described in (Marcus et al.,
2007), contains MRI data obtained from 414 subjects.
This dataset has been divided into three separate sub-
sets for training, validation, and testing, following a
ratio of 314:50:50, respectively. Each subject in the
dataset has T1-weighted scan, as well as segmentation
masks of various regions of the brain. This dataset
incorporates three distinct types of brain segmenta-
tion: a four-label mask, a thirty-five label mask, and
a twenty-four label mask.The 3D T1-weighted scans
and their corresponding masks are of resolution (160,
192, 224). Additionally, the 2D T1-weighted scans
and their corresponding masks have a resolution of
(160, 192).
4.2 SIFT
SIFT (Lindeberg, 2012) was initially chosen for ex-
perimentation due to its reputation in keypoint detec-
tion. Utilizing gradients, SIFT excels in identifying
scale and rotation invariant keypoints. The method
calculates orientation gradients across scales, creat-
ing a distinctive 128-dimensional vector for each key-
point.
Figure 3 shows significant issues we encountered
with keypoint detection using SIFT. Notably, there
was inconsistency in keypoint locations across differ-
ent MRI slices within the brain, reflecting a lack of
repeatability. Our experiments highlighted SIFT’s in-
effectiveness under conditions involving large defor-
mations.
4.3 LoFTR
Due to SIFT’s inability to establish correspondences,
we opted to explore a deep learning method namely
LoFTR (Sun et al., 2021), which stands for detector-
free local feature matching transformer.
Utilizing Radiomic Features for Automated MRI Keypoint Detection: Enhancing Graph Applications
321
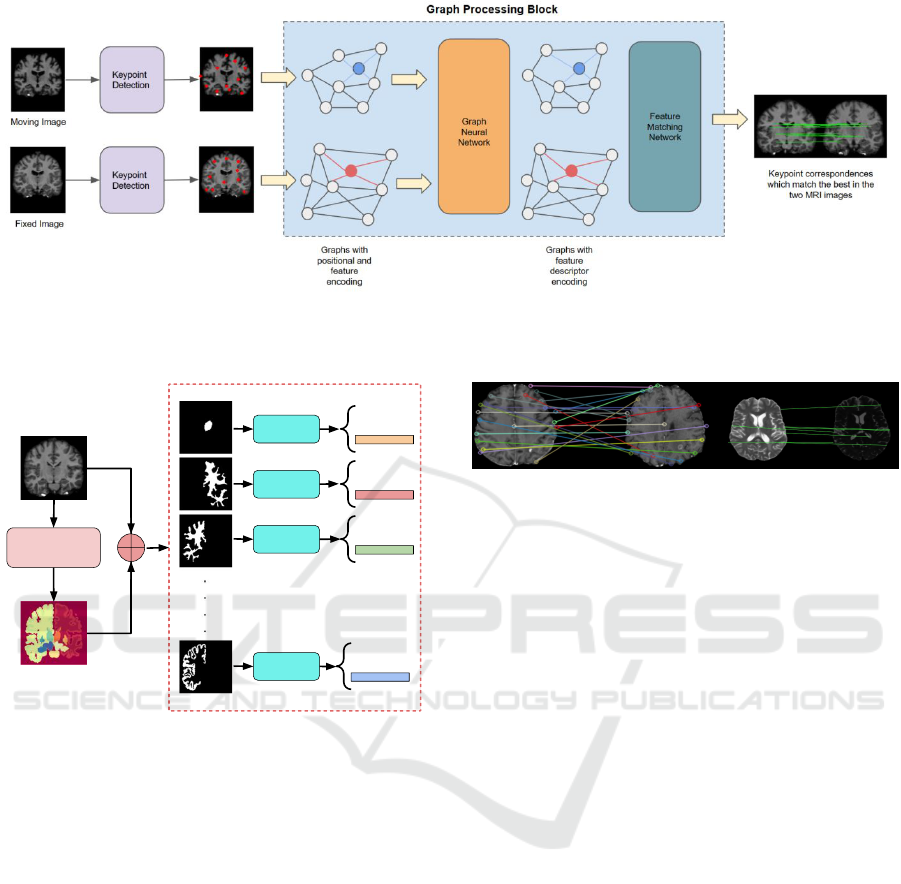
Figure 1: The image matching pipeline, encompassing keypoint detection using neural networks, graph formation from
detected keypoints, graph neural network (GNN) processing to enhance keypoint features, and a dedicated head for keypoint
matching.
Segmentation
Network
Radiomic
Block
Radiomic
Block
Radiomic
Block
Radiomic
Block
.
.
.
.
x1,y1,z1
Features
x2,y2,z2
Features
x3,y3,z3
Features
xn,yn,zn
Features
Input Image
Segmentation
Output
Figure 2: Radiomic features-based Keypoint detection
method.
In the LoFTR approach, a convolutional neural
network (CNN) with an encoder-decoder architecture
extracts both low-resolution and high-level features.
The LoFTR module, incorporating self-attention and
cross-attention blocks, transforms these features, and
a differential matching layer offers two methods: op-
timal transport and dual softmax (Bridle, 1989) .
Our implementation used a pretrained model from
the Aachen Day Night Dataset (Sattler et al., 2018).
LoFTR succeeded in generating keypoint matches for
similar intensity profiles but faced challenges in other
cases similar to SIFT.These findings emphasize the
need for keypoints exhibiting consistency, accurate
matching, and intensity profile invariance across the
dataset.
Figure 3: SIFT (Lindeberg, 2012) and LoFTR (Sun et al.,
2021) performances in keypoint detection. The left pair of
images depicts results for SIFT, while the right one illus-
trates the performance of LoFTR. It is evident that LoFTR
outperforms SIFT in keypoint detection and matching. Nev-
ertheless, LoFTR encounters challenges when the source
and target images exhibit varying intensity distributions.
4.4 Radiomic Features-Based Keypoint
Detection
Having faced challenges with keypoint detection us-
ing SIFT and LoFTR, which proved sensitive to inten-
sity variations, we shifted to our proposed radiomic
features-based method. In this section, we prove the
importance of the detected keypoints by showing our
results on registering brain images among different
subjects. we also showcase the applications of the
dataset containing the original OASIS scans and the
corresponding detected radiomic keypoints on auto-
matic keypoint detection. And finally train a GNN-
based image matcher on the same dataset.
4.4.1 Image Registration
To assess the significance of radiomics keypoints as
landmarks, we integrated them into the loss function
of a registration network (TransMorph (Chen et al.,
2022)). Our findings revealed that incorporating the
keypoints’ loss led to a notable 3% enhancement in
the registration performance.
Vision transformers, excelling in capturing long-
range spatial relationships, prove effective in medical
image tasks due to their large receptive fields. Trans-
Morph (Chen et al., 2022), a hybrid Transformer-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
322

ConvNet model, utilizes these advantages for volu-
metric medical image registration. The encoder di-
vides input volumes into 3D patches, projecting them
to feature representations through linear layers. Se-
quential patch merging and Swin Transformer blocks
follow. The decoder, with upsampling and convolu-
tional layers, connects to the encoder stages via skip
connections, producing the deformation field. We
contributed by designing a customized loss function
for keypoints, utilizing Gaussian-blurred keypoints to
create a ground truth heatmap. Combining Dice and
inverted Dice losses addressed imbalanced masks, re-
sulting in a 3% Dice score improvement over the OA-
SIS test dataset. TransMorph achieved a dice score
of 0.89 with keypoint loss, compared to 0.86 with-
out, underscoring keypoints’ role in enhancing reg-
istration performance. A potential avenue for future
research involves developing a loss function that con-
siders both the feature descriptors of keypoints and
the disparity between the locations of registered key-
points and their counterparts in the target image.
4.4.2 Automated Keypoint Detection
SuperRetina, introduced in (Liu et al., 2022) is an ap-
daptive version of SuperPoint model (DeTone et al.,
2018) for identifying important keypoints in retinal
images. Utilizing a semi-supervised learning frame-
work, SuperRetina maximizes the utility of limited
labeled retinal image data by combining both super-
vised and unsupervised techniques. Yet, its utilization
requires an initial set of ground truth keypoints to ini-
tiate the process, subsequently increasing the detected
keypoints iteratively. In our approach, we use our ra-
diomic keypoints as the initial sets for OASIS images.
LK-SuperRetina (Almahfouz Nasser et al., 2023)
which is a modified version of SuperRetina consists
of an encoder for downsampling, along with two de-
coders—one for keypoint detection and another for
descriptor generation. Keypoint detection utilizes a
mix of labeled and unlabeled data, while descriptor
training employs self-supervised learning.
Following the U-Net (Ronneberger et al., 2015)
design, LK-SuperRetina’s shallow encoder begins
with a single convolutional layer, followed by three
blocks containing two convolutional layers, a 2 × 2
max-pooling layer, and ReLU activation. The key-
point decoder has three blocks with two convolutional
layers, ReLU activation, and concatenation block.
The detection map (P) is generated through a convo-
lutional block with three convolutional layers and a
sigmoid activation.
The loss function combines the detector and the
descriptor losses. The detection loss consists of the
classification loss and the geometric loss as shown in
Equation 1.
l
det
= l
cl f
+ l
geo
(1)
The classification loss component (l
cl f
) is defined
in Equation 2, where
˜
Y represents the smoothed ver-
sion of the binary ground truth labels Y of the key-
points after blurring them with a 2D Gaussian.
l
cl f
(I;Y ) = 1 −
2.
∑
i, j
(P ◦
˜
Y )
i, j
∑
i, j
(P ◦ P)
i, j
+
∑
i, j
(
˜
Y ◦
˜
Y )
i, j
(2)
Where ◦ denotes element-wise multiplication.
The detector generates a heatmap as its output.
Coordinates where the intensity value exceeds a spec-
ified threshold (subject to non-maximum suppression)
are regarded as the keypoint coordinates. When feed-
ing both the image I and its augmented version I
′
to
the network, two tensors for the descriptors D and
D
′
are obtained. For each keypoint (i, j) in the non-
maximum suppressed keypoint set
˜
P, two distances
are computed: Φ
rand
i, j
between the descriptors of (i, j)
in the set
˜
P and a random point from the registered
heatmap H(
˜
P), and Φ
hard
i, j
representing the minimal
distance, as depicted in Equation 3.
l
des
(I, H) =
∑
(i, j)∈
˜
P
max
0, m + Φ
i, j
−
1
2
Φ
rand
i, j
+ Φ
hard
i, j
(3)
For more in-depth information on the loss func-
tion, please refer to the SuperRetina paper (Liu et al.,
2022). Figure 4 shows the results obtained from
LK-SuperRetina. As demonstrated, the number of
additionally identified keypoints meets expectations,
showcasing the network’s proficiency in capturing
good new keypoints. The network successfully de-
tects both the ground truth keypoints and extra key-
points during the testing phase.
Figure 4 presents two instances demonstrating the
resilience of the keypoint detection model against de-
formations. The images were deformed randomly by
an affine deformation. We passed the original image
(target) and the deformed image (reference) seper-
ately to LK-SuperRetina. The model successfully
identified corresponding keypoints in both images, as
indicated by the number of good matches. We ad-
justed the thresholds of LK-SuperRetina to detect a
smaller set of keypoints for the clarity of the visual-
ization, but in practice, the model can detect over 300
good keypoints.
4.4.3 Image Matching
Following the detection of keypoints within the brain
images, we proceed to construct graphs to be used as
Utilizing Radiomic Features for Automated MRI Keypoint Detection: Enhancing Graph Applications
323

Table 1: A comparison between brute force matcher and SuperGlue. SuperGlue outperforms the brute force matcher in terms
of both evaluation metrics: the average number of good matches and the average confidence score across the entire test dataset.
Method Avg. No. Good Matches Confidence Score
BF (Lowe, 2004) 7 0.449 ± 0.007
SuperGlue 20 0.988 ± 0.010
Figure 4: Two examples show the robustness of the key-
point detection model. In both rows, the sequence of im-
ages, from left to right, includes: the reference image, the
target image, and the registration output. The good matches
contribute to aligning the images effectively.
inputs of the GNN, for accomplishing specific tasks
such as matching in our study. Within this paper,
we demonstrate our success in training a GNN-based
matcher (SuperGlue (Sarlin et al., 2020)) using the
graphs formed from the detected keypoints.
SuperGlue designed for matching two sets of lo-
cal features by identifying correspondences and fil-
tering non-matchable points. Using attention-based
graph neural networks, it integrates context aggrega-
tion, matching, and filtering within a unified architec-
ture. SuperGlue employs self-attention to enhance the
receptive field of local descriptors and cross-attention
for cross-image communication. The network han-
dles partial assignments and occluded points by solv-
ing an optimal transport problem. With superior per-
formance over other learned approaches, SuperGlue
achieves state-of-the-art results in pose estimation for
challenging real-world indoor and outdoor environ-
ments.
Figure 5 and Table 1 show a performance compar-
ison between the brute force matcher and SuperGlue
across the test dataset. SuperGlue enhances the fea-
tures of the detected keypoints which improves the
matching performance. The data presented in the ta-
ble suggests that exploring the application of GNNs
for keypoint matching is a promising endeavor. To
pursue this, it is essential to identify keypoints in im-
ages and format the data to serve as appropriate input
for GNNs. Having introduced a technique for detect-
ing significant keypoints in brain MRI images, these
keypoints can now function as nodes in graphs, pro-
viding data for GNNs. This development paves the
way for a multitude of graph-based applications in the
analysis of brain MRI images.
CV2 BF Matcher - Correct Matches = 7, Incorrect = 4, Failed = 9
CV2 BF Matcher - Correct Matches = 4, Incorrect = 6, Failed = 10 GNN Matcher - Good Matches = 20, Failed = 0, Avg. Confidence = 0.9526
GNN Matcher - Good Matches = 20, Failed = 0, Avg. Confidence = 0.9932
—— 0.90
—— 0.80
—— 0.70
—— 0.55
—— 0.40
—— 0.25
—— 0.10
Avg. Confidence Score Colour Spectrum
Figure 5: A comparison of the SuperGlue and brute force
matcher performance in matching detected keypoints on
brain images.
5 CONCLUSION
To sum up, our radiomic keypoint detection algorithm
provides a solution for automated keypoint detection
in MRI scans, overcoming challenges encountered by
traditional and other deep learning methods. The lim-
ited set of radiomic keypoints facilitates training Su-
perRetina for increased keypoint detection. These
keypoints are consistent and deformation resilient.
Our approach paves the way for the application of
GNN-based models on brain images, offering a faster
and more parameter-efficient alternative compared to
CNNs and transformers. Moreover, the detection of
keypoints contributes to various tasks, including reg-
istration as justified in this work.
The limitation of this study is the necessity for
segmenting regions of interest to acquire radiomic
features, which are subsequently utilized as the initial
keypoints for the keypoint detection algorithm, LK-
SuperRetina.
REFERENCES
Almahfouz Nasser, S., Gupte, N., and Sethi, A. (2023). Re-
verse knowledge distillation: Training a large model
using a small one for retinal image matching on lim-
ited data. arXiv e-prints, pages arXiv–2307.
Baheti, B., Waldmannstetter, D., Chakrabarty, S., Akbari,
H., Bilello, M., Wiestler, B., Schwarting, J., Cal-
abrese, E., Rudie, J., Abidi, S., et al. (2021). The brain
tumor sequence registration challenge: establishing
correspondence between pre-operative and follow-up
mri scans of diffuse glioma patients. arXiv preprint
arXiv:2112.06979.
Balntas, V., Lenc, K., Vedaldi, A., and Mikolajczyk, K.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
324

(2017). Hpatches: A benchmark and evaluation of
handcrafted and learned local descriptors. In Proceed-
ings of the IEEE conference on computer vision and
pattern recognition, pages 5173–5182.
Bridle, J. (1989). Training stochastic model recognition al-
gorithms as networks can lead to maximum mutual in-
formation estimation of parameters. Advances in neu-
ral information processing systems, 2.
Chen, J., Frey, E. C., He, Y., Segars, W. P., Li, Y., and Du,
Y. (2022). Transmorph: Transformer for unsupervised
medical image registration. Medical image analysis,
82:102615.
Christiansen, P. H., Kragh, M. F., Brodskiy, Y., and
Karstoft, H. (2019). Unsuperpoint: End-to-end unsu-
pervised interest point detector and descriptor. arXiv
preprint arXiv:1907.04011.
DeTone, D., Malisiewicz, T., and Rabinovich, A. (2018).
Superpoint: Self-supervised interest point detection
and description. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition
workshops, pages 224–236.
Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. R.,
and Xu, D. (2021). Swin unetr: Swin transformers
for semantic segmentation of brain tumors in mri im-
ages. In International MICCAI Brainlesion Workshop,
pages 272–284. Springer.
He, X., Hooi, B., Laurent, T., Perold, A., LeCun, Y., and
Bresson, X. (2023). A generalization of vit/mlp-mixer
to graphs. In International Conference on Machine
Learning, pages 12724–12745. PMLR.
Lindeberg, T. (2012). Scale invariant feature transform.
Liu, J., Li, X., Wei, Q., Xu, J., and Ding, D. (2022). Semi-
supervised keypoint detector and descriptor for retinal
image matching. In Computer Vision–ECCV 2022:
17th European Conference, Tel Aviv, Israel, October
23–27, 2022, Proceedings, Part XXI, pages 593–609.
Springer.
Lowe, D. G. (2004). Distinctive image features from scale-
invariant keypoints. International journal of computer
vision, 60:91–110.
Marcus, D. S., Wang, T. H., Parker, J., Csernansky, J. G.,
Morris, J. C., and Buckner, R. L. (2007). Open ac-
cess series of imaging studies (oasis): cross-sectional
mri data in young, middle aged, nondemented, and
demented older adults. Journal of cognitive neuro-
science, 19(9):1498–1507.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18, pages 234–241.
Springer.
Sarlin, P.-E., DeTone, D., Malisiewicz, T., and Rabinovich,
A. (2020). Superglue: Learning feature matching
with graph neural networks. In Proceedings of the
IEEE/CVF conference on computer vision and pattern
recognition, pages 4938–4947.
Sattler, T., Maddern, W., Toft, C., Torii, A., Hammarstrand,
L., Stenborg, E., Safari, D., Okutomi, M., Pollefeys,
M., Sivic, J., et al. (2018). Benchmarking 6dof out-
door visual localization in changing conditions. In
Proceedings of the IEEE conference on computer vi-
sion and pattern recognition, pages 8601–8610.
Sun, J., Shen, Z., Wang, Y., Bao, H., and Zhou, X. (2021).
Loftr: Detector-free local feature matching with trans-
formers. In Proceedings of the IEEE/CVF conference
on computer vision and pattern recognition, pages
8922–8931.
Truong, P., Apostolopoulos, S., Mosinska, A., Stucky, S.,
Ciller, C., and Zanet, S. D. (2019). Glampoints:
Greedily learned accurate match points. In Proceed-
ings of the IEEE/CVF International Conference on
Computer Vision, pages 10732–10741.
Van Griethuysen, J. J., Fedorov, A., Parmar, C., Hosny, A.,
Aucoin, N., Narayan, V., Beets-Tan, R. G., Fillion-
Robin, J.-C., Pieper, S., and Aerts, H. J. (2017).
Computational radiomics system to decode the radio-
graphic phenotype. Cancer research, 77(21):e104–
e107.
Utilizing Radiomic Features for Automated MRI Keypoint Detection: Enhancing Graph Applications
325
