
Fine-Tuning of Conditional Transformers Improves the Generation of
Functionally Characterized Proteins
Marco Nicolini
1 a
, Dario Malchiodi
1 b
, Alberto Cabri
1 c
, Emanuele Cavalleri
1 d
,
Marco Mesiti
1 e
, Alberto Paccanaro
5 f
, Peter N. Robinson
3 g
, Justin Reese
4 h
,
Elena Casiraghi
1,2 i
and Giorgio Valentini
1,2 j
1
AnacletoLab, Dept. of Computer Science, University of Milan, Italy
2
ELLIS European Laboratory for Learning and Intelligent Systems
3
Berlin Institute of Health at Charit
´
e (BIH), Germany
4
Environmental Genomics and Systems Biology Bioscience, Lawrence Berkeley National Laboratory, U.S.A.
5
School of Applied Mathematics (EMAp) - FGV, Rio de Janeiro, Brazil
Keywords:
Large Language Models, Protein Language Models, Conditional Transformers, Protein Design and Modeling.
Abstract:
Conditional transformers improve the generative capabilities of large language models (LLMs) by process-
ing specific control tags able to drive the generation of texts characterized by specific features. Recently, a
similar approach has been applied to the generation of functionally characterized proteins by adding specific
tags to the protein sequence to qualify their functions (e.g., Gene Ontology terms) or other characteristics
(e.g., their family or the species which they belong to). In this work, we show that fine tuning conditional
transformers, pre-trained on large corpora of proteins, on specific protein families can significantly enhance
the prediction accuracy of the pre-trained models and can also generate new potentially functional proteins
that could enlarge the protein space explored by the natural evolution. We obtained encouraging results on the
phage lysozyme family of proteins, achieving statistically significant better prediction results than the original
pre-trained model. The comparative analysis of the primary and tertiary structure of the synthetic proteins
generated by our model with the natural ones shows that the resulting fine-tuned model is able to generate bi-
ologically plausible proteins. Our results confirm and suggest that fine-tuned conditional transformers can be
applied to other functionally characterized proteins for possible industrial and pharmacological applications.
1 INTRODUCTION
Recent years witnessed remarkable developments in
natural language models, greatly enhancing capabil-
ities in natural language processing (NLP) and ma-
chine translation. Particularly noteworthy are genera-
tive models, which excel in creating text that is both
structurally and semantically coherent (Bommasani
a
https://orcid.org/0009-0008-5137-2361
b
https://orcid.org/0000-0002-7574-697X
c
https://orcid.org/0000-0003-1373-8402
d
https://orcid.org/0000-0003-1973-5712
e
https://orcid.org/0000-0001-5701-0080
f
https://orcid.org/0000-0001-8059-1346
g
https://orcid.org/0000-0002-0736-9199
h
https://orcid.org/0000-0002-2170-2250
i
https://orcid.org/0000-0003-2024-7572
j
https://orcid.org/0000-0002-5694-3919
et al., 2021). A key milestone in this field was the
introduction of the transformer architecture (Vaswani
et al., 2017), which constitutes a foundational ele-
ment for many advanced language models, including
the widely recognized BERT (Devlin et al., 2019) and
GPT (Brown et al., 2020; OpenAI, 2023) models.
One of the key advantages of transformers in NLP
is their ability to learn representations that capture
both syntactic (grammatical arrangement of words)
and semantic (meaning of words) information. The
self-attention mechanism enables the model to weigh
the importance of different words or tokens in the in-
put sequence, considering their contextual relation-
ships. This attention-based approach has shown re-
markable performance in tasks such as machine trans-
lation, sentiment analysis, text summarization, and
question-answering (Wolf et al., 2020). Transform-
ers models are pre-trained on vast amounts of textual
Nicolini, M., Malchiodi, D., Cabri, A., Cavalleri, E., Mesiti, M., Paccanaro, A., Robinson, P., Reese, J., Casiraghi, E. and Valentini, G.
Fine-Tuning of Conditional Transformers Improves the Generation of Functionally Characterized Proteins.
DOI: 10.5220/0012567900003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 561-568
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
561

data, enabling them to learn rich linguistic patterns
and structures using self-supervised learning tech-
niques. The pre-training phase involves predicting
masked tokens or next tokens in a sequence, enabling
the model to acquire knowledge about syntax, gram-
mar, and semantic relationships.
The scope of large language models (LLMs) goes
well beyond linguistic applications, as exemplified by
their use in protein modeling, indicating their expan-
sive potential and transformative role in scientific in-
quiry (Valentini et al., 2023). Indeed, both text and
proteins rely on a vocabulary. In text, words serve
as the basic units of meaning, forming an alphabet of
sorts. Similarly, proteins are encoded by amino acids,
which can be viewed as an alphabet of building blocks
to be combined to create diverse protein sequences.
In text, phrases are sequences of words, whereas, in
the realm of molecules, proteins are sequences of
amino acids. Just as different phrases convey differ-
ent ideas or sentiments, different protein sequences
result in unique molecular structures. The relation-
ship between meaning and structure can be observed
in both text and proteins. In text, the meaning of a
sentence arises from the arrangement and interaction
of words. Similarly, in proteins, patterns, domains,
and more in general the structure of the molecule de-
termines its function and meaning within a biological
context. The folding and arrangement of amino acids
in a protein sequence contribute to its structural prop-
erties, which in turn govern its functional characteris-
tics.
Several protein language models have been re-
cently proposed to model and generate proteins, by
training transformers on large corpora of proteins
from public databases (Ferruz and H
¨
ocker, 2022).
Several works showed that fine tuning pre-trained
language models, by using relatively small well-
focused data, enhance their predictive and generative
power (Devlin et al., 2019). Moreover, conditional
transformer architectures, enabling the use of key-
words to direct the generation of specific types of text
(Keskar et al., 2019), recently paved the way to sim-
ilar models for the generation of functionally charac-
terized protein sequences (Madani et al., 2023).
In this work, we show that by combining a pre-
trained conditional transformer and transfer learning
we can fine tune a model to boost the generation
of specific functionally characterized protein fami-
lies. This is of paramount importance for the auto-
matic generation of proteins for specific applications
in pharmacology and precision medicine (Moor et al.,
2023).
2 PROTEIN GENERATIVE
MODELS
During the last decade, advancements in protein
generative models revolutionized protein engineer-
ing (Ferruz et al., 2022a). By leveraging machine
learning techniques, these models offer new opportu-
nities to design proteins with desired properties, over-
coming the limitations of traditional methods (Valen-
tini et al., 2023).
In recent years, deep neural networks, specifically
generative architectures, have emerged as promis-
ing tools for protein science and engineering (Shin
et al., 2021; Jumper et al., 2021; Ferruz et al., 2022b;
Das et al., 2021; Kilinc et al., 2023). These mod-
els, such as attention-based models trained on pro-
tein sequences, have shown remarkable success in
classification and generation tasks relevant to artifi-
cial intelligence-driven protein design. They have the
potential to learn complex representations and effec-
tively utilize vast amounts of unaligned protein se-
quence data from public databases such as Pfam and
UniProt (The UniProt Consortium, 2022).
Protein language models (PLMs) offer a robust
framework for learning from extensive collections of
amino acid sequences within various protein fami-
lies, facilitating the generation of diverse and realistic
protein sequences. These language models leverage
the power of natural language processing techniques
to comprehend and extract meaningful patterns from
vast sequences of data. ProGen (Madani et al., 2023),
ProteinGPT2 (Ferruz et al., 2022b), and IgLM (Shuai
et al., 2022) are all PLMs decoder-only models devel-
oped in the last few years. It is noteworthy that both
ProGen and IgLM generate sequences conditioned
on prefix(es) at the start of the sentences, providing
additional constraints during the generation process.
IgLM is specifically trained for unpaired antibody se-
quence modeling.
By employing PLMs, researchers can generate
protein sequences that exhibit well-folded structures,
despite their divergence in sequence space. This ca-
pability is achieved by capturing relationships and
dependencies within the sequence data. To tailor
PLMs for specific protein families of interest, a fine-
tuning approach can be adopted, where the models are
trained on a subset of relevant proteins. This targeted
training allows the PLMs to learn the specific char-
acteristics associated with the desired protein family,
enhancing the quality and specificity of the generated
sequences.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
562

24/09/23, 19:15Untitled Diagram.drawio
Page 1 of 1https://app.diagrams.net/
Desired Arguments
Homo Sapiens
Acting-Binding
Cytoplasm
Cardiac disease
YMIQEE
Organism:
Function:
Location:
Process:
Amino Acid:
Inferred Result
Function
Structure
Controlled Sequence Generation
ProGen
Generated Sequences
YMIQEE + FZEWDR...
YMIQEE + FWDECK...
YMIQEE + EWDRLL...
YMIQEE + XZEGNL...
Figure 1: An example of ProGen protein generation using multiple control sequences and an amino acid prefix (in desired
argument box). The generation process can create multiple output sequences with the same Input (controlled sequence gener-
ation box). For each generated sequence, it is possible to compute its structure with tools like AlphaFold-2.
3 THE PROGEN MODEL
ProGen (Madani et al., 2023) uses a LLM to gener-
ate novel protein sequences that are not present in any
database; specifically, it implements and uses the con-
ditional transformer architecture (CTRL) proposed in
(Keskar et al., 2019) that relies on the use of keywords
to guide the generation of texts. Instead of training
the whole model from scratch, the weights in ProGen
were initialized to those of a trained CTRL model.
Examples of ProGen protein generations are shown
in Figure 1.
Madani et al. proposed protein engineering as a
self-supervised sequence generation problem. Us-
ing 280 million protein sequences, the authors trained
ProGen with 1.2 billion parameters.
ProGen processes not only the sequence of amino
acids x = (x
1
, . . . , x
n
), where each x
i
represents an
amino acid, but also includes functional tags during
training. More precisely, its input prefixes one or
more functional tags t to the sequence x of amino
acids. The functional tags represent a protein family
or a Gene Ontology term, or whatever property of the
protein. The objective of conditional protein language
modeling is to acquire knowledge about the probabil-
ity distribution p(x) given a functional tag t. Given
that (t, x) represents a sequence of amino acids pre-
fixed by a functional tag, using an approach similar
to (Bengio et al., 2000), it is reasonable to factorize
the conditional probability p(x|t) using the chain rule
of probability:
p(x|t) =
n
∏
i=1
p(x
i
|x
<i
,t) , (1)
in which p(x
i
|x
<i
,t) denotes the conditional probabil-
ity of x
i
given all the preceding elements x
1
, . . . , x
i−1
and the functional tag t.
This decomposes protein language modeling into
next-amino-acid prediction. Hence we can train a
deep neural network with parameters θ to minimize
the negative log-likelihood over a dataset of |D| se-
quences D = {(t, x)
k=1
, . . . , (t, x)
k=|D|
}:
L (D) = −
|D|
∑
k=1
|x
k
|
∑
i=1
log p
θ
(x
k
i
|x
k
<i
,t
k
) , (2)
The functional tag provides a point of control over
the generation process, and it constraints the protein
generation toward proteins having a specific property
t
k
.
Since protein language models acquire knowl-
edge about the conditional probability distribution
p
θ
(x
i
|x
<i
,t), it is possible to generate a new sequence
˜x of length m that is obtained by sequentially sam-
pling its constituent symbols: p
θ
(x
0
|t), p
θ
(x
1
| ˜x
0
,t),
. . . , p
θ
(x
m
| ˜x
<m
,t).
The overall architecture of ProGen is borrowed
from CTRL. The model has internal embedding di-
mension d = 1028, inner dimension f = 512, 36 lay-
ers, and 8 heads per layer. Dropout with probability
0.1 follows the residual connections in each layer. To-
ken embeddings are tied with the embeddings of the
final output layer.
4 FINE-TUNING OF THE
PRE-TRAINED PROGEN
MODEL
The objective is to harness the knowledge ProGen
has acquired from millions of sequences, and trans-
fer its “learned knowledge” (represented by the model
weights) to the task of generating a specific family of
proteins. We selected the family of phage lysozymes,
Fine-Tuning of Conditional Transformers Improves the Generation of Functionally Characterized Proteins
563
i.e., enzymes that can act as anti-microbials through
the hydrolysis of the peptidoglycan component of the
cell wall.
To specialize the model on phage lysozyme data
we downloaded 19473 sequences from the Pfam API
(hosted by InterPro) (Mistry et al., 2020). We ran-
domly split the data in a test set (2000 sequences,
about 10% of the total data), and in a training set
(17473 sequences, roughly 90% of the data) for fine-
tuning the model. The average sequence length of the
phage lysozyme dataset is 201.6 amino acids.
During the learning process, since proteins are in-
variant to the temporal notion of sequence generation,
each amino acid sequence has a certain probability of
being flipped, allowing the model to receive both the
direct sequence and its reverse. Additionally, the in-
put sequence for fine-tuning may sometimes (with a
certain probability) lose its keyword that represents
the phage family, to allow for generation also without
an initial keyword. In total, a training sequence can
be transformed into four distinct training inputs: the
sequence with its family keyword, its reverse with the
keyword, the sequence without the family keyword,
and its reverse without the keyword.
The fine-tuning process involved different param-
eters, described below.
Flip Probability. A feature was introduced to ran-
domly flip the amino acid sequence with a 0.2 prob-
ability. In other words, there’s a 20% chance that the
sequence will be read from the end to the beginning,
acting as a data augmentation technique.
Omitted Keyword Probability. We introduced a
probability with which the phage family keyword can
be dropped from the sequence. Specifically, the over-
all probability of dropping the keyword while us-
ing different transformation objects for data loading
stands at 0.13, similarly to the implementation of
Madani et al.
Maximum Sequence Length for Training. The
sequence length was limited to 512 tokens, includ-
ing the keywords. This is because, during the initial
training phase, the model was not trained to generate
inputs longer than 512 tokens, due to inherent limita-
tions in the model size and architecture.
Adam Optimizer. The optimization algorithm that
computes adaptive learning rates for each param-
eter used is Adam (Adaptive Moment Estima-
tion) (Kingma and Ba, 2014).
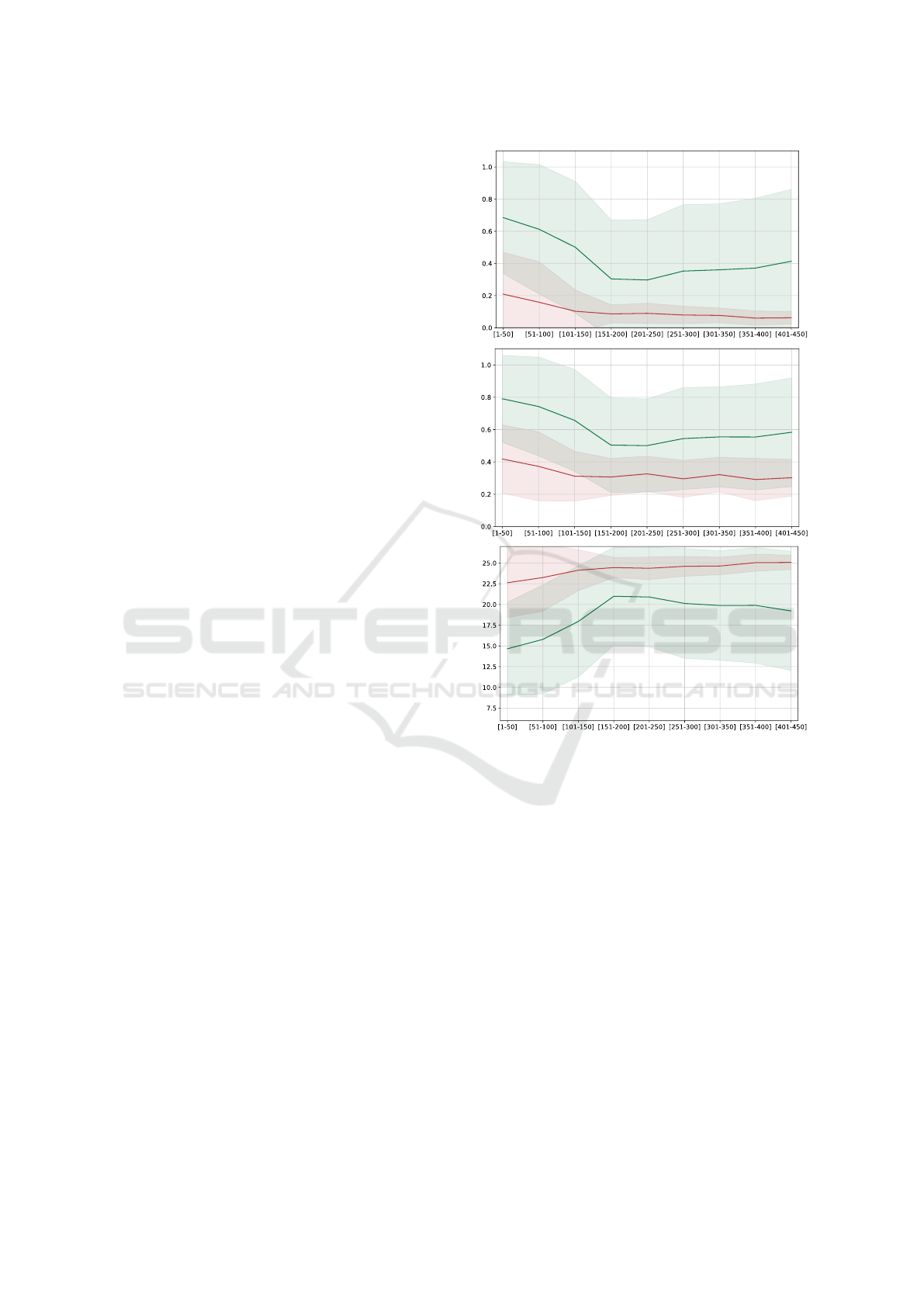
Figure 2: Accuracy (top), soft accuracy (middle) and per-
plexity (bottom) comparison of the fine-tuned and general
ProGen models on the phage lysozyme family (PF00959).
Results are computed on different ranges of 50 amino
acids with standard deviation represented by shaded re-
gions. Fine-tuned and general model results are in green
and red, respectively.
Learning Rate. The learning rate determines the
step size at each iteration while moving towards a
minimum of the loss function. In our experiments, we
tested two distinct learning rates: 0.0001 and 0.001.
Batch Size and Epochs. The batch size, set at 2,
represents the number of training examples utilized
in one iteration. A smaller batch size often provides
a regularizing effect and lower generalization error.
The training process was conducted over 4 epochs,
meaning the entire dataset was passed forward and
backward through the model four times.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
564

Gradient Norm Clipping. The gradients were
clipped with a norm of 0.25, to avoid the “exploding
gradient” issue that afflicts deep neural networks.
Warmup Iteration. This parameter, set at 100 in
our experiments, defines the number of iterations over
which the learning rate will be gradually increased.
The code for the experiments is available from
https://github.com/AnacletoLAB/ProGen. We
used PyTorch libraries and data from InterPro and
UniProt. For training and testing the models we used
two multi-processor servers equipped with 128 GB of
RAM and NVIDIA A100 GPU accelerators.
4.1 Testing the Fine-Tuned Model on
Phage Lysozymes
In this section, we evaluate the performance of the
ProGen model that has been fine-tuned for the phage
lysozyme family (PF00959). Classification here in-
volves predicting the next amino acid in a sequence
based on the current sequence input.
Figure 2 shows that the fine-tuned model sig-
nificantly outperforms the general model (Wilcoxon
rank-sum test α = 0.01) on the test set. Results are
averaged across the 2000 phage lysozyme proteins
of the test set, with the top-k parameter fixed to 1
(i.e., we predicted the amino acid with the highest
predicted probability), repetition penalty set to 0 and
keywords usage. Data are tested up to input length
512.
4.2 Generating New Functionally
Characterized Proteins
In this section, we show the results of the application
of our fine-tuned model to the generation of synthetic
protein sequences with functional characteristics that
closely resemble those of natural proteins.
We considered two generation processes: a) gen-
eration from scratch using only the input keyword;
b) prefix generation, i.e., generation from an initial
sequence of amino acids. For this second genera-
tion process we selected three phage lysozymes in-
volved in the degradation of peptidoglycans and in
the programmed host cell lysis: RddD (UniProt entry
P78285), P1 (UniProt entry Q37875), and T4 (Uni-
Prot entry P00720). We started the generation from
the 25, 50, and 75% amino acid position for each pro-
tein.
To assess the quality of the generated proteins we
compared:
1. the primary structure (sequence) of the generated
proteins versus the natural ones of the family of
the phage lysozymes;
2. the phylogenetic relationships of generated se-
quences versus the natural ones;
3. the tertiary (three-dimensional) structure of the
generated sequences versus the natural ones.
4.2.1 Comparison by Sequence Alignment
We initially compared the sequences newly gener-
ated by our fine-tuned model with those of the phage
lysozyme family. More precisely, we searched for
similar natural proteins by using NCBI BLAST+ inte-
grated into Galaxy to estimate similarity scores (Cock
et al., 2015). BLAST searches were conducted versus
the whole phage lysozyme dataset.
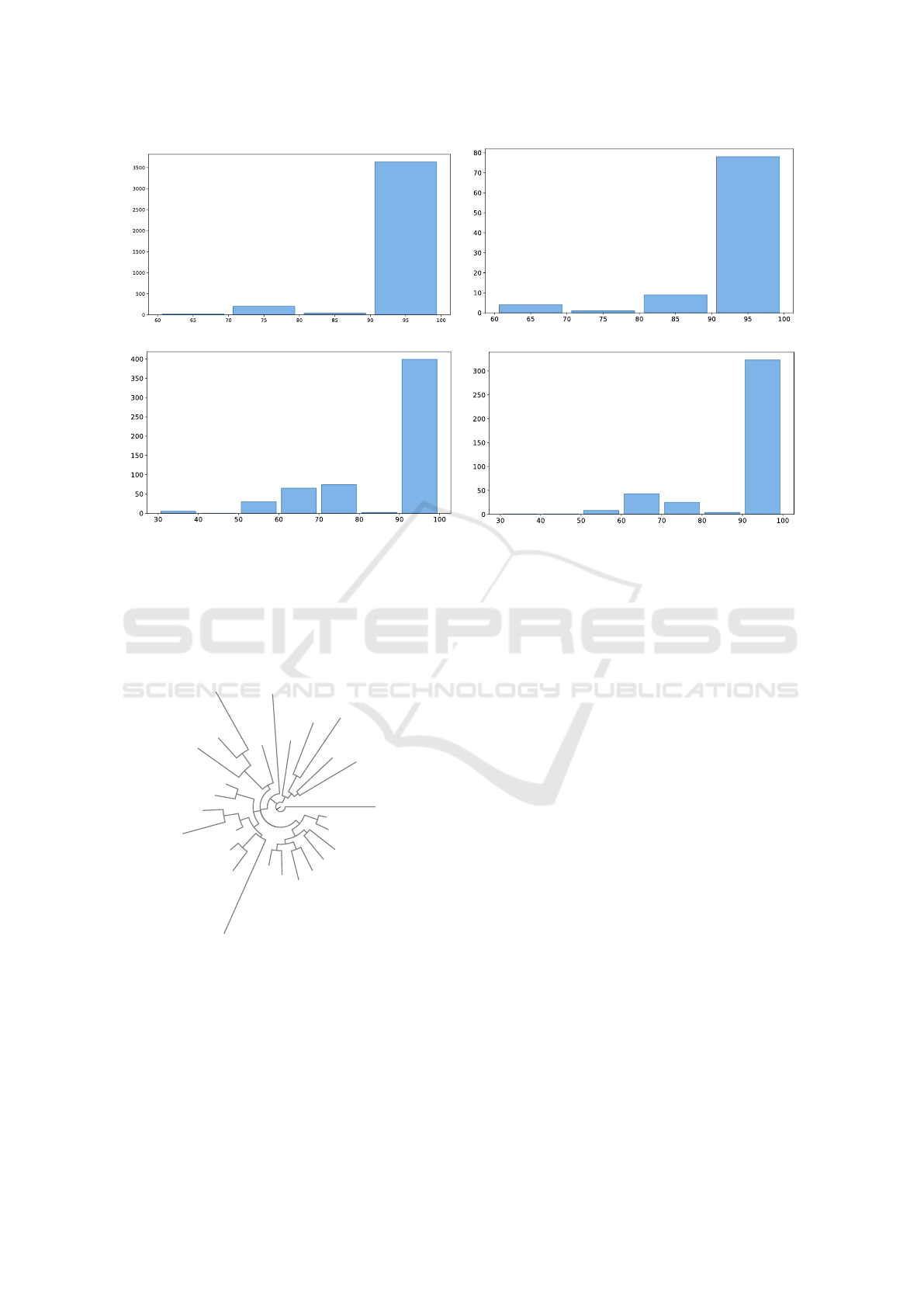
Figure 3 shows that most of the sequences gener-
ated from scratch or starting from a prefix sequence
have a relevant sequence similarity with respect to
the natural proteins belonging to the phage lysozyme
family. Nevertheless, several newly generated se-
quences show only a partial similarity, showing that
our model can explore protein spaces unexplored by
the natural evolution.
4.2.2 Phylogenetic Tree Construction
We initially used CD-HIT (Fu et al., 2012) to cluster
protein sequences generated by our model, in order
to reduce redundancy in the generated sequence data
(several generated sequences are very similar, data not
shown) and to optimize the subsequent multiple align-
ment analysis and phylogenetic tree construction. To
this end we employed CLUSTAL-W (Larkin et al.,
2007) for the multiple alignment of the centroid se-
quences found by CD-HIT, and FAST-TREE (Price
et al., 2009), a maximum likelihood algorithm de-
signed to construct phylogenetic trees using a multi-
ple alignment of the sequences. This analysis allows
us to discern the evolutionary relationships within the
group of representative sequences generated by our
fine-tuned model. Figure 4 shows the evolutionary
relationships found by FAST-TREE between Q37875
and the new proteins generated by our model start-
ing from half of protein Q37875 itself. Only the
newly generated sequences considered representative
by CD-HIT are shown.
4.2.3 Tertiary Structure Comparison
We finally conducted a comparison of the tertiary
structure of the generated proteins versus those of
the family of natural phage lysozymes. To this end,
we compared the three-dimensional structure of the
Fine-Tuning of Conditional Transformers Improves the Generation of Functionally Characterized Proteins
565
(a) (b)
(c) (d)
Figure 3: Evaluation of the sequence similarity between proteins generated by the fine-tuned model and proteins from the
phage lysozyme family. Histograms display the distribution of BLAST Max-ID for data generated from the fine-tuned model
versus the phage lysozyme family. (a) Generation from scratch using only the lysozyme keyword as input. (b) Generation
from half of the P78285 protein. (c) Generation from half of the P00720 protein. (d) Generation from half of the Q37875
protein.
13/12/2023, 22:48
Galaxy59-%5BNewick_Display_on_data_28__Tree_Graph%5D.svg
file:///Users/nicorota/Desktop/Galaxy59-%5BNewick_Display_on_data_28__Tree_Graph%5D.svg
1/1
Sequence 160
Sequence 293
Sequence 376
Sequence 89
Sequence 299
Sequence 109
Sequence 55
Sequence 181
Sequence 12
Sequence 388
Sequence 85
Sequence 99
Sequence 199
Sequence 329
Sequence 207
Sequence 144
Sequence 277
Sequence 7
Sequence 172
Sequence 43
Sequence 93
Original
Sequence 149
Sequence 209
Sequence 255
Sequence 230
Sequence 426
0 2 4 6
Figure 4: Circular phylogenetic tree representing the evolu-
tionary relationships among protein sequences generated by
our model from Q37875 (the original sequence is the root).
In the tree the 27 sequences selected as representative by
CD-HIT are shown. Each branch of the tree corresponds
to a sequence, with the length of the branch indicating the
degree of divergence from the ancestral sequence. The tree
provides a graphical representation of the sequence similar-
ity and evolutionary distance between the sequences.
newly generated proteins (obtained by AlphaFold-
2 (Jumper et al., 2021)) with that of the reference
lysozyme (obtained from the X-ray crystallography
protein folding of the swissProt database).
We conducted a comparative analysis by aligning
several of the AlphaFold-2 generated protein struc-
tures, represented as PDB files, with the correspond-
ing structures of the original molecules available in
the UniProt database. To achieve this, we employed
PyMOL (Schr
¨
odinger, LLC, 2023), a molecular visu-
alization and analysis tool used in structural biology.
PyMOL supports the alignment of protein structures,
enabling a detailed examination of the similarities and
differences between the predicted structures and their
experimentally determined counterparts.
More precisely, after being folded, the structures
were compared and aligned with the original natu-
ral protein structures from which they were generated
using PyMOL. In addition to these alignments, we
also calculated the alignment with the tertiary struc-
ture of the natural phage lysozyme with the highest
match (if the structure was available in UniProt and if
a match was found). This step was performed for the
sequences 144 of Q37875, and 59 and 78 of P78285
generated by our model. The resulting Root Mean
Square Deviation (RMSD) values from these pro-
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
566
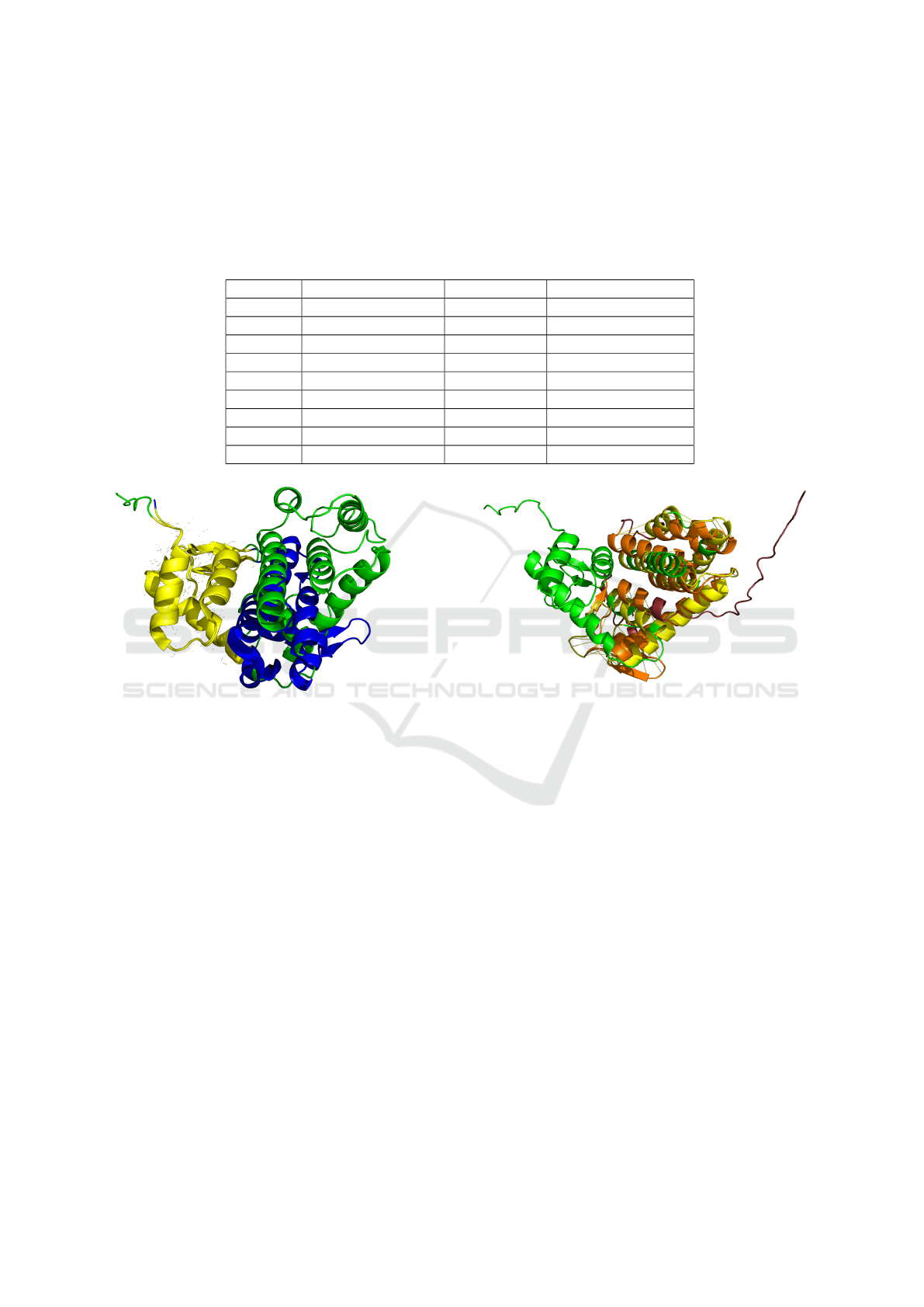
Table 1: RMSD values from PyMOL three-dimensional alignments, using selected sequences generated by the fine-tuned
ProGen and folded using AlphaFold-2. The alignments compare these sequences with the structures of the natural proteins
obtained from X-ray crystallography or AlphaFold-2 predictions. The natural proteins used are: a) proteins used as prefixes
in the model, and b) Max-ID matches found by BLAST (identified by the lines marked with the symbol “†”). The “Protein”
column lists the protein identifiers, “Original pdb ID” indicates the file identifier in UniProt of the 3D structure among with the
method used to obtain the 3D structure inside the brackets (“X-ray” stands for X-ray crystallography or “AF” for AlphaFold-2
predictions), “Generated ID” refers to the sequence number generated for the related batch with the fine-tuned model, and
“RMSD (Atoms)” contains the RMSD value along with the number of atoms used for the alignment.
Protein Original pdb ID Generated ID RMSD (atoms)
P78285 4ZPU (X-ray) 59 20.545 (2043 atoms)
P78285† A0A854AIC3 (AF) 59 0.642 (812 atoms)
P78285 4ZPU (X-ray) 78 11.356 (1491 atoms)
P78285† A0A3Y2C086 (AF) 78 0.276 (491 atoms)
P00720 102L (X-ray) 186 1.324 (581 atoms)
P00720 102L (X-ray) 636 2.598 (665 atoms)
Q37875 1XJT (X-ray) 144 0.506 (525 atoms)
Q37875† A0A2G6EIY6 (AF) 144 7.544 (537 atoms)
Q37875 1XJT (X-ray) 209 1.020 (598 atoms)
Figure 5: Alignment of the selected protein from Q37875
ProGen generation (sequence identifier 144) with 1XJT (X-
ray), i.e., the three-dimensional structure of Q37875 taken
from X-ray crystallography. Perfect alignments are in yel-
low, while the backbone of the protein generated by our
model (in green) is superimposed on the experimental X-ray
crystallography structure (in blue), illustrating the degree of
similarity and differences in the folding patterns.
cesses are listed in Table 1, which quantifies the align-
ment quality. The lower the RMSD value, the closer
the generated structure is to the compared protein.
Figure 5 visualizes the 3D alignment of sequence 144,
which was generated from half of the Q37875 pro-
tein, with the three-dimensional structure of Q37875
obtained from X-ray crystallography. This alignment
shows the similarity and differences in the folding
patterns between the generated and original struc-
tures. Additionally, Figure 6 shows the alignment be-
tween sequence 144 and its closest phage lysozyme
match in nature, identified as A0A2G6EIY6.
Figure 6: Alignment of the selected protein from Q37875
ProGen generation (sequence identifier 144) with its best
match found by BLAST in the phage lysozyme dataset, with
identifier A0A2G6EIY6. The three-dimensional structure
of A0A2G6EIY6 was taken from AlphaFold-2 prediction
(taken form uniProt). In the visualization, the alignment
parts of 144 are highlighted in yellow, and the aligned parts
of A0A2G6EIY6 are shown in orange. The backbone of the
protein generated by our model (in green) is superimposed
on the experimental predicted structure of A0A2G6EIY6
(in ruby), illustrating the degree of similarity and differ-
ences in the folding patterns.
5 CONCLUSIONS
We showed that fine-tuning significantly improves the
capabilities of a pre-trained LLM model for protein
generation on specific specialized tasks. The fine-
tuned generative model is able to design new se-
quences that diverge from their natural counterparts
while retaining potential functionality. Additionally,
incorporating control tags related to the protein family
enhances our ability to design novel protein functions
with more refined control. These developments rep-
Fine-Tuning of Conditional Transformers Improves the Generation of Functionally Characterized Proteins
567

resent a significant step towards the goal of custom-
designed proteins well-focused on specific functions.
We outline that fine-tuning the ProGen Condi-
tional Transformer toward specific protein families
can enable the generation of new proteins that retain
and can also expand their functional characteristics,
with possible relevant applications in pharmacology
(e.g., for the design of new anti-microbic drugs), or
in industrial applications (e.g., for the production of
textiles, biofuels or foods).
In perspective, ProGen model can be fine-tuned on
more complex tasks. For instance, the generation of
functionally characterized protein molecules that can
interact with a specific molecular target (i.e., a target
protein). This is a challenging task, but it represents
our next objective.
ACKNOWLEDGEMENTS
This research was supported by the “National
Center for Gene Therapy and Drugs based on
RNA Technology”, PNRR-NextGenerationEU pro-
gram [G43C22001320007].
REFERENCES
Bengio, Y., Ducharme, R., and Vincent, P. (2000). A neu-
ral probabilistic language model. Advances in neural
information processing systems, 13.
Bommasani, R. et al. (2021). On the opportunities and risks
of foundation models. ArXiv, abs/2108.07258.
Brown, T. et al. (2020). Language models are few-shot
learners. Advances in neural information processing
systems, 33:1877–1901.
Cock, P., Chilton, J., Gr
¨
uning, B., Johnson, J., and Soranzo,
N. (2015). Ncbi blast+ integrated into galaxy. Giga-
science, 4(1):s13742–015.
Das, P. et al. (2021). Accelerated antimicrobial discov-
ery via deep generative models and molecular dy-
namics simulations. Nature Biomedical Engineering,
5(6):613–623.
Devlin, J., Chang, M., Lee, K., and Toutanova, K. (2019).
BERT: Pre-training of deep bidirectional transformers
for language understanding. In Proc. of the 2019 Con-
ference of the North American Chapter of the ACL,
Volume 1 (Long and Short Papers), pages 4171–4186.
ACL.
Ferruz, N., Heinzinger, M., Akdel, M., Goncearenco, A.,
Naef, L., and Dallago, C. (2022a). From sequence to
function through structure: deep learning for protein
design. Computational and Structural Biotechnology
Journal.
Ferruz, N. and H
¨
ocker, B. (2022). Controllable protein de-
sign with language models. Nature Machine Intelli-
gence, 4(6):521–532.
Ferruz, N., Schmidt, S., and H
¨
ocker, B. (2022b). Protgpt2
is a deep unsupervised language model for protein de-
sign. Nature communications, 13(1):4348.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). Cd-
hit: accelerated for clustering the next-generation se-
quencing data. Bioinformatics, 28(23):3150–3152.
Jumper, J. et al. (2021). Highly accurate protein structure
prediction with alphafold. Nature, 596(7873):583–
589.
Keskar, N., McCann, B., Varshney, L., Xiong, C., and
Socher, R. (2019). CTRL: A conditional transformer
language model for controllable generation. arXiv
preprint arXiv:1909.05858.
Kilinc, M., Jia, K., and Jernigan, R. (2023). Improved
global protein homolog detection with major gains in
function identification. Proceedings of the National
Academy of Sciences, 120(9):e2211823120.
Kingma, D. and Ba, J. (2014). Adam: A method for
stochastic optimization. CoRR, abs/1412.6980.
Larkin, M. et al. (2007). Clustal w and clustal x version 2.0.
Bioinformatics, 23(21):2947–2948.
Madani, A. et al. (2023). Large language models generate
functional protein sequences across diverse families.
Nature Biotechnology, pages 1–8.
Mistry, J. et al. (2020). Pfam: The protein families database
in 2021. Nucleic Acids Research, 49(D1):D412–
D419.
Moor, M., Banerjee, O., Shakeri, Z., Krumholz, H.,
Leskovec, J., Topol, E., and Rajpurkar, P. (2023).
Foundation models for generalist medical artificial in-
telligence. Nature, 616:259–265.
OpenAI (2023). GPT-4 Technical Report. arXiv.
Price, M., Dehal, P., and Arkin, A. (2009). Fasttree: com-
puting large minimum evolution trees with profiles in-
stead of a distance matrix. Molecular biology and evo-
lution, 26(7):1641–1650.
Schr
¨
odinger, LLC (2023). The PyMOL molecular graphics
system, version 2.5.
Shin, J. et al. (2021). Protein design and variant prediction
using autoregressive generative models. Nature com-
munications, 12(1):2403.
Shuai, R., Ruffolo, J., and Gray, J. (2022). Generative lan-
guage modeling for antibody design. bioRxiv.
The UniProt Consortium (2022). UniProt: the Universal
Protein Knowledgebase in 2023. Nucleic Acids Re-
search, 51(D1):D523–D531.
Valentini, G., Malchiodi, D., Gliozzo, J., Mesiti, M., Soto-
Gomez, M., Cabri, A., Reese, J., Casiraghi, E., and
Robinson, P. (2023). The promises of large language
models for protein design and modeling. Frontiers in
Bioinformatics, 3:1304099.
Vaswani, A. et al. (2017). Attention is all you need. Ad-
vances in neural information processing systems, 30.
Wolf, T. et al. (2020). Transformers: State-of-the-art natural
language processing. In Proc. of the 2020 conference
on empirical methods in Natural Language Process-
ing, pages 38–45.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
568
