
Colorectal Image Classification Using Randomized Neural Network
Descriptors
Jarbas Joaci de Mesquita S
´
a Junior
1 a
and Andr
´
e Ricardo Backes
2 b
1
Programa de P
´
os-Graduac¸
˜
ao em Engenharia El
´
etrica e de Computac¸
˜
ao, Brazil
2
Department of Computing, Federal University of S
˜
ao Carlos, Brazil
Keywords:
Randomized Neural Network, Colorectal Database, Texture Analysis.
Abstract:
Colorectal cancer is among the highest incident cancers in the world. A fundamental procedure to diagnose
it is the analysis of histological images acquired from a biopsy. Because of this, computer vision approaches
have been proposed to help human specialists in such a task. In order to contribute to this field of research,
this paper presents a novel way of analyzing colorectal images by using a very discriminative texture signature
based on weights of a randomized neural network. For this, we addressed an important multi-class problem
composed of eight types of tissues. The results were promising, surpassing the accuracies of many methods
present in the literature. Thus, this performance confirms that the randomized neural network signature is an
efficient tool for discriminating histological images from colorectal tissues.
1 INTRODUCTION
Colorectal cancer (CRC) remains among the most
prevalent types of cancer in the global population. It
is considered the third prevalent cancer type for esti-
mated new diagnoses and deaths in United States in
2023 (Siegel et al., 2023). There are several tests
to detect this disease (sigmoidoscopy, colonoscopy,
high-sensitive fecal occult blood test etc.). However,
it is fundamental to analyze histological images ac-
quired from a biopsy to diagnose CRC (Yoon et al.,
2019). These images reveal the complex structure of
the tumor, which is composed of several different tis-
sues, such as clonal tumor cells, stroma cells, necrotic
areas, among others (Kather et al., 2016).
Traditionally, these images are evaluated by hu-
man specialists. Over the years, computer vision ap-
proaches have been proposed to analyze histological
colorectal images in order to help the humans special-
ists with an extra opinion. Moreover, these computa-
tional methods have as an advantage to speed up the
diagnosis process and to measure attributes not de-
tected by the human eye. To cite recent instances,
the paper (Peyret et al., 2018) applies a multispec-
tral multiscale local binary pattern texture method to
extract signatures from colorectal biopsy images. In
a
https://orcid.org/0000-0003-3749-2590
b
https://orcid.org/0000-0002-7486-4253
(Ribeiro et al., 2019), authors use fractal dimension,
curvelet transforms, and co-occurrences matrices to
extract features from two colorectal image datasets.
The paper (dos Santos et al., 2018) combines sample
entropy, multiscale approaches and a fuzzy strategy
to classify histological colorectal images into benign
and malign groups.
This work aims to classify the histological col-
orectal database publicly released by the paper
(Kather et al., 2016) using a very discriminative tex-
ture analysis method based on randomized neural net-
works. To explain our work, this paper is organized as
follows: Section 2 briefly describes how to construct
a randomized neural network, and Section 3 explains
how to adapt this network to build a texture analysis
method. Section 4 shows the details of the database
used and describes the classification procedure. Sec-
tion 5 shows the obtained results and discusses them
in the light of the accuracies obtained by compared
methods. Finally, Section 6 presents some remarks
about this work.
2 RANDOMIZED NEURAL
NETWORK
Randomized neural networks (RNN) (Schmidt et al.,
1992; Pao and Takefuji, 1992; Pao et al., 1994; Huang
800
Sá Junior, J. and Backes, A.
Colorectal Image Classification Using Randomized Neural Network Descriptors.
DOI: 10.5220/0012507200003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 3: VISAPP, pages
800-805
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

et al., 2006), in their simplest architecture, are neural
networks composed of a single hidden layer, whose
weights are determined randomly according to a de-
termined probability distribution. These weights en-
able the hidden layer to project the input feature vec-
tors into another dimensional space aiming to sepa-
rate them more easily, according to Cover’s theorem
(Cover, 1965). The output weights, in turn, can be de-
termined using a closed-form solution whose inputs
are the feature vectors projected by the hidden layer
and the output label vectors.
To provide a brief mathematical explanation de-
scribing a simple variant of a randomized neural
network with bias in the output layer, let X =
[~x
1
, ~x
2
,..., ~x
N
] be a matrix composed of N input fea-
ture vectors, each one having p attributes. After in-
cluding −1 as the first feature of each ~x
i
(for bias), the
output of the hidden layer for all the input features in
X can be obtained by computing φ(WQ), where φ(·) is
a transfer function and W is a matrix of weights whose
dimensions are Q×(p +1). Let Z = [~z
1
,~z
2
,..., ~z
N
] be
a matrix of the feature vectors produced by the hid-
den layer, each one having Q attributes. As a final
step, this matrix Z, after the inclusion of −1 as the
first feature for each ~z
i
(for bias), can be used to build
the matrix of weights of the output neuron layer, as
follows
M = DZ
T
(ZZ
T
+ λI)
−1
, (1)
where D =
h
~
d
1
,
~
d
2
,...,
~
d
N
i
is a matrix of label
vectors, Z
T
(ZZ
T
)
−1
is the Moore-Penrose pseudo-
inverse (Moore, 1920; Penrose, 1955), and λI is the
term for Tikhonov regularization (Tikhonov, 1963;
Calvetti et al., 2000) (I is the identity matrix and λ
is a regularization parameter).
3 IMPROVED RANDOMIZED
NEURAL NETWORK
SIGNATURE
An improved version of the randomized neural sig-
nature (S
´
a Junior and Backes, 2016) is presented in
(S
´
a Junior and Backes, ) and used in this work. This
version proposes three different signatures as well as
their combinations. In the first signature
~
α(Q), for a
determined neighborhood in a window K × K, each
neighboring pixel is used as a label to fill a line vec-
tor D and the remainder as an input vector to fill an
input matrix X. In the second signature
~
β(Q), the X
obtained in
~
α(Q) for each K × K window is also used
as labels (X = D). In this case, it is necessary to com-
pute the mean of the weights because there is more
than one neuron in the output layer. The third sig-
nature
~
γ(Q) is similar to
~
α(Q), but the matrix X and
D are exclusive for each window K × K. Thus, we
obtain the signature
~
γ(Q) by computing the mean of
the weights of all windows K × K. The signatures
~
α(Q),
~
β(Q) and
~
γ(Q) are built by using K = {3, 5, 7}.
In this work, we use the signature
~
Ω(Q), which is
~
Ω(Q) = [
~
α(Q),
~
β(Q),
~
γ(Q)]. More details on these
signatures can be found in (S
´
a Junior and Backes, ).
4 EXPERIMENTS
4.1 Colorectal Database
The colorectal database used in our experiments is
presented in the paper (Kather et al., 2016) and avail-
able at DOI:10.5281/zenodo.53169. It contains 5,000
images divided into 8 classes (625 images per class).
The classes are: Tumor epithelium; Simple stroma;
Complex stroma; Immune cells; Debris; Normal mu-
cosal glands; Adipose tissue; and Background. Figure
1 shows one sample for each class.
4.2 Experimental Setup
To classify the texture signatures, we applied a
Radial-Basis SVM (Boser et al., 1992; Cortes and
Vapnik, 1995) with C = 20 by using LIBSVM (Chang
and Lin, 2011). The validation strategy adopted was
5-fold, that is, the signature database is even divided
into five groups, one group used for testing and the re-
mainder for training. Thus procedure is repeated five
times (each time with a different group for testing).
The mean accuracy is the performance measure. We
computed three signatures
~
Ω(39),
~
Ω(39,49,59) and
FUSRNN (Fusion of RNN signatures with other tex-
ture analysis methods) in the colorectal images con-
verted into grayscale. The texture signatures concate-
nated in FUSRNN were:
• Complex Network Texture Descriptor (CNTD)
(Backes et al., 2013): Complex Network Theory
is employed to represent the pixels of a grayscale
image as a dynamic complex network. Topologi-
cal features of the network are then calculated to
create a feature vector that characterizes the orig-
inal image. The parameters utilized include a net-
work modeling radius of r = 3 and 36 threshold
values for generating dynamic transformations,
i.e., how connections between vertices change. A
total of 108 descriptors, comprising energy, en-
Colorectal Image Classification Using Randomized Neural Network Descriptors
801

(a) (b) (c) (d)
(e) (f) (g) (h)
Figure 1: The colorectal database: (a) Tumor epithelium, (b) Simple stroma, (c) Complex stroma, (d) Immune cells, (e)
Debris, (f) Normal mucosal glands, (g) Adipose tissue, (h) Background (Source: paper (Kather et al., 2016) and DOI:10.
5281/zenodo.53169).
tropy, and contrast measurements from each trans-
formation, were computed.
• Fourier descriptors (Weszka et al., 1976): In
this approach, the input image undergoes the bi-
dimensional Fourier transform, and subsequently,
the shifting operator is applied to compute spec-
trum descriptors. The descriptor is obtained by
summing the absolute coefficients located at the
same radial distance from the image’s center. By
following this process, a total of 74 descriptors are
generated.
• Gabor Filter (Daugman and Downing, 1995): A
set of filters was generated using the mathematical
procedure and the parameter values described in
(Manjunath and Ma, 1996), that is, scales S = 4,
rotations K = 6, and upper and lower frequencies
Uh = 0.4 and Ul = 0.05, respectively. Next, mean
and standard-deviation were computed from these
filters, thus resulting in 48 features.
• Gray Level Dependence Matrix (GLDM)
(Weszka et al., 1976): In this method, the
probability-density function is calculated for
pairs of pixels with a given distance and inter-
sample space, representing a specific absolute
difference in intensity. For the experiments, four
distances ((0,d),(−d,d),(d,0), and (−d, −d))
and three intersample spaces (d = 1, 2,5) were
used. From each probability-density function,
five measurements (angular second moment,
entropy, contrast, mean, and inverse difference
moment) were computed. This process yields a
total of 60 descriptors.
• Local Binary Patterns (LBP) (Ojala et al., 2002):
this texture descriptor characterizes the local
structure of an image by comparing the values of a
central pixel with the intensities of its neighboring
pixels. LBP encodes this comparison result into a
binary pattern, where each bit represents whether
the neighboring pixel is greater or smaller in in-
tensity compared to the central pixel. This en-
ables LBP to capture texture information, such as
edges, corners, and texture regularity. For the ex-
periments, we used the LBP histogram computed
using (P,R) = (8,1), resulting in 10 descriptors.
• Joint Adaptive Median Binary Patterns (Hafiane
et al., 2015): This approach employs a combina-
tion of Local Binary Pattern (LBP) and Median
Binary Pattern (MBP) techniques, enhanced by
adaptive threshold selection, to extract local pat-
terns from an image. As a result, a feature vector
consisting of 320 descriptors is obtained, effec-
tively representing the intricate local microstruc-
ture of the texture.
5 RESULTS AND DISCUSSION
In Table 1, we compared the accuracies of signatures
~
Ω(39),
~
Ω(39,49,59) and FUSRNN with the results
showed in the paper (Wang et al., 2017). The com-
parison demonstrates that the descriptor in
~
Ω(39) sur-
passes almost all compared approaches. In turn, the
accuracy of the signature
~
Ω(39,49,59) (92.46% ±
0.44%) presents a result equivalent to the best per-
formance in (Wang et al., 2017) (92.6% ± 1.2%) and
the result of FUSRNN surpasses all the compared ap-
proaches in Table 1.
We also compared the randomized network de-
scriptors as well as their fusion with other texture
analysis methods with the results in (Nanni et al.,
2019). The result of
~
Ω(39), which was the signa-
ture used in (S
´
a Junior and Backes, ) for compari-
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
802
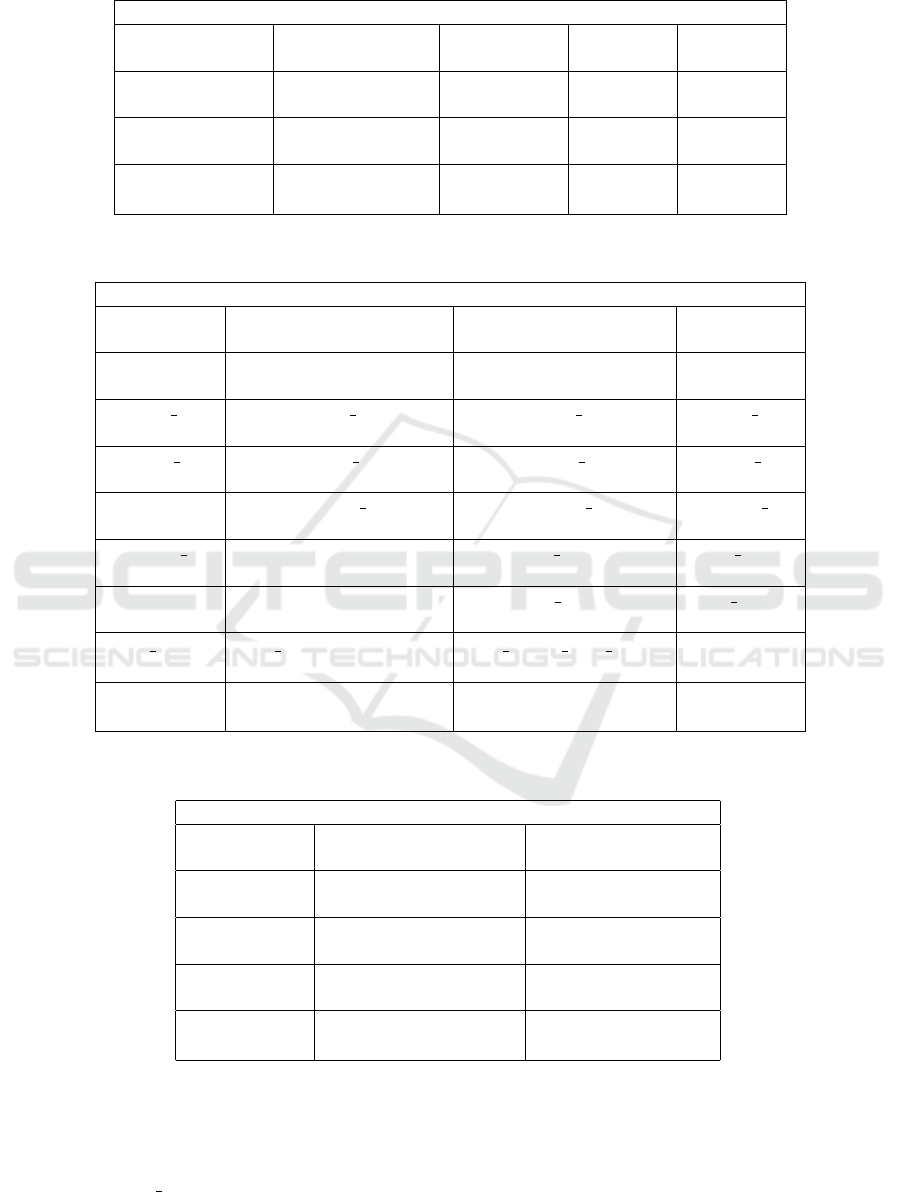
Table 1: Comparison of performance of the randomized neural networks descriptors (
~
Ω(39) and
~
Ω(39,49,59)) and its fusion
with other methods (FUSRNN) with other approaches. The compared results were obtained from (Wang et al., 2017).
Approaches and accuracies (%)
Histogram-lower Histogram-higher LBP GLCM Gabor
80.8 72.4 76.2 71.9 63.1
Perception Best2 Best3 Best4 Best5
62.9 85.8 86.0 86.5 87.4
All-6 CNN CNN-H CNN-E BCNN
87.4 90.2 ± 3.1 89.2 ± 1.1 86.1 ± 1.4 92.6 ± 1.2
~
Ω(39)
~
Ω(39,49,59) FUSRNN - -
91.34 ± 0.81 92.46 ± 0.44 94.36 ± 0.30 - -
Table 2: Comparison of performance of the randomized neural networks descriptors (
~
Ω(39) and
~
Ω(39,49,59)) and its fusion
with other methods (FUSRNN) with other approaches. The compared results were obtained from (Nanni et al., 2019).
Approaches and accuracies (%)
LTP MLPQ CLBP RICLBP
87.6 92.48 90.38 87.88
GOLD HOG AHP FBSIF
84.46 65.60 92.28 92.30
FUS 1 FUS 2 FUS 3 FUS 4
92.24 93.26 93.54 93.74
CLM 1 CLM 2 CLM 3 CLM 4
87.70 86.52 78.02 86.32
CLM CLoVo 1 CLoVo 2 CLoVo 3
85.08 87.26 62.72 86.80
CLoVo 4 CLoVo FUS CLM3 FUS CLM
89.26 89.08 84.82 88.50
DeepOutput DeepScores FUS ND(2) FUS ND(3)
91.90 94.44 92.72 93.02
FUS D(0.5) FUS ND(3)+DeepOutput FUS ND(3) FUS D(0.5)
~
Ω(39)
94.44 93.24 93.98 91.34 ± 0.81
~
Ω(39,49,59) FUSRNN - -
92.46 ± 0.44 94.36 ± 0.30 - -
Table 3: Comparison of performance of the randomized neural networks descriptors (
~
Ω(39) and
~
Ω(39,49,59)) and its fusion
with other methods (FUSRNN) with other approaches. The compared results were obtained from (Paladini et al., 2021).
Approaches and accuracies (%)
LPQ, SVM BSIF, SVM LPQ+BSIF, SVM
68.12 68.10 74.10
LPQ, NN BSIF, NN LPQ+BSIF, NN
69.02 71.04 74.22
ResNet-101 ResNeXt-50 Inception-v3
95.92 95.74 93.98
DenseNet-161 Mean-Ensemble-CNNs NN-Ensemble-CNNs
95.60 96.16 96.14
~
Ω(39)
~
Ω(39,49,59) FUSRNN
91.34 ± 0.81 92.46 ± 0.44 94.36 ± 0.30
son with other approaches, surpasses the performance
of several compared methods in Table 2. The re-
sult of
~
Ω(39,49,59), in turn, is only surpassed by
more sophisticated approaches based on ensembles
(for instance, FUS 2) or Convolutional Neural Net-
works (for instance, DeepScores). On the other hand,
our proposed signature FUSRNN provided a result
(94.36% ± 0.30%) virtually equivalent to the best per-
formance in Table 2 (DeepScores, with 94.44%).
In Table 3, we performed a comparison with the
Colorectal Image Classification Using Randomized Neural Network Descriptors
803

results showed in (Paladini et al., 2021). This table
shows that the three signatures
~
Ω(39),
~
Ω(39,49,59)
and FUSRNN largely outperformed all the hand-
crafted methods. On the other hand, almost all
CNN-based approaches surpassed FUSRNN (excep-
tion for Inception-v3), with NN-Ensemble-CNNs pre-
senting an advantage of 1.78%. To explain this
difference in performance, it is important to stress
that the authors in (Paladini et al., 2021) affirm
that ResNet-101, ResNeXt-50, Inception-v3, and
DenseNet-161 are “four of the most powerful CNN
architectures”(Paladini et al., 2021) and that they
used pre-trained models from the ImageNet Chal-
lenge Database. Moreover, the best CNN-based ap-
proach (NN-Ensemble-CNNs) is an ensemble com-
bining the four mentioned CNN architectures.
Finally, the signature
~
Ω(39,49,59) provides an
accuracy equivalent to ARA-CNN (Raczkowski et al.,
2019) (92.24 ± 0.82%), and the signature FUSRNN
overcomes it. Thus, based on our results, it is pos-
sible to affirm that randomized neural network de-
scriptors (
~
Ω(39) and
~
Ω(39,49,59)) have high perfor-
mance, surpassing several texture analysis methods.
Also, when associated with other descriptors (FUS-
RNN), it provided accuracies slightly inferior to the
best CNN architectures. Such performance suggests
that novel improvements in the RNN signature as well
as its association with other descriptors equally dis-
criminative may result in even higher accuracies when
applied to colorectal images.
6 CONCLUSION
This paper presented an application of a highly
discriminative texture descriptor based on weights
of randomized neural network on a very important
multi-class problem, which consists of discriminat-
ing colorectal images into eight classes, according to
the image database provided by (Kather et al., 2016).
The results of the randomized neural network signa-
ture were promising, surpassing several texture analy-
sis methods. When the neural neural descriptors were
associated with other texture analysis methods, this
fusion signature was capable of providing accuracies
similar or slightly inferior to that of several powerful
convolutional neural networks, which are known for
having a high number of parameters to tune. Thus,
ground on our results, we believe that our proposed
applied approach has potential to provide even better
results and adds a valuable tool to the computer vision
research in colorectal images.
ACKNOWLEDGEMENTS
This study was financed in part by the Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
– Brasil (CAPES) – Finance Code 001. Andr
´
e R.
Backes gratefully acknowledges the financial sup-
port of CNPq (Grant #307100/2021-9). Jarbas
Joaci de Mesquita S
´
a Junior thanks Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
(CAPES, Brazil) for the financial support of this
work.
REFERENCES
Backes, A. R., Casanova, D., and Bruno, O. M. (2013). Tex-
ture analysis and classification: A complex network-
based approach. Information Sciences, 219:168 – 180.
Boser, B. E., Guyon, I. M., and Vapnik, V. N. (1992). A
training algorithm for optimal margin classifiers. In
Proceedings of the Fifth Annual Workshop on Compu-
tational Learning Theory, pages 144–152. ACM.
Calvetti, D., Morigi, S., Reichel, L., and Sgallari, F. (2000).
Tikhonov regularization and the l-curve for large dis-
crete ill-posed problems. Journal of Computational
and Applied Mathematics, 123(1):423–446.
Chang, C.-C. and Lin, C.-J. (2011). LIBSVM: A library
for support vector machines. ACM Transactions on
Intelligent Systems and Technology, 2:27:1–27:27.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine Learning, 20(3):273–297.
Cover, T. M. (1965). Geometrical and statistical properties
of systems of linear inequalities with applications in
pattern recognition. IEEE Transactions on Electronic
Computers, EC-14(3):326–334.
Daugman, J. and Downing, C. (1995). Gabor wavelets for
statistical pattern recognition. In Arbib, M. A., edi-
tor, The Handbook of Brain Theory and Neural Net-
works, pages 414–419. MIT Press, Cambridge, Mas-
sachusetts.
dos Santos, L. F. S., Neves, L. A., Rozendo, G. B., Ribeiro,
M. G., do Nascimento, M. Z., and Tosta, T. A. A.
(2018). Multidimensional and fuzzy sample entropy
(SampEnMF) for quantifying H&E histological im-
ages of colorectal cancer. Computers in Biology and
Medicine, 103:148 – 160.
Hafiane, A., Palaniappan, K., and Seetharaman, G. (2015).
Joint adaptive median binary patterns for texture clas-
sification. Pattern Recognition, 48(8):2609–2620.
Huang, G.-B., Zhu, Q.-Y., and Siew, C.-K. (2006). Extreme
learning machine: theory and applications. Neuro-
computing, 70(1):489–501.
Kather, J. N., Weis, C.-A., Bianconi, F., Melchers, S. M.,
Schad, L. R., Gaiser, T., Marx, A., and Zollner,
F. G. (2016). Multi-class texture analysis in colorec-
tal cancer histology. Scientific Reports, 6(27988).
doi:10.1038/srep27988.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
804

Manjunath, B. S. and Ma, W.-Y. (1996). Texture features
for browsing and retrieval of image data. IEEE Trans.
Pattern Anal. Mach. Intell, 18(8):837–842.
Moore, E. H. (1920). On the reciprocal of the general alge-
braic matrix. Bulletin of the American Mathematical
Society, 26:394–395.
Nanni, L., Brahnam, S., Ghidoni, S., and Lumini, A. (2019).
Bioimage classification with handcrafted and learned
features. IEEE/ACM Transactions on Computational
Biology and Bioinformatics, 16(3):874–885.
Ojala, T., Pietik
¨
ainen, M., and M
¨
aenp
¨
a
¨
a, T. (2002). Mul-
tiresolution gray-scale and rotation invariant texture
classification with local binary patterns. IEEE Trans.
Pattern Anal. Mach. Intell, 24(7):971–987.
Paladini, E., Vantaggiato, E., Bougourzi, F., Distante,
C., Hadid, A., and Taleb-Ahmed, A. (2021). Two
ensemble-CNN approaches for colorectal cancer tis-
sue type classification. Journal of Imaging, 7(3).
Pao, Y.-H., Park, G.-H., and Sobajic, D. J. (1994). Learning
and generalization characteristics of the random vec-
tor functional-link net. Neurocomputing, 6(2):163–
180.
Pao, Y.-H. and Takefuji, Y. (1992). Functional-link net com-
puting: theory, system architecture, and functionali-
ties. Computer, 25(5):76–79.
Penrose, R. (1955). A generalized inverse for matrices.
Mathematical Proceedings of the Cambridge Philo-
sophical Society, 51(3):406–413.
Peyret, R., Bouridane, A., Khelifi, F., Tahir, M. A., and
Al-Maadeed, S. (2018). Automatic classification of
colorectal and prostatic histologic tumor images us-
ing multiscale multispectral local binary pattern tex-
ture features and stacked generalization. Neurocom-
puting, 275:83 – 93.
Raczkowski, L., Mozejko, M., Zambonelli, J., and
Szczurek, E. (2019). ARA: accurate, reliable and
active histopathological image classification frame-
work with Bayesian deep learning. Scientific Reports,
9(14347).
Ribeiro, M. G., Neves, L. A., Nascimento, M. Z., Roberto,
G. F., Martins, A. S., and Tosta, T. A. A. (2019). Clas-
sification of colorectal cancer based on the association
of multidimensional and multiresolution features. Ex-
pert Systems with Applications, 120:262 – 278.
S
´
a Junior, J. J. M. and Backes, A. R. An improved ran-
domized neural network signature for texture classifi-
cation. SSRN.
S
´
a Junior, J. J. M. and Backes, A. R. (2016). ELM based
signature for texture classification. Pattern Recogni-
tion, 51:395–401.
Schmidt, W. F., Kraaijveld, M. A., and Duin, R. P. W.
(1992). Feedforward neural networks with random
weights. In Proceedings., 11th IAPR International
Conference on Pattern Recognition. Vol.II. Confer-
ence B: Pattern Recognition Methodology and Sys-
tems, pages 1–4.
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A.
(2023). Cancer statistics, 2023. CA: A Cancer Journal
for Clinicians, 73(1):17–48.
Tikhonov, A. N. (1963). On the solution of ill-posed prob-
lems and the method of regularization. Dokl. Akad.
Nauk SSSR, 151(3):501–504.
Wang, C., Shi, J., Zhang, Q., and Ying, S. (2017).
Histopathological image classification with bilinear
convolutional neural networks. In 2017 39th Annual
International Conference of the IEEE Engineering in
Medicine and Biology Society (EMBC), pages 4050–
4053.
Weszka, J. S., Dyer, C. R., and Rosenfeld, A. (1976). A
comparative study of texture measures for terrain clas-
sification. IEEE Trans. Syst., Man, Cyb., 6(4):269–
285.
Yoon, H., Lee, J., Oh, J. E., Kim, H. R., Lee, S., Chang,
H. J., and Sohn, D. K. (2019). Tumor identification in
colorectal histology images using a convolutional neu-
ral network. Journal of Digital Imaging, 32(1):131–
140.
Colorectal Image Classification Using Randomized Neural Network Descriptors
805
