
An Improved VGG16 Model Based on Complex Invariant Descriptors
for Medical Images Classification
Mohamed Amine Mezghich, Dorsaf Hmida, Taha Mustapha Nahdi and Faouzi Ghorbel
GRIFT Research Group, CRISTAL laboratory, ENSI, Tunisia
Keywords:
Complex Moments, Invariant Descriptors, Stability, Completeness, Deep Learning, Classification.
Abstract:
In this paper, we intent to present an improved VGG16 deep learning model based on an invariant and complete
set of descriptors constructed by a linear combination of complex moments. First, the invariant features are
studied to highlight it’s stability and completeness properties over rigid transformations, noise and non rigid
transformations. Then our proposed method to inject this family to the well know deep leaning VGG16 model
is presented. Experimental results are satisfactory and the model accuracy is improved.
1 INTRODUCTION
Recently, images have captured great importance be-
cause they are the main source of information. It’s
used in many aspects to share information between
different recipients in various fields such as military
operations, medical imaging, artificial intelligence,
social media and many other applications. Feature
extraction technique consists in computing object de-
scriptors with some interesting properties such as in-
variant to some geometric transformations, complete-
ness which means discrimination between objects
if they are different and finally stability over noise
and shape distortion. It’s the most crucial stage in
every kind of classification system because the ex-
tracted features significantly influence the system per-
formance. It is sometimes necessary to calculate de-
scriptors directly from gray-scale objects when the
external profile of the objects is not sufficiently dis-
criminating. This problem is more complex than the
previous one because the description must take into
account information both on the contour and the in-
ternal texture of objects.
Many shape representation and description tech-
niques have been developed in the past. Early works
for computing invariant shape features were based
on Fourier descriptors of the curve, e.g. (Zahn and
Roskies, 1972) , (Persoon and Fu, 1977). (Ghorbel,
1998) introduced a joint topology and harmonic anal-
ysis formulation for the extraction of global shape de-
scriptors which are invariant under a given group of
geometrical transformations. The second family of
object descriptors are based on geometric and com-
plex moments of the image which take into account
the gray-scale level of pixels. In (Hu, 1962), Hu in-
troduced seven invariant descriptors based on a com-
bination of centred and normalised geometric mo-
ments. This set is not complete which means that
we can’t reconstruct the original object from it’s mo-
ments. Khotanzad and Hong introduced in (Khotan-
zad and Hong, 1990) Zernike moments which are
invariant and complete set to rigid transformations.
Fourier–Merlin moments were introduced in (Sheng
and Shen, 1994) to extract invariant gray-scale fea-
tures. In (Ghorbel, 1994), The author proposed the
Analytical Fourier-Mellin Transform (AFMT) for in-
variant and complete shape description. A set of com-
plete and stable invariants was then proposed and in
(Derrode and Ghorbel, 2001), the authors proposed
three methods to compute this set based on polar,
log-polar and cartesian approximations. Flusser et
al. proposed respectively in (Flusser, 2002) and (Suk
and Flusser, 2003) a new set of descriptors based
on complex moments which are invariant to rota-
tion. In 2006, Ghorbel et al. (Ghorbel et al., 2006)
proposed a systematic method to extract a complete
set of similarity invariants (translation, rotation and
scale), by means of some linear combinations of com-
plex moments. Also the authors proposed an orig-
inal method to reconstruct the image from its com-
plex moments derived from the relationship between
the Discrete Fourier Transform (DFT) and complex
moments of the image. In recent years, many ef-
forts have been made to apply convolutional neural
networks (CNNs) on the medical imaging modality.
In fact CNNs have already shown impressive perfor-
444
Mezghich, M., Hmida, D., Nahdi, T. and Ghorbel, F.
An Improved VGG16 Model Based on Complex Invariant Descriptors for Medical Images Classification.
DOI: 10.5220/0012467800003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 444-452
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

mance on the classification of natural images. The
fusion of CNN with different techniques like local
and global features have proven encourages and best
results. (Sharma and Mehra, 2020) proposed two
machine learning approaches. The first approach is
based on handcrafted features which are extracted
using Hu’s moments, color histogram, and Haralick
textures. The extracted features are then utilized to
train the conventional classifiers, while the second ap-
proach is based on transfer learning where the pre-
existing networks (VGG16, VGG19, and ResNet50)
are utilized as feature extractor and as a baseline
model. (Cao et al., 2020) proposed a ship recogni-
tion method based on morphological watershed im-
age segmentation and Zernike moment to solve the
problem of ship recognition in video images. The fea-
ture of ship image is extracted based on deep learn-
ing convolution neural network (CNN) and Zernike
moments method. (Wang et al., 2020) developed a
deep learning-based model for Drug-Target Interac-
tions (DTIs) prediction. The proteins evolutionary
features are extracted via Position Specific Scoring
Matrix (PSSM) and Legendre Moment (LM). (Moung
et al., 2021) proposed a fusion of a moment invariant
(MI) method and a Deep Learning (DL) algorithm for
feature extraction to address the instabilities in the ex-
isting COVID-19 classification models. (Ren et al.,
2021) proposed a ship recognition approach based on
Hu invariant moments and Convolutional Neural Net-
work (CNN) to solve the issue with automatic recog-
nition of ship images in video surveillance system.
In this work, an improved VGG16 model based on
complex invariant descriptors is proposed for medi-
cal images classification. The proposed invariant fea-
tures are added to the ones extracted by the considered
CNN deep model in order to enhance the accuracy of
classification. Our paper is organized as follows: In
section 2, we introduce the set of invariant complex
descriptors. By section 3, we recall the used VGG16
deep learning model then we present our approach to
improve the accuracy of this model. Section 4 high-
light experimental results. Finally in section 5, we
summarize our work and provides brief insight into
our planned future work.
2 INVARIANT FEATURES
In this paper, we are interested in invariant descrip-
tors computed from image complex moments. It’s a
mathematical tools used in image processing to de-
scribe the intensity distribution in an image and it’s a
generalization of standard geometric moments used to
extract features that are invariant under certain trans-
formations such as translation, rotation and scaling.
First, we recall some basic terms that will be used
to construct the invariant set. Let f (x, y) be an im-
age function having a bounded support and a finite
nonzero integral. The complex moments of f (x, y)
are defined as:
c
f
(p, q) =
ZZ
R
2
(x + iy)
p
(x − iy)
q
f (x, y)dx dy (1)
where p, q ∈ N. Eq.1 in polar coordinates be-
comes:
c
f
(p, q) =
Z
∞
0
Z
2π
0
r
p+q+1
e
i(p−q)θ
f (r, θ)dr dθ (2)
In (Flusser, 2002) and (Flusser, 2006), a set of
complete and independent set of rotation invariant
descriptors is given as follows :
∀p, q ∈ N, Φ
f
(p, q) = c
f
(p
0
− 1, p
0
)
p−q
c
f
(p, q)
(3)
And in (Ghorbel et al., 2006), the authors proposed a
systematic method to extract a complete set of sim-
ilarity invariant features to translation, rotation and
scale by means of some linear combinations of com-
plex moments. The complete formulation I
f
is de-
fined as follows:
∀p, q ∈ N, I
f
(p, q) = Γ
−(p+q+2)
f
e
−i(p−q)Θ
f
c
f
(p, q)
(4)
with Θ
f
= arg(c
f
(1, 0)) and Γ
f
=
q
c
f
(0, 0).
The completeness property is easy obtained by this
formula:
∀p, q ∈ N, c
f
(p, q) = Γ
p+q+2
f
e
i(p−q)Θ
f
I
f
(p, q) (5)
We will be based on this family to study first it’s sta-
bility and robustness, then we will inject it to CNN
based deep learning model in order to improve its ac-
curacy.
3 AN IMPORVED VGG16 MODEL
In this section, we start by describing the CNN archi-
tecture which is utilised in this work , Sect. 3.1 then,
we detail our proposed method in Sect. 3.2.
3.1 Deep Learning VGG16 Model
VGG16 is a convolutional neural network (CNN)
architecture designed for image classification. It
was introduced by (Simonyan and Zisserman, 2014)
An Improved VGG16 Model Based on Complex Invariant Descriptors for Medical Images Classification
445

which participated in the ImageNet Large Scale Vi-
sual Recognition Challenge (ILSVRC) in 2014 and
achieved notable success. It can more accurately ex-
press the characteristics of the data set when identi-
fying and classifying images. This model has a 16
weight layers, which include 13 convolutional layers
and 3 fully connected layers. It represented an ad-
vance on previous models, offering convolution lay-
ers with smaller convolution kernels (3×3) than had
previously been the case.
During model training, the input for the first con-
volution layer is an RGB image of size 224 x 224. For
all convolution layers, the convolution kernel is 3×3.
These convolution layers are accompanied by Max-
Pooling layers, each 2×2 in size, to reduce the size of
the filters during learning.
In the proposed method, the model, as shown in 1,
is inspired by (Moung et al., 2021). So, the last max-
pooling layer was removed and replaced with an av-
erage pooling layer. this will introduce some feature
generalization because this operation will take the av-
erage of pixels to retain fine-grain details in the fi-
nal convolutions. During the classification phase, the
output features from the average pooling layer of the
VGG16 model are fed into the new fully-connected
layers. The new classifier part of the VGG16 model
consists of one flattened layer and two dense lay-
ers, with each generating 100 and 2 outputs, respec-
tively. The first dense layer has a Rectified Linear
Unit (ReLU) activation function with 0.5 dropout.
The output layer, which is the last dense layer, has
a Sigmoid activation function. The feature map size
of the generated VGG16 features used in this work is
25,088 × 1 dimension per input image.
In this work, the proposed approach is built us-
ing a convolutional neural network (CNN) with batch
normalization (BN) which is a popular and effec-
tive technique that consistently accelerates the con-
vergence of deep networks.
3.2 Proposed Method
Our proposed method consists of 3 phases: feature
extraction, feature concatenation and classification.
Figure 2 illustrates the architecture of the proposed
method. We will detail each phase in the next section.
3.2.1 Feature Extraction
In the feature extraction step, two types of features
are extracted: those associated to DL (Deep Learn-
ing) and those relative to MI (moment invariant).
For DL feature extraction, VGG16 is utilised in this
work. Meanwhile, the MI-based features are ex-
tracted using the (Ghorbel et al., 2006) moment in-
variant method. Then, these two features (DL-based
and MI-based features) are concatenated using the
join fusion. Lastly, classification based on the fused
features is performed by using the fully connected
(FC) layers.
VGG16 Network Architecture
In this work, we use a CNN model VGG16 (see Sec-
tion 3.1 to extract essential features. This model was
composed of two parts: convolutional base and classi-
fier. The convolutional base comprises convolutional
and pooling layers to generate features, whereas the
classifier categorises the image based on extracted
features. We initialized the model weights using
Kaiming Initialization for convolutions and normal
initialization for fully connected layers. Additionally,
we did not employ any transfer learning in our exper-
iments. The VGG16 output features of shape 77512
at the final max-poling layer.
Complete Complex Invariant Descriptors
Based on the work of (Ghorbel et al., 2006), the MI-
based features of each CT image are extracted using
complex invariant moments (see Section 2) to trans-
lation, rotation and scale, by means of some linear
combinations of complex moments. We use both the
absolute and phase values of the obtained complex in-
variant features, so no data are lost. More details are
presented in the experimental results.
3.2.2 Feature Concatenation
In (Huang et al., 2020), authors described all the dif-
ferent fusion strategies using deep learning. In this
work, we use the joint fusion as shown in Fig 3.
It’s the process of joining learned feature represen-
tations from intermediate layers of neural networks
with features from other modalities as input to a fi-
nal model. Thus, the features of each batches of im-
ages from the VGG16 model are concatenated with
the Ghorbel moments features into a composite vec-
tor. The features extracted from VGG16 were multi-
dimensional vectors of shape (7x7x512). Thus, it is
flattened to be (25088x1) and the extracted Ghorbel
complex moment invariant values were presented as a
one-dimensional row vector. To be on the same scale
as the deep learning feature, which is a vector in the
R
(25088x1)
, we extracted the module and phase infor-
mation from the Ghorbel complex invariant feature.
Then we did the fusion. Fig3 illustrates the joint fu-
sion process. The size of the final vector will vary
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
446
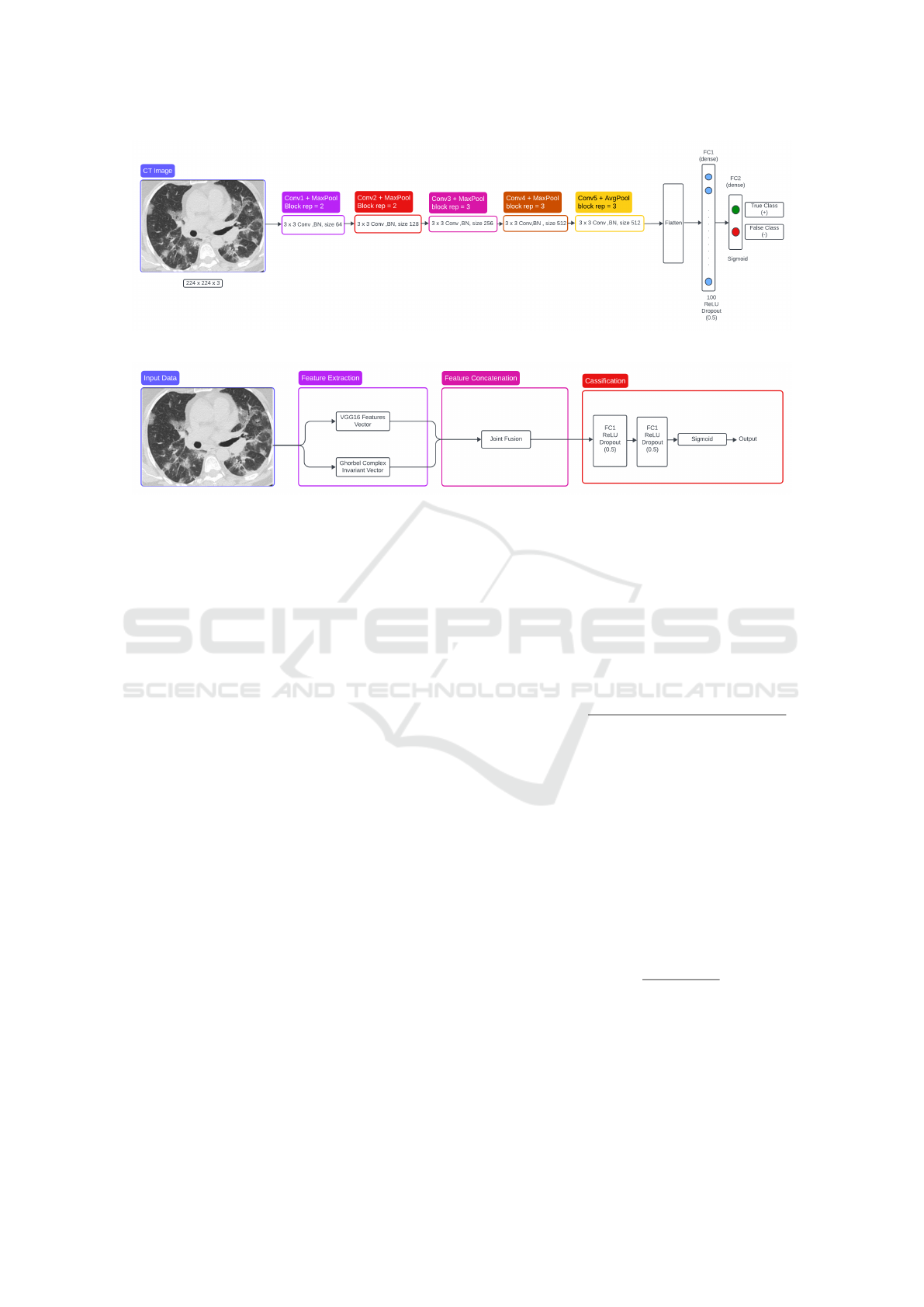
Figure 1: VGG16-based architecture with classifer block.
Figure 2: Overview of proposed method.
depending on the values chosen for p and q to calcu-
late the MI features. This aspect will be discussed in
the experimental section.
3.2.3 Classification Phase
For the classification phase, we followed the same
method used in (Moung et al., 2021). We use a fully
connected network which is built with two fully con-
nected (FC) layers. On model VGG16, the first FC
(dense) layers generates 100 output units with a ReLU
activation function and a dropout of 50% and the sec-
ond FC (dense) layers generates two output units with
a sigmoid activation function.
4 EXPERIMENTAL RESULTS
In order to prove the robustness of our model, first,
a set of experimental results where made in order to
study invariant complex moments under rigid trans-
formations, noise and non rigid ones performed on
columbia coil database(Nene et al., 1996) which is
available on its official website :https://www.cs.col
umbia.edu/CAVE/software/softlib/coil-20.php .
Then, the proposed deep model is validated on SARS-
COV-2 Ct-Scan Dataset.This dataset has been made
public by the authors on 22 April 2020 and can be ac-
cessed via the Kaggle website at the following link:
https://www.kaggle.com/datasets/plamenedua
rdo/sarscov2-ctscan-dataset. The dataset contain
1252 CT scans that are positive for SARS-CoV-2 in-
fection (COVID-19) and 1230 CT scans for patients
non-infected by SARS-CoV-2, 2482 CT scans in to-
tal. The selected data set was manually divided into
training, validation and test sets with a ratio of 82.8
%, 9.1% and 8.1% respectively.
In this paper, we evaluate the performance of our
predictor by calculating accuracy (ACC) for SARS-
CoV-2 infection (COVID-19) .
Accuracy =
Number of Correct predictions
Total no of predictions made
4.1 Stability of the Used Descriptors
In this section, we will study four families of invari-
ant based moments which are Hu (Hu, 1962), Zernike
(Khotanzad and Hong, 1990), Flusser (Flusser, 2002)
and finally the Ghorbel et al. ones (Ghorbel et al.,
2006). For 2D matrix moments (especially Flusser,
Zernike and Ghorbel et al.), we used the zigzag tech-
nique described in (Ghorbel et al., 2006) to obtain 1D
vector and keep the neighborhood. The comparison is
performed according to the relative error as follows :
E
I
1
,I
2
(K) = |
I
1
(k) − I
2
(k)
I
1
(k)
| (6)
We’ll start by applying several rigid transforma-
tions to Lena’s image to demonstrate the stability of
these invariants. Figure 4 shows the image of Lena
on which we’ll be experimenting. For this first set of
experiments, we fix (p = q = 3) for Flusser and Ghor-
bel,(radios = digres = 4) for Zernike.
An Improved VGG16 Model Based on Complex Invariant Descriptors for Medical Images Classification
447
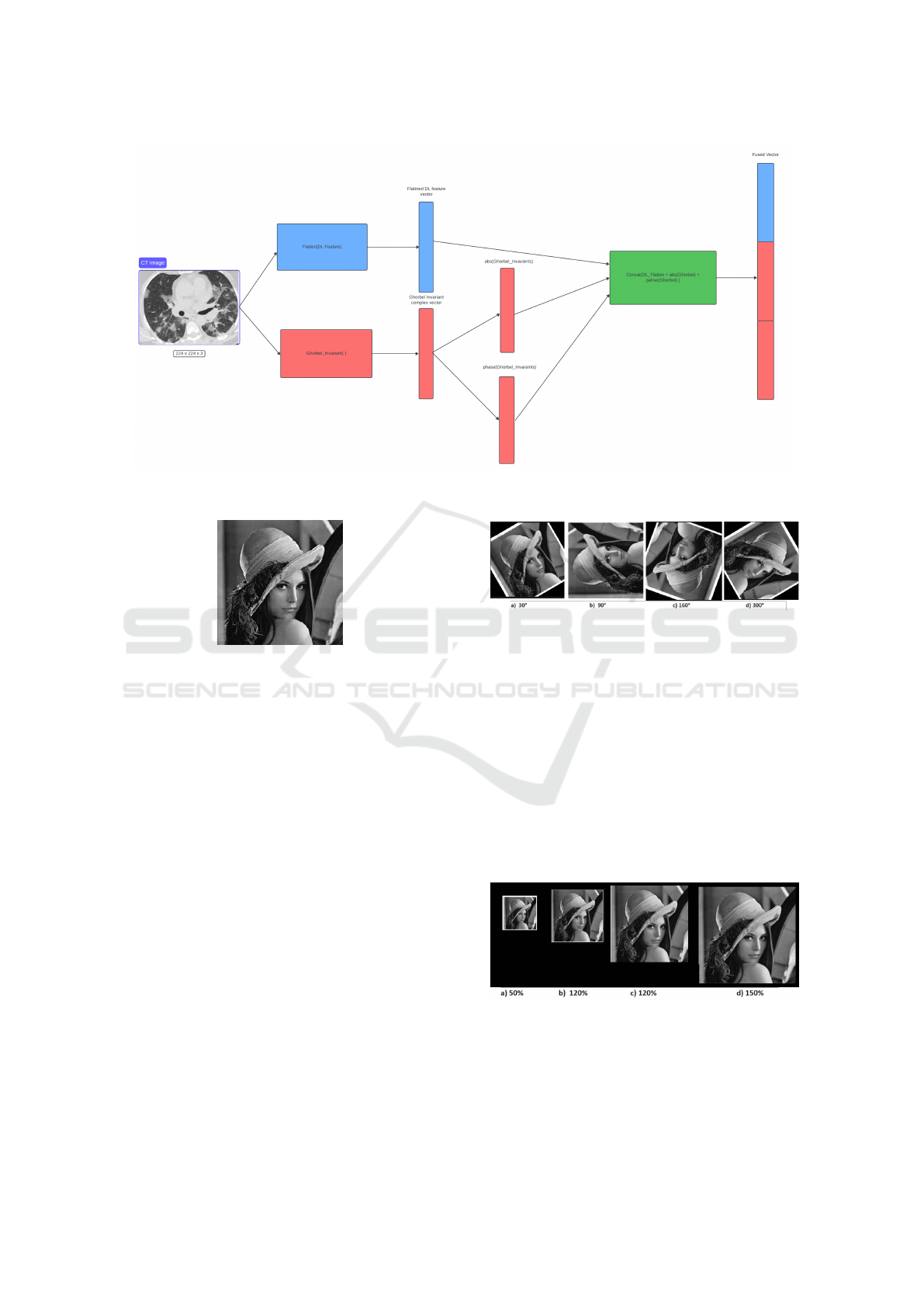
Figure 3: Feature Concatenation Process Between DL and Ghorbel Invariants Feature.
Figure 4: Original image of Lena with size 224x224.
Invariance Against Rotation
To evaluate rotation invariance, we applied rotations
to the image of Lena presented in Fig 4, rotating it
by angles of 30°, 90°, 160° and 300°, as shown in
Fig.5. An experimental demonstration of the stabil-
ity between the invariants of (Flusser, 2006), (Hu,
1962), (Khotanzad and Hong, 1990) and (Ghorbel
et al., 2006) is well illustrated in Fig.9 to Fig. 12. The
results confirm that (Ghorbel et al., 2006) is invariant
to rotation whatever the angle of rotation, compared
with other families which have a higher relative er-
ror. We illustrate by Fig.14 and Fig.13 a summary of
the evolution of relative error under two rotation an-
gles (30° and 60°). It’s clear that the descriptors we
use have low, stable error values, whereas Flusser’s
descriptors give high error values. We also observe
an oscillation in error values for the two families of
Zernike and Hu moments.
Invariance to Scale
To evaluate the robustness against scale factor, we
have scaled the Lena image to scaling factors of 50%,
Figure 5: Several rotations of the original image.
80%, 120% and 150%, as shown in Fig. 6. The plots
in Fig.15 to Fig.18 show the relative errors between
vector of invariants for the original image and those of
the scaled ones. Relative error values for the Ghorbel
et al. descriptors range from [0.1], while those for the
Zernike descriptors range from [0.4] and those for the
Hu’moments from [0.2]. For Flusser’s descriptors,
the error is huge, in fact Flusser’moments are only
invariant under rotation. Also, we observe certain sta-
bility for the error only for Ghorbel et al. descriptors.
This result confirms the robustness of (Ghorbel et al.,
2006) against scale factor.
Figure 6: Several scaled images of the original image.
Invariance Against Noise
To evaluate the robustness against noise, we intro-
duced white Gaussian noise into the Lena image, in-
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
448
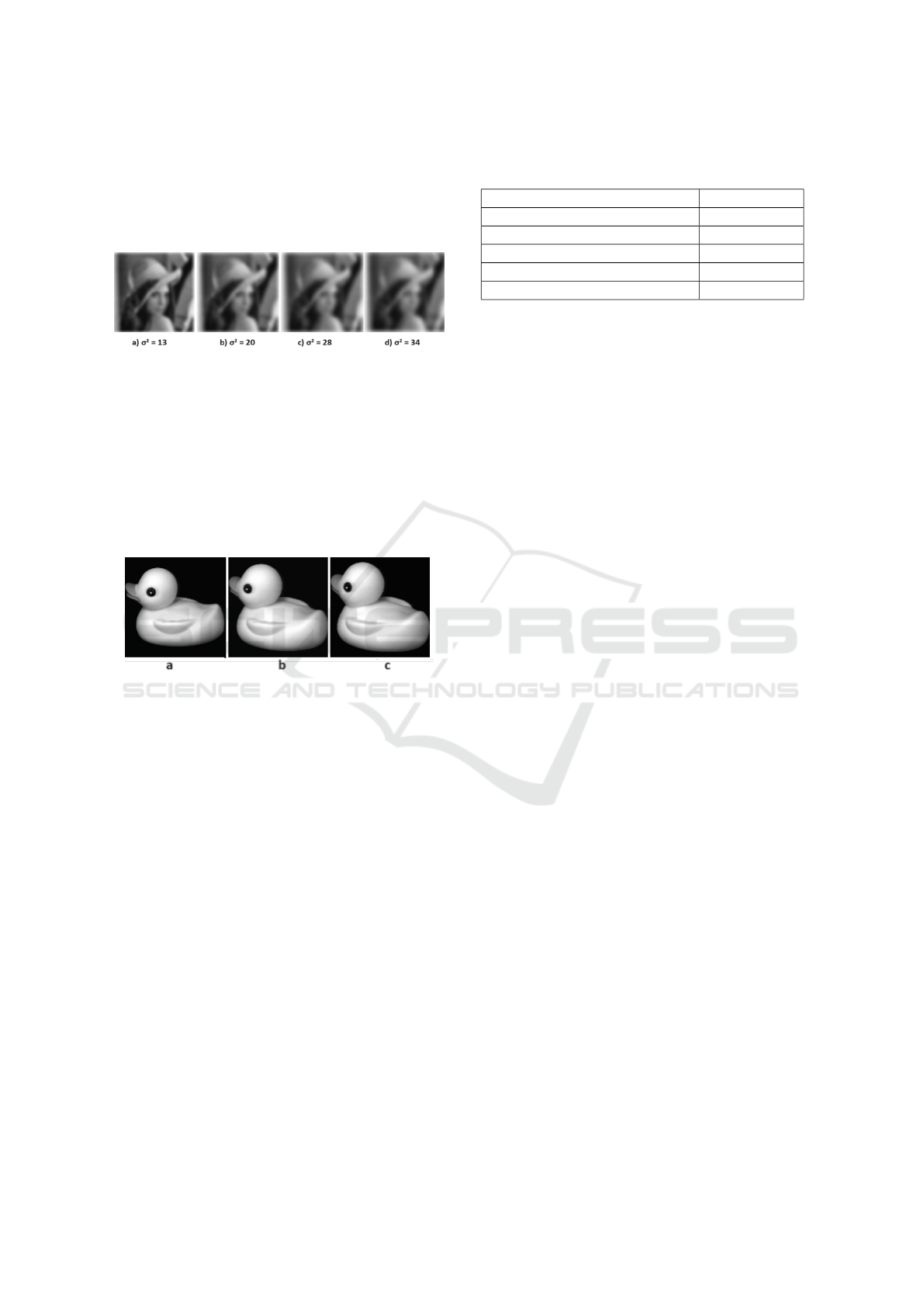
corporating a mean (µ) of 0 and variances (σ
2
) of 13 ,
20, 28, and 34, as shown in Fig.7. The plots in Figs.19
to 22 illustrate the relative errors between the invari-
ant vector of the original image and the invariant vec-
tors of the noisy images.
Figure 7: Several noisy images of Lena.
Invariance to Non-Rigid Transformations
In this section, we extend our experiment to non-rigid
transformations with the images of ducks in Fig. 8
from Columbia Coil data base. The results in graphs
23 and 24 verified that complex invariant moments
are stable to the non-rigid transformation using the
(COIL-20) dataset.
Figure 8: Three test images used in the pattern experiments
(original size 128×128). Fig. (a), (b) and (c) represent the
same duck but with different orientation.
4.2 The Improved VGG16 Model
The performance of the VGG16 model with and with-
out MI is summarized in table 1. In the first part of
the experiments, the CT images are given only ba-
sic pre-processing steps before the training and test
phases, which includes rescaling into 224 × 224 di-
mension using the resize function and we performed
the normalization. All images that were fed into the
VGG16 model for feature extraction retained their
three-dimensional color channels format. However,
the images that were fed into Ghorbel’s invariant mo-
ment method (Ghorbel et al., 2006) were transformed
from color channels to a single channel by taking the
pixel-wise average across channels. We implemented
the model using PyTorch.
We set a learning rate of 0.0004 and the number of
epochs of 100 with an Adam optimiser based on the
work (Moung et al., 2021). The accuracy that we ob-
tained with this architecture is 97.512%. The concate-
Table 1: Performance of VGG16 model with and without
MI on the testing dataset.
Deep learning model Accuracy (%)
VGG16 93.034
VGG16 + 7 Hu’moments 94.527
VGG16 + Zernike descriptors 93.532
VGG16 + Flusser descriptors Not available
VGG16 + Ghorbel descriptors 97.512
nation of the invariant based complex moments in the
VGG16 features improved the informativeness and
discrimination capabilities of the flattened vector. The
model’s recognition ability improves with an increase
in the number of features it learns. Furthermore,
during training, the VGG16 model was exposed to
complex moments, extent and solidity values in addi-
tion to the deep-learning feature vector. The absolute
and phase values of the complex moments influenced
the activations in the network. Feature(VGG16) +
Abs(Ghorbel) + Phase(Ghorbel) showed a significant
increase in accuracy over VGG16 (93.034%), VGG16
+ Hu’moments (94.527%), VGG16 + Zernike de-
scriptors (93.532%) and VGG16 + Flusser descriptors
(Not available). In this case the Flusser invariant was
so large that it resulted in numerical overflow and re-
quired more processing to be mapped to the VGG16
features, so we didn’t train the model with Flusser.
5 CONCLUSION
In this research, we proposed an improved VGG16
deep learning model based on the fusion of CNN fea-
tures and a complete and invariant set of descriptors
based on the image’s complex moments. Several ex-
perimental results were performed in order to demon-
strate the stability of the proposed set of shape de-
scriptors. Compared to other works based on Hu mo-
ments, Zernike and Flusser invariants, the proposed
method achieves the highest accuracy. As future per-
spectives, we plan to study our approach on other
databases with more sophisticated CNN architectures
such as VGG19.
An Improved VGG16 Model Based on Complex Invariant Descriptors for Medical Images Classification
449
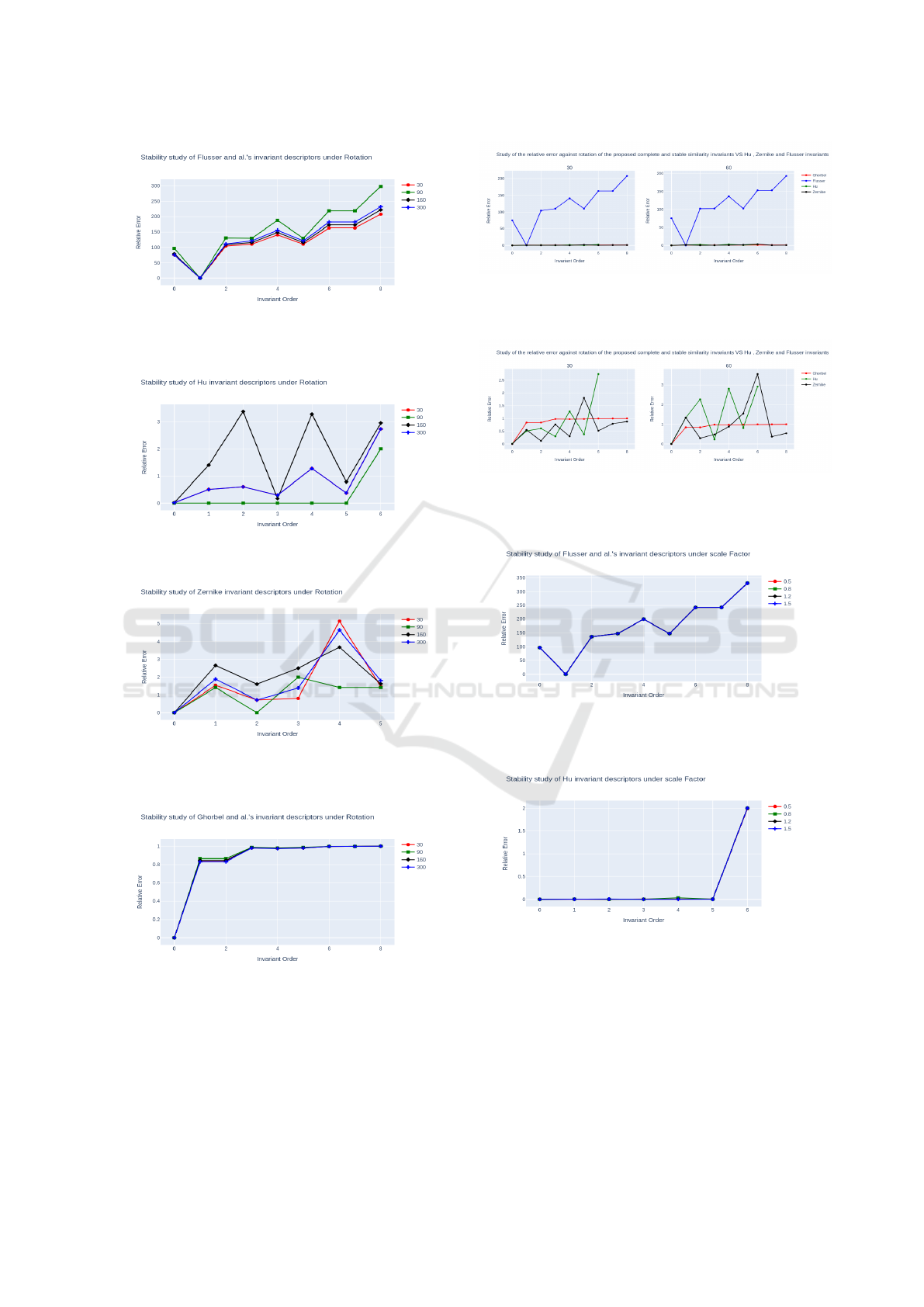
Figure 9: Stability study of Flusser invariants under rota-
tion.
Figure 10: Stability study of Hu invariant under rotation.
Figure 11: Stability study of Zernike invariants under rota-
tion.
Figure 12: Stability study of Ghorbel invariant under rota-
tion.
Figure 13: Relative error against rotation, for the Hu’s,
Flusser’s, Zernike’s and Ghorbel’s set of similarity invari-
ants.
Figure 14: Relative error against rotation, for the Hu’s,
Zernike’s and Ghorbel’s set of similarity invariants (loga-
rithmic scale except for the image).
Figure 15: Stability study of Flusser invariant moments un-
der scale factor.
Figure 16: Stability study of Hu invariant moments under
scale factor.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
450
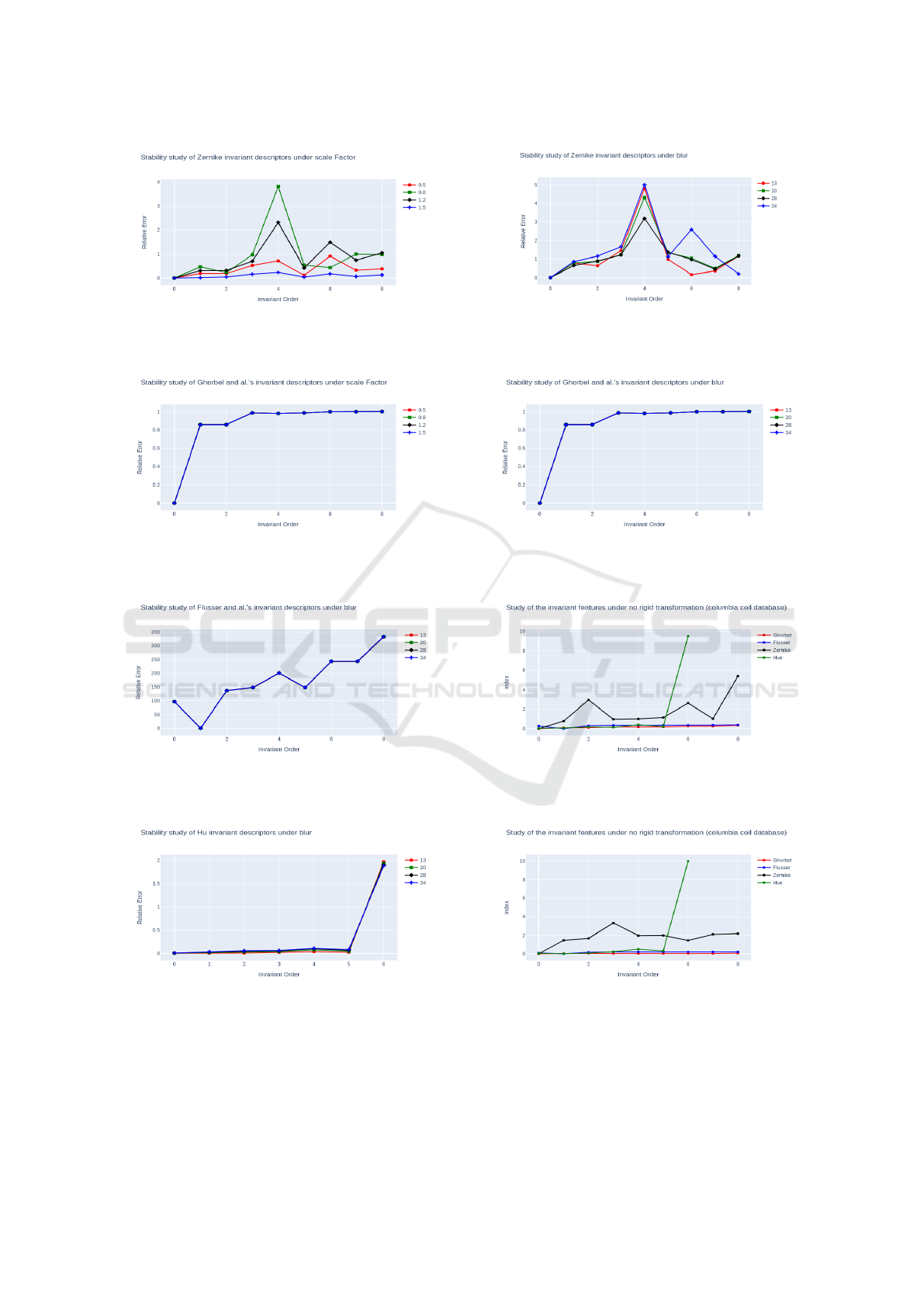
Figure 17: Stability study of Zernike invariant moments un-
der scale factor.
Figure 18: Stability study of Ghorbel invariant moments
under scale factor.
Figure 19: Stability study of Flusser invariant moments un-
der blur.
Figure 20: Stability study of Hu invariant moments under
blur.
Figure 21: Stability study of Zernike invariant moments un-
der blur.
Figure 22: Stability study of Ghorbel invariant moments
under blur.
Figure 23: Study of the invariant features under non-rigid
transformation (duck image) between (a) and (b).
Figure 24: Study of the invariant features under non-rigid
transformation (duck image) between (a) and (c).
An Improved VGG16 Model Based on Complex Invariant Descriptors for Medical Images Classification
451

REFERENCES
Cao, X., Gao, S., Chen, L., and Wang, Y. (2020). Ship
recognition method combined with image segmen-
tation and deep learning feature extraction in video
surveillance. Multimedia Tools and Applications,
79:9177–9192.
Derrode, S. and Ghorbel, F. (2001). Robust and effi-
cient fourier-mellin transform approximations for in-
variant grey-level image description and reconstruc-
tion. Computer Vision and Image Understanding,
83(1):57–78.
Flusser, J. (2002). On the inverse problem of rotation mo-
ment invariants. Pattern Recognition, 35(12):3015–
3017.
Flusser, J. (2006). Moment invariants in image analysis. In
proceedings of world academy of science, engineering
and technology, volume 11, pages 196–201. Citeseer.
Ghorbel, F. (1994). A complete invariant description for
gray-level images by the harmonic analysis approach.
Pattern Recognit. Lett., 15(10):1043–1051.
Ghorbel, F. (1998). Towards a unitary formulation for in-
variant image description: application to image cod-
ing. Annals of telecommunications, 53(5):242–260.
Ghorbel, F., Derrode, S., Mezhoud, R., Bannour, T.,
and Dhahbi, S. (2006). Image reconstruction from
a complete set of similarity invariants extracted
from complex moments. Pattern recognition letters,
27(12):1361–1369.
Hu, M.-K. (1962). Visual pattern recognition by moment
invariants. IRE transactions on information theory,
8(2):179–187.
Huang, S.-C., Pareek, A., Seyyedi, S., Banerjee, I., and
Lungren, M. P. (2020). Fusion of medical imaging and
electronic health records using deep learning: a sys-
tematic review and implementation guidelines. NPJ
digital medicine, 3(1):136.
Khotanzad, A. and Hong, Y. H. (1990). Invariant im-
age recognition by zernike moments. IEEE Trans-
actions on pattern analysis and machine intelligence,
12(5):489–497.
Moung, E. G., Hou, C. J., Sufian, M. M., Hijazi, M. H. A.,
Dargham, J. A., and Omatu, S. (2021). Fusion of mo-
ment invariant method and deep learning algorithm for
covid-19 classification. Big Data and Cognitive Com-
puting, 5(4):74.
Nene, S. A., Nayar, S. K., Murase, H., et al. (1996).
Columbia object image library (coil-20).
Persoon, E. and Fu, K.-S. (1977). Shape discrimination us-
ing fourier descriptors. IEEE Transactions on systems,
man, and cybernetics, 7(3):170–179.
Ren, Y., Yang, J., Zhang, Q., and Guo, Z. (2021). Ship
recognition based on hu invariant moments and con-
volutional neural network for video surveillance. Mul-
timedia Tools and Applications, 80:1343–1373.
Sharma, S. and Mehra, R. (2020). Conventional ma-
chine learning and deep learning approach for multi-
classification of breast cancer histopathology im-
ages—a comparative insight. Journal of digital imag-
ing, 33:632–654.
Sheng, Y. and Shen, L. (1994). Orthogonal fourier–mellin
moments for invariant pattern recognition. JOSA A,
11(6):1748–1757.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Suk, T. and Flusser, J. (2003). Combined blur and affine
moment invariants and their use in pattern recognition.
Pattern Recognition, 36(12):2895–2907.
Wang, Y.-B., You, Z.-H., Yang, S., Yi, H.-C., Chen, Z.-H.,
and Zheng, K. (2020). A deep learning-based method
for drug-target interaction prediction based on long
short-term memory neural network. BMC medical in-
formatics and decision making, 20(2):1–9.
Zahn, C. T. and Roskies, R. Z. (1972). Fourier descriptors
for plane closed curves. IEEE Transactions on com-
puters, 100(3):269–281.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
452
