
A Mutual Information Based Discretization-Selection Technique
Artur J. Ferreira
1,3 a
and M
´
ario A. T. Figueiredo
2,3 b
1
ISEL, Instituto Superior de Engenharia de Lisboa, Instituto Polit
´
ecnico de Lisboa, Portugal
2
IST, Instituto Superior T
´
ecnico, Universidade de Lisboa, Portugal
3
Instituto de Telecomunicac¸
˜
oes, Lisboa, Portugal
fi
Keywords:
Bit Allocation, Classification, Explainability, Feature Discretization, Feature Selection, Machine Learning,
Mutual Information, Supervised Learning.
Abstract:
In machine learning (ML) and data mining (DM) one often has to resort to data pre-processing techniques to
achieve adequate data representations. Among these techniques, we find feature discretization (FD) and fea-
ture selection (FS), with many available methods for each one. The use of FD and FS techniques improves the
data representation for ML and DM tasks. However, these techniques are usually applied in an independent
way, that is, we may use a FD technique but not a FS technique or the opposite case. Using both FD and FS
techniques in sequence, may not produce the most adequate results. In this paper, we propose a supervised
discretization-selection technique; the discretization step is done in an incremental approach and keeps infor-
mation regarding the features and the number of bits allocated per feature. Then, we apply a selection criterion
based upon the discretization bins, yielding a discretized and dimensionality reduced dataset. We evaluate our
technique on different types of data and in most cases the discretized and reduced version of the data is the
most suited version, achieving better classification performance, as compared to the use of the original fea-
tures.
1 INTRODUCTION
In machine learning (ML) and data mining (DM)
when dealing with large amounts of data, one may
need to apply data pre-processing methods to obtain a
more suitable representation (Ram
´
ırez-Gallego et al.,
2017). For the data pre-processing stage, there are
many available techniques, among which we find fea-
ture discretization (FD) and feature selection (FS)
techniques (Duda et al., 2001; Guyon et al., 2006;
Guyon and Elisseeff, 2003).
Both FD and FS are vast research fields with
many techniques available in the literature. How-
ever, many efforts continue to be developed on those
fields (Alipoor et al., 2022; Chamlal et al., 2022;
Huynh-Cam et al., 2022; Jeon and Hwang, 2023).
Learning on high-dimensional (HD) data is a chal-
lenge, due to the curse of dimensionality (Bishop,
1995), which poses many difficulties to the problem
of finding the best features and their best represen-
tation, among a large set of features. It is known
from the literature that the use of FD and FS im-
a
https://orcid.org/0000-0002-6508-0932
b
https://orcid.org/0000-0002-0970-7745
proves the performance of ML and DM tasks (Witten
et al., 2016). Often, researchers develop independent
FD or FS techniques without addressing their joint or
combined use. However, it is expected that an ade-
quate combination of discretization and selection pro-
cedures would provide better results than the indepen-
dent use of these techniques.
In this paper, we propose the mutual information
discretization-selection (MIDS) algorithm, which is a
hybrid discretization-selection technique. An FS fil-
ter is applied on the discretized data, guided by the
discretization stage details, yielding a discretized and
reduced dataset suitable for learning. The variable
number of discretization bins assigned to each fea-
ture provides a hint on the explainability and on the
importance of each feature.
The remainder of this paper is organized as fol-
lows. In Section 2, we overview related work and
techniques for FS and FD. The proposed approach
and its key insights are described in Section 3. The
experimental evaluation procedure is reported in Sec-
tion 4. Finally, Section 5 ends the paper with conclud-
ing remarks and directions of future work.
436
Ferreira, A. and Figueiredo, M.
A Mutual Information Based Discretization-Selection Technique.
DOI: 10.5220/0012467300003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 436-443
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

2 RELATED WORK
We briefly review some related work regarding key
aspects of FS and FD techniques over the past years.
In Section 2.1, we describe the key notation fol-
lowed in this paper. Section 2.2 overviews the use of
FS techniques with emphasis on filter techniques ad-
dressed in the experimental evaluation. Finally, Sec-
tion 2.3 overviews FD techniques with some details
about the techniques considered in this work.
2.1 Notation and Terminology
In this paper, we use the following terminology. Let
X = {x
1
, . . . , x
n
} be a dataset, with n patterns. Each
pattern denoted as x
i
is a d−dimensional vector, with
d being the number of features. Each dataset X is
represented by a n × d matrix; the rows hold the pat-
terns, while the columns are the features, denoted as
X
i
. We denote the number of distinct class labels as C,
with c
i
∈ {1, . . . ,C} being the class of pattern i and
y = {c
1
, . . . , c
n
} is the set of class labels.
2.2 Feature Selection
FS techniques bring many benefits to ML and DM
tasks, the accuracy of a classifier is often improved
mitigating the effects of the curse of dimensionality
and training is faster (Guyon et al., 2006; Guyon and
Elisseeff, 2003). Over the past years, many different
FS algorithms have been proposed. These algorithms
are usually placed into one of four categories: filter,
wrapper, embedded, and hybrid.
Filter methods check the adequacy of a feature
subset using characteristics of that subset, without
resorting to any learning algorithm and keep some
of the features, discarding others. Thus, filter ap-
proaches are agnostic in the sense that they do not re-
sort to any learning algorithm. In some big-data and
high-dimensional datasets, filters are often the only
suitable category of FS methods to be used. One
of the most successful filters is the fast correlation-
based filter (FCBF), under the relevance-redundancy
(RR) framework for the FS task (Yu and Liu, 2004).
FCBF computes the feature-class and feature-feature
association. It selects a set of features highly related
with the class. In the first step, these features are
called predominant and the correlation is assessed by
the symmetrical uncertainty (SU), defined as
SU(U,V ) =
2MI(U;V )
H(U) + H(V)
, (1)
where H denotes the Shannon entropy and MI denotes
the mutual information (MI) (Cover and Thomas,
2006), where U and V are feature vectors or class la-
bel vectors. SU is zero for independent random vari-
ables and one for deterministically dependent random
variables. In the second step, a redundancy detection
procedure finds redundant features among the pre-
dominant ones. These redundant features are further
split, keeping the ones that are the most relevant to
the class. For recent surveys on FS techniques, please
see (Pudjihartono et al., 2022; Dhal and Azad, 2022).
2.3 Feature Discretization
FD is a research field with many available unsuper-
vised and supervised techniques, following different
criteria. We briefly review some aspects of FD meth-
ods with emphasis on supervised techniques.
Many datasets have real-valued features; however,
some classification algorithms can only deal with dis-
crete/categorical features, and for this reason a dis-
cretization procedure is necessary. FD techniques aim
at finding a representation of each feature that con-
tains enough information for the learning task at hand,
while ignoring minor fluctuations that may be irrele-
vant for that task. In a nutshell, FD seeks more com-
pact and better representations of the data for learning
purposes. As a consequence, these discrete features
usually lead to both better accuracy and lower train-
ing time, as compared to the use of the original fea-
tures (Dougherty et al., 1995; Biba et al., 2007; Tsai
et al., 2008; Garc
´
ıa et al., 2013; Witten et al., 2016;
Ram
´
ırez-Gallego et al., 2017). Supervised FD ap-
proaches use class label information to compute the
cut-points in the discretization process.
The information entropy maximization (IEM)
method (Fayyad and Irani, 1993) is one of the old-
est and most used FD techniques. It assumes that the
most informative features to discretize are the most
compressible ones, which is an entropy minimization
heuristic. It works in a recursive approach computing
the discretization cut-points in such a way that it min-
imizes the number of bits to represent each feature.
The static class-attribute interdependence maxi-
mization (CAIM) algorithm (Kurgan and Cios, 2004)
aims to maximize the class-attribute interdependence
and to generate a (possibly) minimal number of dis-
crete intervals.
The class-attribute contingency coefficient
(CACC) (Tsai et al., 2008) is based on the maximiza-
tion of a modification of the contingency coefficient,
overcoming the key drawbacks of earlier schemes
such as CAIM.
The mutual information discretization (MID) al-
gorithm was proposed by Ferreira and Figueiredo
(2013). It performs supervised FD with q bits per fea-
A Mutual Information Based Discretization-Selection Technique
437
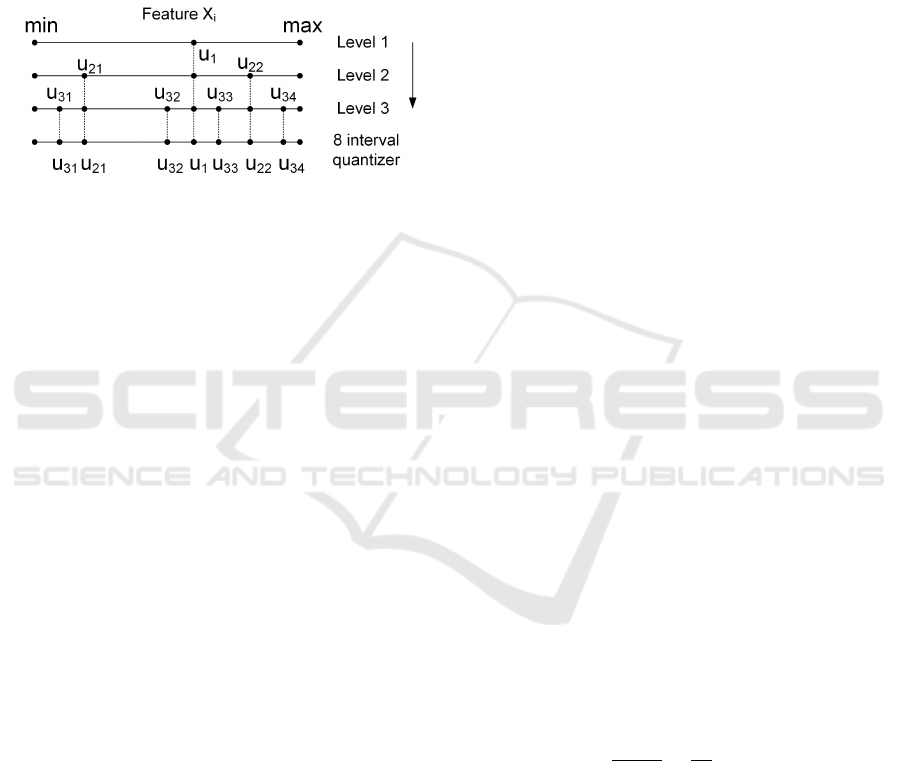
ture, based on the assessment of the MI between the
discretized feature and the class label vector y. MID
searches for discretization intervals such that the re-
sulting discrete feature has the highest MI as possible
with the class label vector. Each feature is discretized
individually, by successively breaking its range of val-
ues into intervals with boundaries u
i j
, as depicted in
Figure 1 for a given feature, say X
i
, with q = 3 bits
(8-intervals), yielding the discretized feature
e
X
i
.
Figure 1: Mutual Information Discretization (MID) of one
feature with q = 3 bits, for a 8-interval discretized feature,
proposed by Ferreira and Figueiredo (2013).
The maximum value for MI(
e
X
i
;y) depends on
both the number of bits used to discretize X
i
and
the number of classes C. If we discretize X
i
with b
i
bits, its maximum entropy is H
max
(
e
X
i
) = b
i
bit/symbol; the maximum value of the class entropy
is H
max
(y) = log
2
(C) bit/symbol, which corresponds
to C equiprobable classes. Thus, the maximum value
of the MI between the class label and a discretized
feature (with b
i
bits) is upper bounded as
max{MI
(
e
X
i
;y)
} ≤ min{b
i
, log
2
(C)}. (2)
To ensure that the discretization process attains the
maximum possible value for the MI, one must choose
the maximum number of bits q following this expres-
sion. Thus, we have q ≥ ⌈log
2
(C)⌉, where ⌈.⌉ is the
ceiling operator.
3 PROPOSED APPROACH
In this section, we describe the key ideas of our ap-
proach and present them with an algorithm in Sec-
tion 3.1. We also show some insights on the dis-
cretization behavior for different datasets, in Sec-
tion 3.2.
3.1 Key Ideas and Algorithms
First, we state the key ideas of our proposal and then
we present it in an algorithmic style. For a dataset,
with n instances and d features, we use a static dis-
cretizer @disc, a relevance function @rel, and a fea-
ture selection technique @ f s, as follows:
• Use of the static discretizer @disc technique over
the training data, with an increasing number of
bits per feature.
• Discretize each feature starting with one bit (a bi-
nary feature) up to a maximum number of bits.
• For each feature, find the minimum number of bits
such that maximizes @rel between the discretized
feature and the class label vector.
• For each feature, obtain its discretized version
with the minimum number of bits.
• Apply a feature selection technique @ f s on the
discretized features, resorting to the discretization
information.
We use the MID algorithm as @disc and @rel as
the MI between the discretized feature vector and the
class label vector. Algorithm 1 presents our proposal,
named as mutual information discretization-selection
(MIDS).
After using MIDS on a training set, we get a dis-
cretized and dimensionality reduced version. On the
FS procedure @ f s, we use filter techniques, namely:
• A relevance-based approach, which we denote as
MIDS, for simplicity.
• A relevance-redundancy approach, denoted as
MIDSred, performing a redundancy analysis af-
ter the discretization step.
On the relevance-based approach, we use a thresh-
old parameter to select the top-m features, from the
original set of d discretized features. We use a cu-
mulative relevance (CR) criterion as follows. Let
r
i
1
, ..., r
i
d
be the sorted relevance values and
c
l
=
l
∑
f =1
r
i
f
, (3)
be the CR of the top l most relevant features. We
select the number of features as the lowest value m
that satisfies the condition
m
∑
f =1
r
i
f
d
∑
i=1
r
i
=
c
m
c
d
≥ T
h
, (4)
where T
h
is a threshold (e.g., 0.95), leading to a frac-
tion of the top-m ranked features. The relevance is
the MI value computed on the discretization stage.
On the relevance-redundancy approach, we follow the
ideas by Ferreira and Figueiredo (2012). After com-
puting the sorted relevance values, we assess the re-
dundancy between the most relevant features. At the
end, we keep features with high relevance and low re-
dundancy, below some threshold, named as maximum
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
438

Algorithm 1: Supervised mutual information discretization-selection (MIDS).
Input: X, n patterns of a d-dimensional training set.
y, n-length vector with class labels.
q
m
, the maximum number of bits per feature.
@ f s, a feature selection procedure.
Output:
e
X
′
, the discrete and reduced training set with n patterns and m dimensions.
Q
1
b
1
, ..., Q
m
b
m
, set of m quantizers (one per feature).
b, m-length vector with the number of bits per feature.
1: For all features X
i
, with i ∈ {1, . . . , d}, compute their q
m
discretized versions with q bits per feature, with q ∈ {1, . . . , q
m
},
using MID as described in Figure 1.
2: Compute the MI matrix, M, with dimensions q
m
× d. Each element of M, denoted as M
ji
, holds the MI between feature
X
i
discretized with j bits, and the class label vector y.
3: For each feature, identify the minimum number of bits that yields the maximum MI, denoted as b
j
. For each column of
M, locate the first row (from top to bottom) that achieves the maximum MI. Create a d-dimensional vector b with the
minimum number of bits per feature, b
j
.
4: Quantize each feature X
i
using MID with b
j
bits,
e
X
i
= Q
i
b
j
(X
i
).
5: Apply the FS procedure @ f s to
e
X, reducing its dimensionality from d to m, yielding
e
X
′
.
6: Return the discretized/reduced training set
e
X
′
, the quantizers Q
i
b
j
, and the vector with the number of bits per feature b.
similarity (M
S
). The redundancy is assessed with the
MI between two feature vectors.
3.2 Discretization Analysis
We now observe the contents of the M matrix defined
in Algorithm 1. Figure 2 shows the increase of the
MI between each feature X
i
and the class label vec-
tor y, as a function of the number of bits per fea-
ture, q ∈ {1, . . . , 6}, for the Wine dataset with d = 13
features and C = 3 distinct labels. On the left-hand-
side, we have an image with the MI value with a color
scale; the image rows are the number of bits per fea-
ture q and the columns correspond to the d features.
For most features, we have an increment on the MI
value, as we increase q, stopping at some point. For
feature number 7, we observe a clear increase on the
MI when discretizing up to 4 bits. For feature number
3, we have the opposite behavior, with no increase on
the MI, regardless of the increase in q. On the right-
hand-side, the top plot shows the maximum MI after
discretization and the bottom plot exhibits the mini-
mum number of bits that yields maximum MI; no fea-
ture requires more than 4 bits and most features use 3
bits. There is a large variability on the MI values for
all the features.
Figure 3, left-hand-side, shows the histogram of
the final MI values after discretization. On the right-
hand-side, we display the histogram of the b vector
elements. This assessment is done with the Colon
dataset with d = 2000 features and C = 2 distinct la-
bels. The MI values range from 0.0103 to 0.5954,
with a peak located around 0.1. Requiring 1, 2, 3, and
4 bits per feature, we have 86, 1134, 761, and 19 fea-
tures, respectively. No feature requires more than 4
bits for discretization.
We now check for the association between the
highest MI values and the minimum number of bits
per feature, in vector b, using scatter plots. Figure 4
depicts the value of MI as a function of the number
of bits per feature, for the Wine and Colon datasets,
with up to q = 6 bits per feature. No feature requires
more than 4 bits. There is no strong correlation be-
tween the MI value and the number of bits per fea-
ture. We have many features that achieve a high value
of MI with q = 1. Other features achieve higher MI
with q = 3 than with q = 4, which means that 3 bits is
sufficient to achieve the maximum MI implying that
many features achieve their highest MI value with a
small number of bits.
4 EXPERIMENTAL EVALUATION
We now report the evaluation of our method. Sec-
tion 4.1 describes the datasets as well as the evalua-
tion metrics. In Section 4.2, we check the MIDS bit
allocation and compare it with other FD algorithms.
In Section 4.3, we check for the sensitivity of MIDS
with its input parameters. A FS assessment is pro-
vided in Section 4.4. Finally, Section 4.5 discusses
the findings of the experimental evaluation.
4.1 Datasets and Evaluation Metrics
Table 1 presents the datasets used in this work, with
different problems and diverse types of data. The
datasets are available at public repositories such as
University of California at Irvine (UCI) https://arch
A Mutual Information Based Discretization-Selection Technique
439
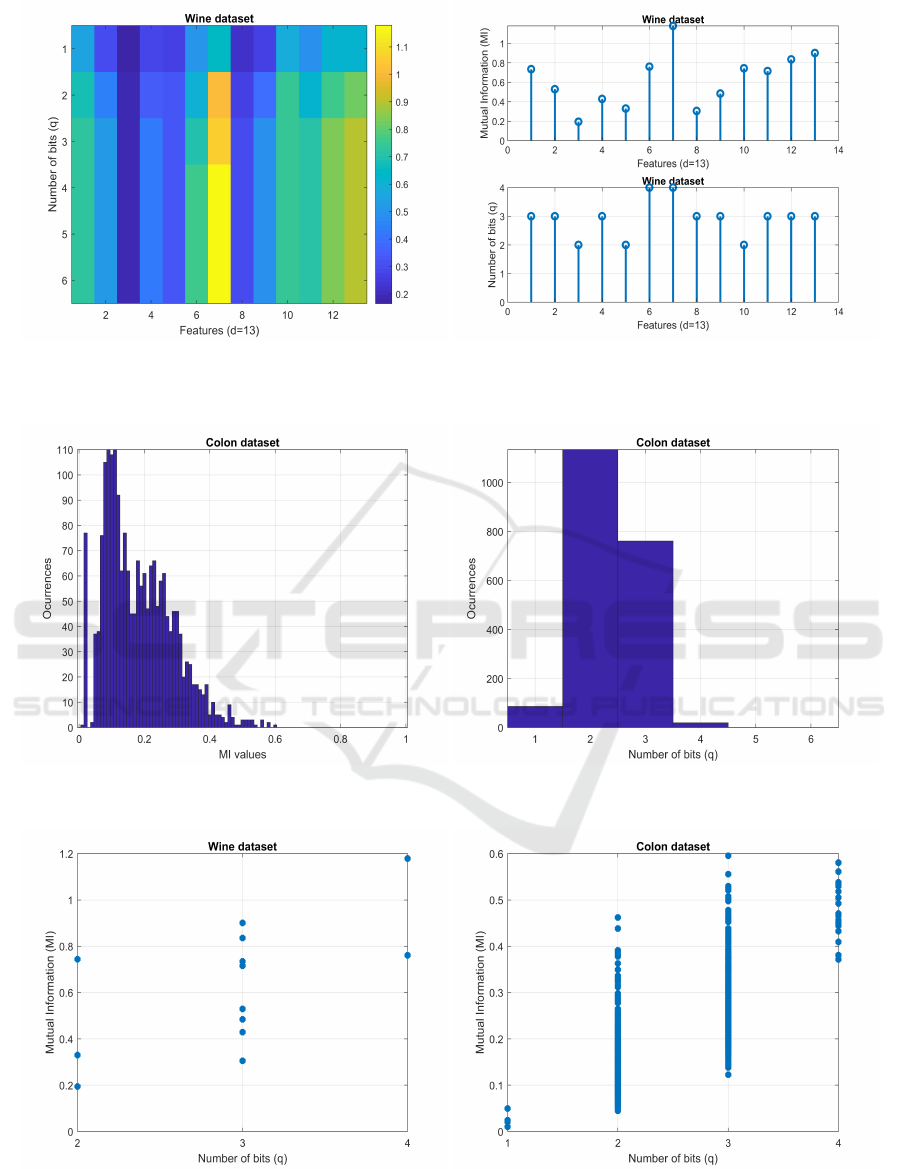
Figure 2: Discretization of the Wine dataset. Left: MI as a color scale image for the d = 13 features (image columns), as
functions of the number of bits per feature, q ∈ {1, . . . , 6} (image rows). Right: on top, the maximum MI between each
discretized feature and the class label vector; on bottom, the minimum number of bits that achieves the maximum MI.
Figure 3: Discretization of the Colon dataset. Left: histogram of the maximum MI values attained for the d = 2000 features.
Right: histogram of the number of bits allocated per feature.
Figure 4: Discretization of the Wine (left) and Colon (right) datasets. Scatter plot showing the MI value on the yy-axis as a
function of the number of bits per feature on the xx-axis.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
440
ive.ics.uci.edu/ml/index.php, the knowledge extrac-
tion evolutionary learning (KEEL), https://sci2s.ugr.
es/keel/datasets.php, and the ones available at https:
//csse.szu.edu.cn/staff/zhuzx/Datasets.html and https:
//jundongl.github.io/scikit-feature/datasets.html.
We report the test set error rate of 10-fold cross
validation for classification with the linear support
vector machines (SVM) classifier from Waikato en-
vironment for knowledge analysis (WEKA) (Frank
et al., 2016).
4.2 Bit Allocation per Feature
We assess the number of bits allocated to each feature
by the MIDS algorithm, setting T
h
= 1, that is, using
only discretization without performing feature selec-
tion. We aim to identify, for each dataset, the number
of bits allocated to each feature and the test error rate
of MIDS, compared with the original features and the
features discretized by the IEM, CAIM, and CACC
methods. Table 2 reports these results.
For most datasets, we observe that discretization
stops at q = 4 bits. Many datasets end up with many
features discretized with one bit; this may imply that
for these features it is mostly important if they are
present or absent (based on a given threshold), regard-
less of their exact value. The majority of features is
discretized with 2, 3, or 4 bits.
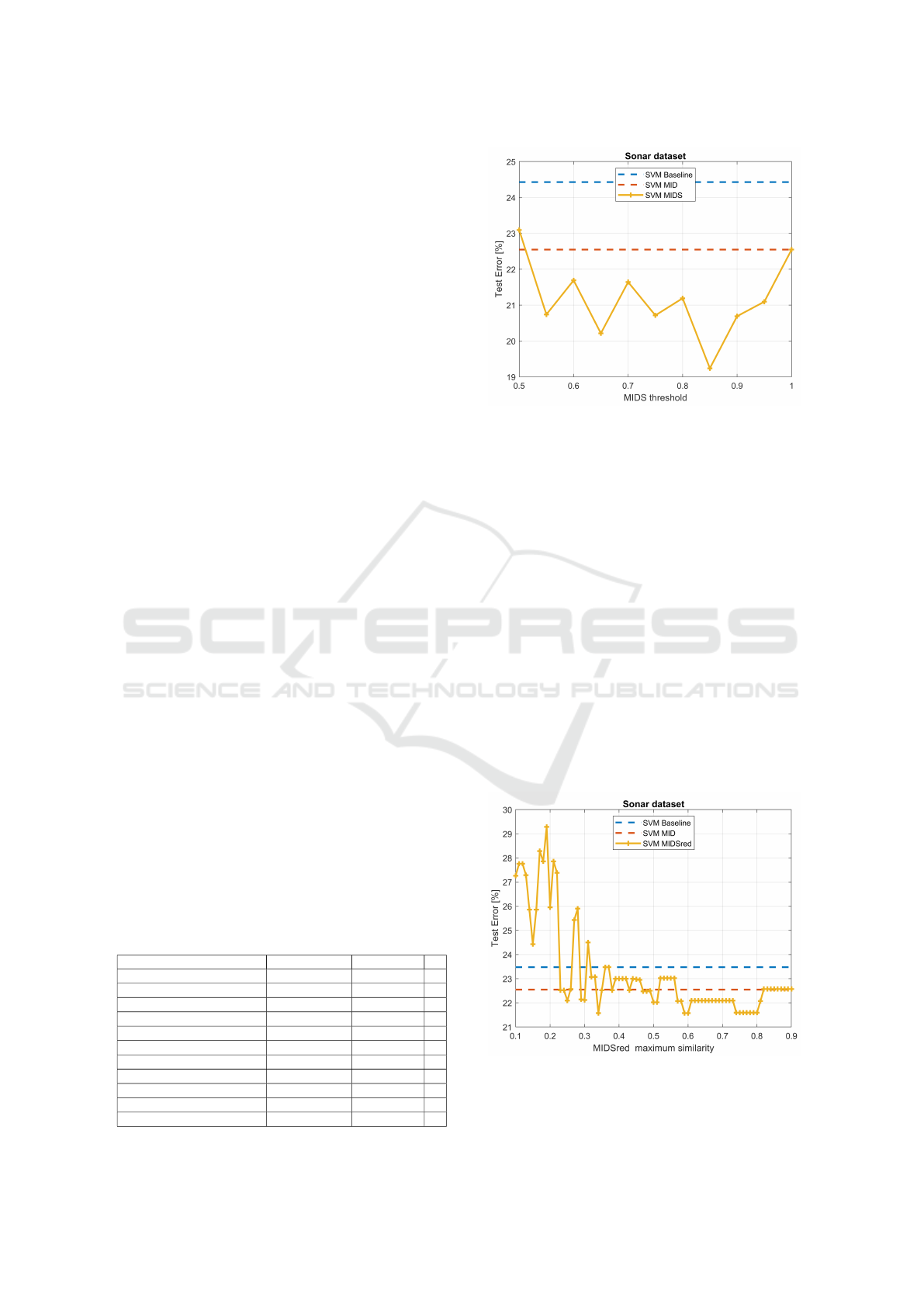
4.3 MIDS Parameter Sensitivity
We analyze the sensitivity of MIDS with the thresh-
old T
h
, for a fixed number of bits per feature. Figure 5
shows the test set error rate (10-fold CV), as a func-
tion of the T
h
parameter ranging from 0.5 to 1, with
q = 4, for the Sonar dataset, with the SVM classifier.
For this dataset, the optimal threshold value is
T
h
= 0.95, which yields the lowest error rate. We also
analyze how MIDSred behaves as a function of its
Table 1: Datasets with n instances (per class), d features
(numeric + nominal), and C classes.
∗
For the Dermatol-
ogy, SRBCT, and Wine datasets, the instance distribution
per class is 358 = 111 + 60 + 71 + 48 + 48 + 20, 83 =
29 + 25 + 11 + 18, and 178 = 59 + 71 + 48.
Dataset name and task n d C
Australian - credit card 690
(307+383)
14
(8+6)
2
Basehock - text classification 1993
(994+999)
4862
(4862+0)
2
Colon - cancer detection 62
(22+40)
2000
(2000+0)
2
Dermatology - skin disease 358
∗
34
(34+0)
6
DLBCL - cancer detection 77
(58+19)
5469
(5469+0)
2
Heart - coronary disease 270
(150+120)
13
(13+0)
2
Hepatitis - detection 155
(123+32)
19
(19+0)
2
Spambase - email spam 4601
(2788+1813)
54
(54+0)
2
Sonar - signals 208
(97+111)
60
(60+0)
2
SRBCT - cancer detection 83
∗
2308
(2308+0)
4
Wine - cultivar classification 178
∗
13
(13+0)
3
Figure 5: Test set error rate (10-fold CV) for the SVM clas-
sifier, on the Sonar dataset, with the original features, MIDS
(q = 4, T
h
= 1, no feature selection), and MIDS (q = 4 and
T
h
ranging from 0.5 to 1).
maximum similarity parameter, M
S
, for a fixed num-
ber of bits per feature. Figure 6 shows the test set
error rate (10-fold CV), as a function of the M
S
pa-
rameter ranging from 0.1 to 0.9, with q = 4, for the
Sonar dataset, with the SVM classifier. We find that
the optimal maximum similarity value is in the range
from 0.57 to 0.81, achieving the lowest error rate.
4.4 Discretization and Selection
We now report the experimental results regarding the
test set error rate for 10-fold CV, after feature dis-
cretization and selection with MIDS and MIDSred,
on Table 3, with the SVM classifier. We also ap-
ply the FCBF filter over the original and the dis-
cretized/selected data representation, to have a bench-
Figure 6: Test set error rate (10-fold CV) for the SVM clas-
sifier, on the Sonar dataset, with the original features, MIDS
(q = 4, T
h
= 1, no feature selection), and MIDSred (q = 4
and M
S
ranging from 0.1 to 0.9).
A Mutual Information Based Discretization-Selection Technique
441

Table 2: The average test error rate (%) with the linear SVM classifier with 10-fold CV, for the original features and the IEM,
CAIM, CACC, and MIDS discretized features. For MIDS we use T
h
= 1 (no FS) and we report the histogram of the number
of bits allocated to each feature, on the 10 folds, with q ∈ {1, . . . , 6}. The best result (lower error rate) is in boldface.
Error rate MIDS (T
h
= 1) allocated bits
Dataset Baseline IEM CAIM CACC MIDS q = 1 q = 2 q = 3 q = 4 q = 5 q = 6
Australian 14.49 14.49 44.49 14.49 14.49 69 33 25 13 0 0
Basehock 4.06 2.16 1.96 1.51 2.16 46408 2198 14 0 0 0
Colon 15.71 22.62 17.62 16.19 16.43 1029 11408 7403 160 0 0
Dermatology 2.23 2.52 5.31 2.23 1.39 330 0 10 0 0 0
DLBCL 2.50 5.18 5.18 2.50 2.50 4234 30421 19686 349 0 0
Heart 16.30 15.56 22.59 16.30 15.19 80 15 30 5 0 0
Hepatitis 19.88 18.54 19.83 19.83 16.63 132 27 26 5 0 0
Spambase 10.11 6.41 6.67 6.50 6.39 83 189 212 56 0 0
Sonar 22.12 21.12 22.10 21.64 16.31 18 181 339 62 0 0
SRBCT 0.00 0.00 1.25 1.25 0.00 73 3057 14531 5410 9 0
Wine 1.14 3.37 2.25 2.84 1.70 0 26 84 20 0 0
Average 9.86 10.17 13.56 9.57 8.47 - - - - - -
Table 3: The average test error rate (Err, %) and the average number of features (m) with the linear SVM classifier with 10-fold
CV, using the original features, MID discretization, MIDS discretization/selection, and MIDSred discretization/selection. We
also apply the FCBF filter over the original and discretized data. The best result (lower error and fewer features) is in boldface.
Baseline MID MIDS MIDSred FCBF MID-FCBF MIDS-FCBF MIDSred-FCBF
Dataset Err d Err m Err m Err m Err m Err m Err m Err m
Australian 14.49 14 14.64 14 14.64 8 14.49 7 14.49 6 14.49 6 14.49 5 14.49 5
Basehock 4.37 4862 2.61 4862 2.86 2597 2.71 2916 10.64 58 6.78 60 6.98 58 6.88 60
Colon 14.29 2000 17.62 2000 17.62 1490 17.38 970 15.95 14 22.86 22 22.86 22 17.86 21
Dermatology 2.51 34 1.40 34 1.40 27 4.20 19 3.92 13 6.17 11 6.16 8 7.83 6
DLBCL 2.68 5469 3.93 5469 3.93 4043 3.93 3141 6.61 63 3.93 101 3.93 101 2.68 103
Heart 15.93 13 14.81 13 13.70 9 15.56 6 17.41 5 15.56 6 15.56 5 17.04 4
Hepatitis 21.92 19 17.83 19 16.63 13 18.63 10 17.42 6 17.25 7 17.25 7 17.92 6
Spambase 10.13 54 6.37 54 6.67 41 7.39 31 13.19 13 7.98 13 8.09 11 8.74 10
Sonar 23.52 60 18.74 60 19.24 49 20.74 35 24.93 9 26.31 11 26.36 10 27.33 8
SRBCT 0.00 2308 0.00 2308 0.00 1865 0.00 1204 1.25 72 1.25 117 1.25 116 0.00 94
Wine 1.67 13 3.37 13 2.81 11 2.78 6 2.22 9 2.22 10 2.78 8 3.33 5
Average 10.14 1349 9.21 1349 9.04 923 9.80 758 11.64 24 11.34 33 11.43 32 11.28 29
mark comparison. We have chosen the FCBF filter,
because it is a successful technique for different types
of data. For MIDS and MIDSred we set the T
h
and
M
S
parameters as
(T
h
;M
S
) =
(0.95;0.70), if d < 100
(0.85;0.60), if d ≥ 100.
(5)
We have a generic trend that the discretized ver-
sions of the data usually lead to lower test set error,
as compared to the original representation. Moreover,
the MIDS discretized versions of the data with feature
selection usually attain better results than FS applied
over the original data representation.
4.5 Discussion
Our experimental evaluation was carried out on quite
different types of data to provide an overview on how
the proposed technique performs under different sce-
narios of binary and multi-class problems. We find
that the supervised discretization techniques are use-
ful for the majority of the datasets considered in these
experiments. The MID based discretization provides
comparable or better results as compared to existing
FD techniques. The use of selection right after dis-
cretization, using the maximum MI and the minimum
bits is an adequate criterion for most types of data. In
most cases with dense and sparse data, this approach
is preferable as compared to applying feature selec-
tion directly over the original data.
5 CONCLUSIONS
Feature discretization and feature selection tech-
niques often improve the performance of machine
learning algorithms. In this paper, we have proposed
a discretization-selection method, in which the selec-
tion criterion is based upon the discretization steps,
yielding a discretized and lower dimensionality ver-
sion of the data.
Our algorithm by itself or combined with other
techniques attains better results than feature selec-
tion algorithms applied directly on the original data.
These results show that discretization is an important
step to pre-process the data for accurate classification.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
442

On different types of data, our method shows in most
cases that the discretized and reduced version of the
data is suited for better classification performance.
The proposed technique allocates a variable num-
ber of bits per feature, showing that many features
reach its maximum possible mutual information with
the class label vector, using only a few bits. Thus, our
method is also suitable for explainability purposes as-
sessing the importance of a feature, given by the allo-
cated number of bits per feature. Some features only
require a binary representation (presence or absence
information) while other features demand more bits
for their accurate representation to maximize the mu-
tual information with the class label.
As future work directions, we aim to fine tune our
method to specific types of data. We also plan to ex-
plore R
´
enyi and Tsallis definitions of entropy and mu-
tual information and to fine tune their free parameters.
REFERENCES
Alipoor, G., Mirbagheri, S., Moosavi, S., and Cruz, S.
(2022). Incipient detection of stator inter-turn short-
circuit faults in a doubly-fed induction generator using
deep learning. IET Electric Power Applications.
Biba, M., Esposito, F., Ferilli, S., Di Mauro, N., and Basile,
T. (2007). Unsupervised discretization using kernel
density estimation. In International Joint Conference
on Artificial Intelligence (IJCAI), pages 696–701.
Bishop, C. (1995). Neural Networks for Pattern Recogni-
tion. Oxford University Press.
Chamlal, H., Ouaderhman, T., and Rebbah, F. (2022). A hy-
brid feature selection approach for microarray datasets
using graph theoretic-based method. Information Sci-
ences, 615:449–474.
Cover, T. and Thomas, J. (2006). Elements of information
theory. John Wiley & Sons, second edition.
Dhal, P. and Azad, C. (2022). A comprehensive survey on
feature selection in the various fields of machine learn-
ing. Applied Intelligence, 52(4):4543–45810.
Dougherty, J., Kohavi, R., and Sahami, M. (1995). Super-
vised and unsupervised discretization of continuous
features. In Proceedings of the International Confer-
ence Machine Learning (ICML), pages 194–202.
Duda, R., Hart, P., and Stork, D. (2001). Pattern classifica-
tion. John Wiley & Sons, second edition.
Fayyad, U. and Irani, K. (1993). Multi-interval discretiza-
tion of continuous-valued attributes for classification
learning. In Proceedings of the International Joint
Conference on Uncertainty in AI, pages 1022–1027.
Ferreira, A. and Figueiredo, M. (2012). Efficient feature
selection filters for high-dimensional data. Pattern
Recognition Letters, 33(13):1794 – 1804.
Ferreira, A. and Figueiredo, M. (2013). Relevance and mu-
tual information-based feature discretization. In Pro-
ceedings of the International Conference on Pattern
Recognition Applications and Methods (ICPRAM),
Barcelona, Spain.
Frank, E., Hall, M., and Witten, I. (2016). The WEKA Work-
bench. Online Appendix for ”Data Mining: Practical
Machine Learning Tools and Techniques”. Morgan
Kaufmann, fourth edition.
Garc
´
ıa, S., Luengo, J., S
´
aez, J. A., L
´
opez, V., and Herrera,
F. (2013). A survey of discretization techniques: Tax-
onomy and empirical analysis in supervised learning.
IEEE Transactions on Knowledge and Data Engineer-
ing, 25:734–750.
Guyon, I. and Elisseeff, A. (2003). An introduction to vari-
able and feature selection. Journal of Machine Learn-
ing Research (JMLR), 3:1157–1182.
Guyon, I., Gunn, S., Nikravesh, M., and Zadeh (Editors), L.
(2006). Feature extraction, foundations and applica-
tions. Springer.
Huynh-Cam, T.-T., Nalluri, V., Chen, L.-S., and Yang, Y.-
Y. (2022). IS-DT: A new feature selection method for
determining the important features in programmatic
buying. Big Data and Cognitive Computing, 6(4).
Jeon, Y. and Hwang, G. (2023). Feature selection with
scalable variational gaussian process via sensitivity
analysis based on L2 divergence. Neurocomputing,
518:577–592.
Kurgan, L. and Cios, K. (2004). CAIM discretization al-
gorithm. IEEE Transactions on Knowledge and Data
Engineering (TKDE), 16(2):145–153.
Pudjihartono, N., Fadason, T., Kempa-Liehr, A., and
O’Sullivan, J. (2022). A review of feature selection
methods for machine learning-based disease risk pre-
diction. Frontiers in Bioinformatics, 2:927312.
Ram
´
ırez-Gallego, S., Krawczyk, B., Garc
´
ıa, S., Wo
´
zniak,
M., and Herrera, F. (2017). A survey on data pre-
processing for data stream mining: Current status and
future directions. Neurocomputing, 239:39–57.
Tsai, C.-J., Lee, C.-I., and Yang, W.-P. (2008). A discretiza-
tion algorithm based on class-attribute contingency
coefficient. Information Sciences, 178:714–731.
Witten, I., Frank, E., Hall, M., and Pal, C. (2016). Data min-
ing: practical machine learning tools and techniques.
Morgan Kauffmann, 4th edition.
Yu, L. and Liu, H. (2004). Efficient feature selection via
analysis of relevance and redundancy. Journal of Ma-
chine Learning Research (JMLR), 5:1205–1224.
A Mutual Information Based Discretization-Selection Technique
443
