
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for
Classifying Olive Diseases in Images
Emna Guermazi
1,2,3
, Afef Mdhaffar
1,3
, Mohamed Jmaiel
1,3
and Bernd Freisleben
4
1
ReDCAD Laboratory, ENIS, University of Sfax, B. P. 1173 Sfax, Tunisia
2
National School of Electronics and Telecommunications of Sfax, University of Sfax, 3018 Sfax, Tunisia
3
Digital Research Center of Sfax, 3021 Sfax, Tunisia
4
Department of Mathematics and Computer Science, Philipps-Universit
¨
at, Marburg, Germany
Keywords:
Olive Disease Detection, Knowledge Distillation, Incremental Learning.
Abstract:
We present LIDL4Oliv, a novel lightweight incremental deep learning model for classifying olive diseases
in images. LIDL4Oliv is first trained on a novel annotated dataset of images with complex background.
Then, it learns from a large-scale deep learning model, following a knowledge distillation approach. Finally,
LIDL4Oliv is successfully deployed as a cross-platform application on resource-limited mobile devices, such
as smartphones. The deployed deep learning can detect olive leaves in images and classify their states as
healthy or unhealthy, i.e., affected by one of the two diseases “Aculus Olearius” and “Peacock Spot”. Our
mobile application supports the collection of real data during operation, i.e., the training dataset is continu-
ously augmented by newly collected images of olive leaves. Furthermore, our deep learning model is retrained
in a continuous manner, whenever a new set of data is collected. LIDL4Oliv follows an incremental update
process. It does not ignore the knowledge of the previously deployed model, but it (1) incorporates the current
weights of the deployed model and (2) employs fine-tuning and knowledge distillation to create an enhanced
incremental lightweight deep learning model. Our conducted experiments show the impact of using our com-
plex background dataset to improve the classification results. They demonstrate the effect of using knowledge
distillation in enhancing the performance of the deployed model on resource-limited devices.
1 INTRODUCTION
Agricultural crops have a significant nutritional role
in our lives. However, plant diseases threaten agricul-
tural yields and pose a considerable risk to the produc-
tion of food. Indeed, about 20% of global crop losses
stem from plant diseases (Savary et al., 2012; Ney
et al., 2013). This percentage has increased in the past
decade due to the influence of pollution and shifts in
climate patterns. To tackle this problem, researchers
have made significant efforts to identify and detect
plant disease at an early stage. The proliferation of
smart farming technologies has fostered the develop-
ment of innovative technologies to automatically de-
tect plant diseases, such as smartphone-assisted plant
disease diagnosis.
To build an effective mobile device solution for
plant disease detection, several researchers have pro-
posed to use appropriate deep learning models, ei-
ther (a) by hosting a deep learning model on a cloud
server and implementing a mobile application that in-
teracts with the deep learning model using a com-
munication protocol, or (2) by embedding the deep
learning model directly into a mobile application run-
ning on a mobile device. The first approach supports
the use of large-scale deep learning models running
on a server with substantial storage capacity and ex-
tensive computational resources. Moreover, this ap-
proach is not suitable for real-time agricultural appli-
cations. The second approach supports real-time agri-
cultural data analysis, but creating and deploying a
lightweight high-quality neural network is not an easy
task. Several researchers have created automated mo-
bile solutions that leverage the power of deep learn-
ing across various plant types. Some of these solu-
tions are specialized to detecting diseases in tomato
leaves (Aishwarya et al., 2023; Chandan et al., 2022).
Others identify diseases in corn plants (Hidayat et al.,
2019). Additionally, there are efforts to detect ter-
restrial plants in the Philippines using similar tech-
niques (Valdoria et al., 2019). Furthermore, some
researchers proposed solutions for disease detection
Guermazi, E., Mdhaffar, A., Jmaiel, M. and Freisleben, B.
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images.
DOI: 10.5220/0012466900003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 2, pages 583-594
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
583

of a variety of plants (Ahmed and Reddy, 2021; Gu-
rav et al., 2022). However, all these approaches do
not take into account the need to adapt the deployed
model to the changes seen in real-life images. More-
over, up to this point, a smart deep learning based mo-
bile application specifically designed to detect olive
leaf diseases is still absent. Detecting olive leaf dis-
eases is particular relevant for the Mediterranean and
Aegean regions that collectively contribute to about
95% of the world’s olive production (Soyyigit and
Yavuzaslan, 2018). This justifies the need for a smart
deep learning-based mobile application dedicated to
detecting olive diseases.
In this paper, we propose a novel lightweight in-
cremental mobile deep learning model to detect and
classify olive diseases in images. For classification,
we use the object detection model YOLOv5-n to de-
tect an olive leaf in an image and then proceed with
disease classification. We also create and annotate a
complex background dataset. This dataset improves
the capabilities of the deep learning model to rec-
ognize diseases under various settings and in images
with different realistic backgrounds. We use knowl-
edge distillation (KD) for object detection to enhance
the performance of YOLOv5-n. Next, we deploy our
lightweight neural network in a mobile application
dedicated to olive farm management and disease pre-
diction. Our approach supports sustainable data col-
lection and offers an incremental update process. This
process enables the model to always be up-to-date to
the features observed in real-life images.
This paper is structured as follows. Section 2 dis-
cusses related work. In Section 3, we present our ap-
proach, called LIDL4Oliv. Section 4 sketches imple-
mentation details. Section 5 presents the conducted
experiments and discusses the obtained results. Sec-
tion 5 concludes the paper and outlines some areas for
future research.
2 RELATED WORK
Detecting plant diseases is among the primary appli-
cations of smart agriculture. Deep learning models
are widely used for the detection and classification of
plant leaf diseases.
Several studies have shown a dedicated interest
in the development of deep learning models for the
recognition of olive leaf diseases. (Alshammari et al.,
2023) presented an approach to analyze olive leaves.
The authors used the Whale Optimization Algorithm
(WOA) to choose necessary features. Then, they clas-
sified the leaves using an optimized artificial neural
network (ANN). (Alshammari and Alkhiri, 2023) pre-
sented an approach that combines a recurrent neu-
ral network architecture with an ant colony optimiza-
tion algorithm. The ant colony optimization algo-
rithm was used to extract relevant features. Then, dis-
ease classification was performed using RNN models.
(Osco-Mamani and Chaparro-Cruz, 2023) employed
a modified VGG16 architecture for the classification
of olive leaf diseases in Tacna. (Mamdouh and Khat-
tab, 2022) used a deep learning model based on the In-
ceptionV3 architecture. (Ksibi et al., 2022) presented
MobiRes-Net, a neural network that combines the ar-
chitectures of ResNet50 and MobileNet. Although
these methods propose efficient deep learning mod-
els to classify olive leaf diseases, the complexity of
the developed deep learning models hinders their de-
ployment on resource-limited mobile devices, such as
smartphones. Moreover, none of these methods in-
volves a mobile application-based approach to detect
olive diseases automatically.
Research on mobile application approaches that
offer automated deep learning-based detection of
plant diseases can be classified into two categories.
The first category uses cloud servers to host deep
learning models (Ahmed and Reddy, 2021; Chan-
dan et al., 2022; Hidayat et al., 2019), while the
second category involves deploying a deep learning
model within the mobile application itself (Aishwarya
et al., 2023; Gurav et al., 2022; Valdoria et al., 2019).
(Chandan et al., 2022) developed a plant disease de-
tection system. This system integrated an Internet-
of-Things (IoT) platform to collect various measure-
ments, including humidity and temperature. Then,
these measurements were uploaded to a cloud server
for processing. Leaf images were also transmitted to
the cloud platform for analysis. Then, the authors em-
ployed a mobile app to display measurement values,
send SMS alerts, and provide notifications. (Hidayat
et al., 2019) developed an Android application for di-
agnosing corn diseases using deep learning. Here,
farmers select an image from their gallery, which is
then sent to a cloud server for processing. A convo-
lutional neural network (CNN) predicts the disease,
and the diagnosis is subsequently displayed in the mo-
bile application. (Ahmed and Reddy, 2021) proposed
a cross-platform mobile application designed to de-
tect infected plants. The mobile app enables taking
a picture of the leaf via the camera or uploading an
existing image from the smartphone. Then, a cloud
server uses a CNN model for disease detection. Sub-
sequently, the diagnosis of the disease is displayed
in the app. The methods belonging to the first cate-
gory successfully achieve automatic plant disease de-
tection, but their approaches necessitate the availabil-
ity of extensive computing resources and cannot be
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
584
Complex-background
dataset
Initial lightweight
model
Deployment on
mobile-app
Data-collection
Incremental update
process
Large-scale
model
Triggers
Incremented model
1
2
3
4
5
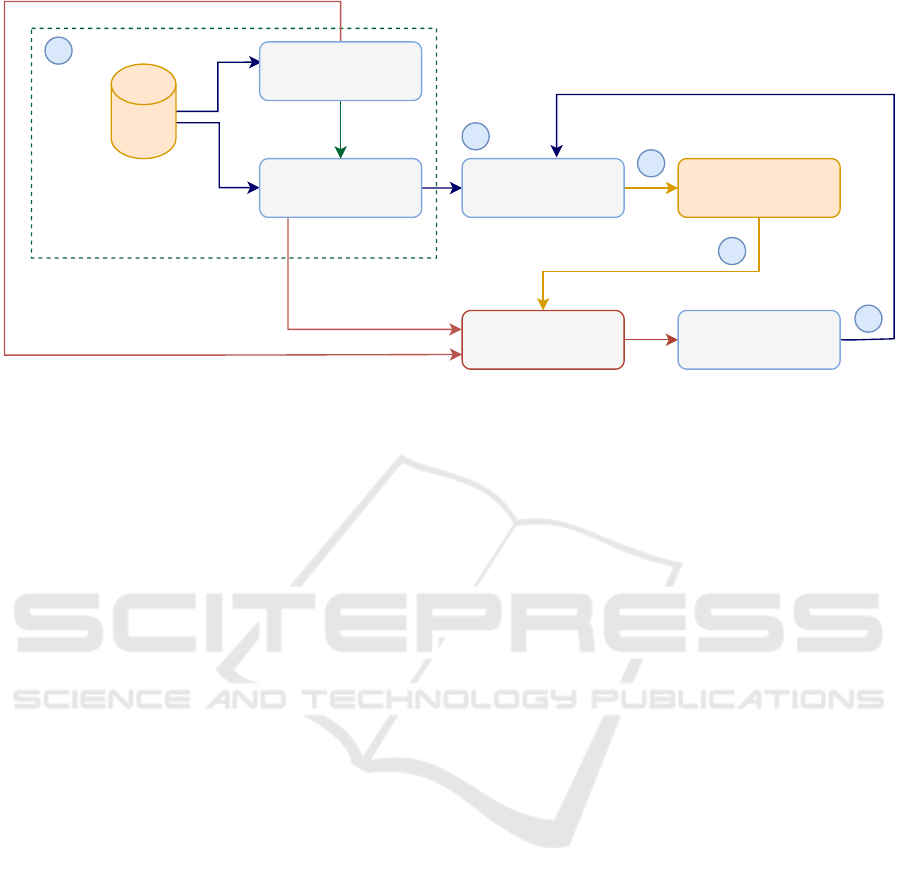
Figure 1: LIDL4Oliv.
easily deployed on resource-limited mobile devices.
The second category of approaches addresses this
issue by developing a lightweight neural network and
deploying it directly in a mobile application. (Valdo-
ria et al., 2019) proposed a mobile application named
“iDahon”. This app aims to improve terrestrial dis-
ease classification and diagnosis in the Philippines us-
ing a deep learning model. (Aishwarya et al., 2023)
suggested using a custom CNN model to classify
tomato leaf diseases. The application uses a Flask
API to establish communication between the user in-
terface and the underlying model. (Gurav et al., 2022)
presented an Android application that offers different
functionalities. First, it enables farmers to buy and
sell saplings. Moreover, it enables the plants’ dis-
ease detection via employing a lightweight Tensor-
Flow Lite based deep learning model. It also en-
ables the farmers to establish an agricultural com-
munity. Additionally, it offers up-to-date weather
forecasts for the duration of one week and delivers
real-time GPS tracking by utilizing satellite informa-
tion sourced from OpenWeatherMap. Although the
methods in the second category effectively integrate
lightweight neural network models into mobile ap-
plications, they fail to take into account the models’
need to accommodate changes occurring in real-life
images.
In contrast to the previously mentioned state-of-
the-art methods, we present an incremental update
process that allows us to maintain an updated deep
learning model that is well-adapted to the features of
real-life images. Furthermore, we ensure sustainable
data collection, as opposed to other mobile disease di-
agnosis solutions.
3 LIDL4Oliv
Our approach, called LIDL4Oliv, is based on an incre-
mental lightweight deep learning model that detects
and classifies olive leaf diseases via a cross-platform
mobile app. Figure 1 illustrates LIDL4Oliv (i.e.,
Lightweight Incremental Deep Learning model for
classifying Olive diseases). Our approach involves
five phases. The first phase includes several steps:
(i) Initially, we identify our deep learning model that
will be used for deployment. (ii) Then, we create
our complex background dataset. (iii) Next, we iden-
tify a large-scale deep learning model that is used as
our teacher model. (iv) Subsequently, we use knowl-
edge distillation to optimize the performance of our
student model. In the second phase, we deploy our
lightweight deep learning model as a cross-platform
mobile application. The third phase consists of con-
tinuously enriching our original dataset by newly pro-
cessed images. This ensures the creation of a sustain-
able dataset of olive leaf images. Upon accumulat-
ing 30% of new data relative to the original dataset,
the fourth phase is initiated where the deep learning
model undergoes an incremental update process to
enhance its performance, guaranteeing that it is con-
stantly adapted to the new features of the collected
images. In the fifth phase, we deploy our incremented
deep learning model in the mobile app. Each phase is
further detailed in the following sections.
3.1 Deep Learning Model Identification
Classification of olive diseases in images is a chal-
lenging task. In this paper, we use olive leaves to de-
tect and identify olive diseases. To solve this task,
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images
585

(a) Aculus Olearius (b) Aculus Olearius (c) Aculus Olearius (d) Peacock Spot
(e) Peacock Spot (f) Peacock Spot (g) Healthy (h) Healthy
Figure 2: Different olive leaf states (Uguz,S., 2020).
we employ an object detection model instead of a
classification model. This decision is motivated by
three reasons. First, real-life images usually contain
different backgrounds. Thus, it is more suitable to
make a distinction between the background and the
region of interest itself (i.e., the olive leaf) before
identifying the disease. Object detection first local-
izes the object and then proceeds with classification.
By following this process, we have a higher chance
of classifying the disease. Second, deploying a single
model and using it for both localization and classifi-
cation is more practical and resource-conserving than
deploying multiple models (initially a deep learning
model to eliminate the background, followed by an-
other model to classify the leaf disease). Third, an
object detection model can detect multiple objects per
image. However, in this case, we use it on images
where each image includes only one leaf. This is
due to the non-existence of an open real-life anno-
tated olive leaf dataset containing multiple leaves per
image. Thus, we decided to primarily create a dataset
where each image includes a background and solely
one leaf. This serves as the first set of training data
to train our initial model. Then, after deploying the
model as a mobile app, it is possible to collect realis-
tic images and annotate them on the basis of experts’
knowledge. Subsequently, we are able to adjust our
model accordingly to localize and classify multiple
leaves in an image.
We conducted a search for annotated olive leaf
images and identified an open dataset established by
(Uguz,S., 2020). This dataset was created for olive
leaf disease classification. It involves olive leaf im-
ages that correspond to three states (i.e., ‘Healthy” or
affected with “Aculus Olearius” or “Peacock Spot”).
However, it is composed of images having a simple
and unrealistic background, as illustrated in Figure 2.
Thus, relying on such a dataset would yield a deep
learning model that is not suitable for real-life olive
leaf disease detection. Given these circumstances, we
used the Uguz dataset (Uguz,S., 2020) and generated
a complex background dataset with bounding box an-
notations to localize a leaf and identify its state. The
next section explains the process of generating the
complex background dataset with bounding box an-
notations.
We use the YOLO architecture as our object de-
tection model. Indeed, contrary to two-stage detec-
tors like Faster R-CNN, YOLO’s architecture enables
us to detect multiple objects in a single pass through
the neural network, making it faster. Additionally,
YOLO achieves a good balance between speed and
accuracy, qualifying it for usage scenarios requiring
real-time processing. Moreover, YOLO is available
in different lightweight architectures, such as YOLO-
s and YOLO-n, which make it adequate for our use
case. We identified YOLOv5-n, following the work
of (Ye et al., 2023), as our model to use for deploy-
ment.
3.2 Generation of an Annotated
Complex Background Dataset
To create a dataset including leaf images with a com-
plex background, we followed two steps. First, we
constructed a dataset with a white background and
created the corresponding bounding box labels. Then,
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
586

we used an overlapping technique to generate im-
ages with complex background. These intricate back-
grounds correspond to images captured in the garden,
incorporating elements such as sand, hands, etc. Al-
though these backgrounds may not perfectly match
the context of photographing in the field, they allow
the model to detect the leaf within images containing
backgrounds. Thus, our model differentiates between
the background and the leaf, and identifies the state of
the leaf accurately. This dataset allows us to deploy
an initial model in real life (i.e., ready to be used by
farmers).
3.2.1 Generation of an Annotated White
Background Dataset
Our newly produced white background dataset is
based on Uguz’s dataset (Uguz,S., 2020). It incorpo-
rates bounding box annotations to train deep learning
models for object detection and classification, con-
trary to Uguz’s dataset that is solely used for im-
age classification. The original dataset is composed
of two folders: one for training and the second one
for testing. Within each folder, there are three sub-
folders. Each of these subfolders contains images that
correspond to a specific class of an olive leaf. This
class can be either “Healthy” or affected with either
”Aculus Olearius” or ”Peacock Spot”. In the remain-
der of this paper, we refer to the “bounding box an-
notations” by “annotations”, and to the true labels of
each class by “ground truth labels”.
To construct our dataset, we first applied data aug-
mentation techniques to Uguz’s dataset. This allowed
us to introduce greater image variations.
Then, we used the Canny method (Canny, 1986)
as a contour detection algorithm to annotate 31%
(1,479 images) of our dataset. We also used several
preprocessing techniques such as Gaussian blur, his-
togram equalization, and median filtering to create an-
notations. We developed a Tkinter (Steen Lumholt
and Guido van Rossum, 2009) interface to support the
selection of images with accurate bounding boxes, as
shown in Figure 3.
The Tkinter interface is used for validating the an-
notation of each image. For the rest of the images that
were not annotated, we used YOLOv5-m to complete
the annotation process. We employed the annotated
images, which represent 31% of the dataset, to train
the model. Then, we used the trained model to pro-
duce the annotations of the remaining images. YOLO
allowed us to annotate around 66% of the dataset, i.e.,
leaving 3% (i.e., 150 images) not annotated. The re-
maining images were manually annotated, via the use
of LabelImg (Tzutalin, 2015), as shown in Figure 4.
We selected LabelImg due to its user-friendly simplic-
Figure 3: The Tkinter interface used to validate the bound-
ing box annotations.
ity.
Figure 4: Number of images annotated using different
methods.
3.2.2 Overlapping Process
The second step involves changing the white back-
ground of the dataset to a more complex one. To
guarantee the retention of the established annotations,
we undertook a meticulous overlapping process. This
process involves two steps. The first one consists of
generating masks from the white background dataset
using a pre-trained U2-Net model (Qin et al., 2020).
The generated masks are binary images where the
background is represented by black pixels, and the re-
gion of interest, which is the leaf, is represented by
white pixels. The employed U2-net model was pre-
trained on the DUTS dataset (Wang et al., 2017). Af-
ter the masks’ generation, we moved on to the sec-
ond step. This step involved combining the gener-
ated masks with other images that represent a com-
plex background. The resulting complex background
dataset enables the YOLO model to accurately detect
and classify the state of the olive leaves in intricate
and varied settings, paving the way for improved clas-
sification performance in real-life situations.
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images
587

(a) Splash screen (b) Prediction page (c) Prediction results (d) Trees’ monitoring page
Figure 5: Selection of some pages of the mobile application.
3.3 Initial Training Process
To enhance the performance of our lightweight neu-
ral network, i.e., YOLOv5-n, we incorporated knowl-
edge distillation into the training process. This tech-
nique is based on knowledge transfer from a large-
scale neural network referred to as teacher to a
lightweight neural network named student to boost
its performance. Its initial application was in image
classification tasks (Hinton et al., 2015; Gou et al.,
2023; Tian et al., 2020), subsequently expanding to
encompass object detection in images (Chen et al.,
2017; Kruthiventi et al., 2017). The integration of
knowledge distillation into object detection tasks sig-
nificantly boosted the performance of the employed
models (Li et al., 2023; Yadikar et al., 2023). How-
ever, it is still a challenging task.
Some approaches are based on global distillation
where the student tries to mimic the whole feature
maps extracted from the middle layers of the teacher.
However, relying on all the feature maps might lead
to learning irrelevant knowledge that would not help
the student in its task. Therefore, some researchers
showed interest in local feature distillation (Wang
et al., 2019) to arrive at detector distillation, propos-
ing the distillation of backbone features extracted
from local regions surrounding objects. We use this
approach to distill knowledge from YOLOv5-m to
YOLOv5-n.
3.4 Cross-Platform Mobile Application
We deployed our model as a cross-platform mobile
application (i.e., compatible with both Android and
iOS). This application is designed to enable automatic
olive leaf disease prediction while also establishing an
olive farm management system. It is called “Olive
Leaf Disease Detector”, as illustrated in Figure 5a.
The splash screen of the application allows users to ei-
ther explore or sign in. It provides them with multiple
functionalities based on their roles. A user can have
the role of an admin, a farmer, or an expert. An admin
assumes responsibility for managing other users and
the existing trees on the property. Indeed, the admin
adds trees to the system. Each added tree corresponds
to an existing tree in the field. Our application en-
ables the farmer to predict the state of an olive leaf
based on the prediction of a deployed deep learning
model. A farmer can either take a picture using the
camera of his or her smartphone or import it from his
or her gallery. Then, (s)he chooses the tree to which
the leaf is associated. The prediction page is shown in
Figure 5b. Subsequently, (s)he can either accept the
model’s prediction or seek an expert’s evaluation, as
illustrated in Figure 5c. Additionally, the farmer can
recommend an expert to the admin through a provided
form within the mobile application. The expert is re-
sponsible for answering the farmers’ requests and giv-
ing his or her opinion regarding the condition of spe-
cific leaves. Furthermore, (s)he is able to go through
all the leaves existing in the database via the mobile
application, enabling him or her to offer insights on
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
588
Student1
Dataset1
KD: Fine-grained
feature imitation
Dataset2
Fine-Tune
Student2
Fine-Tune
KD: Fine-grained
feature imitation
Teacher3
Fine-Tune
Student3
Fine-Tune
KD: Fine-grained feature
imitation
Incremental Update Process
Dataset3
Teacher1
Teacher2
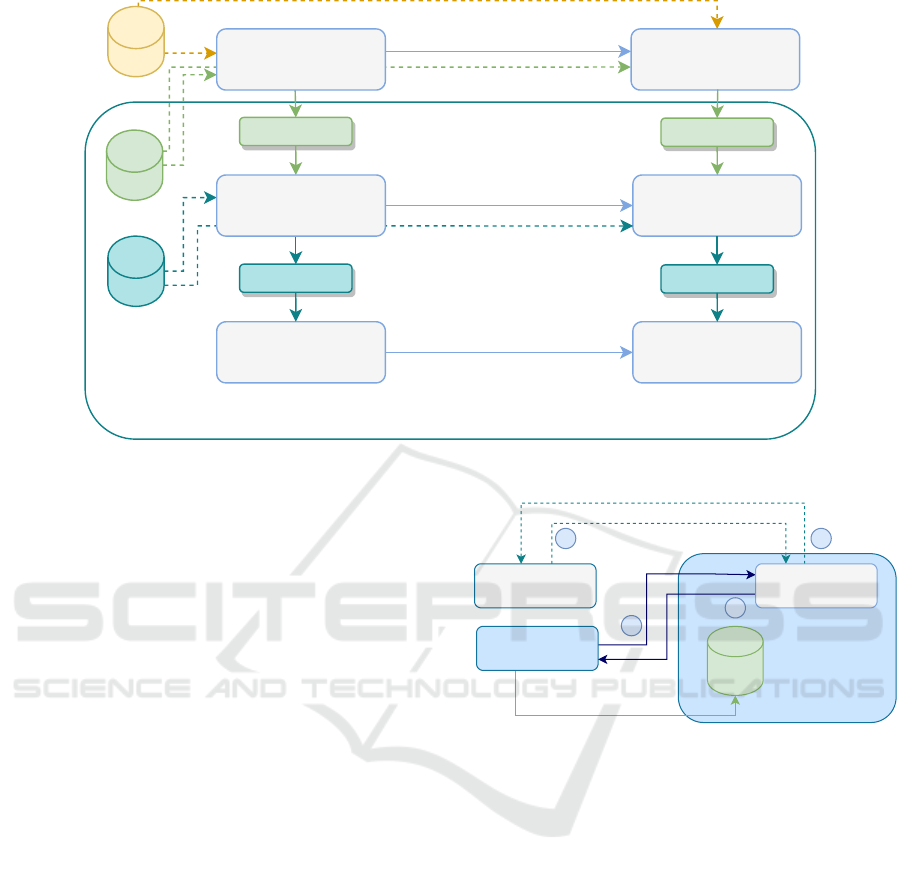
Figure 6: Incremental update process.
any leaf status. When a leaf has two predictions, one
provided by the deep learning model and the other
given by an expert, the status attributed to it will be
based on the expert’s opinion.
Furthermore, our application enables the monitor-
ing of the health of the existing trees. By default, all
trees are considered to be in a healthy state. How-
ever, when a leaf state is changed to “Aculus Olear-
ius” or “Peacock Spot”, the status of that correspond-
ing tree changes to sick. After the identification of an
unhealthy tree, it receives proper treatment. Then, a
farmer can access the mobile application and change
the status of the tree to healthy again. (S)he can also
delete all its leaves since they hold no meaning after
its status has changed. Furthermore, the farmer can
also delete the leaves after the season changes. Figure
5d presents the trees’ monitoring page. This appli-
cation also ensures a sustainable data collection. In-
deed, whenever a leaf status is predicted, the image is
saved with its corresponding label and bounding box
annotations in a remote server. Figure 7 illustrates
the communication process between the mobile app
screens (i.e., front-end) and the back-end, while also
representing the data collection process.
3.5 Incremental Update Process
To ensure the adaptability of our deployed model to
the features of the real-life images, we propose to use
an incremental update process. This process gives the
model the ability to evolve after gathering new data.
It involves a number of iterations as illustrated in Fig-
DL-powered
mobile app
Atlas Mongo DB
Distant Server
Node js Backend
Server
1
23
4
Data Collection
Figure 7: Global overview of the mobile application.
ure 6. Each iteration is initiated when the images of
new data reach 30% of the initial data. We retrain the
insights of the pre-trained teacher and the pre-trained
student by incorporating their pertained weights into
the update process. In each iteration, we fine-tune the
pre-trained teacher on dataset
i+1
, which represents
the newly gathered dataset, at iteration
i
. Thus, we ob-
tain a fine-tuned teacher. This teacher is referred to
as teacher
i+1
. We also apply fine-tuning on the pre-
trained student. The produced student is referred to as
student
i+1
. Then, we apply the same knowledge dis-
tillation method that we used in the initial step (Wang
et al., 2019) (i.e., fine-grained feature imitation). This
incremental update process reaches its end when a
student
i+1
(i.e., produced in iteration
i
) achieves good
performance when evaluated on a dataset
i+2
. It means
that this student model is well adapted to the features
of real-life images. This incremental process will be
implemented and experimentally validated upon col-
lecting real-life images through the mobile app and
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images
589
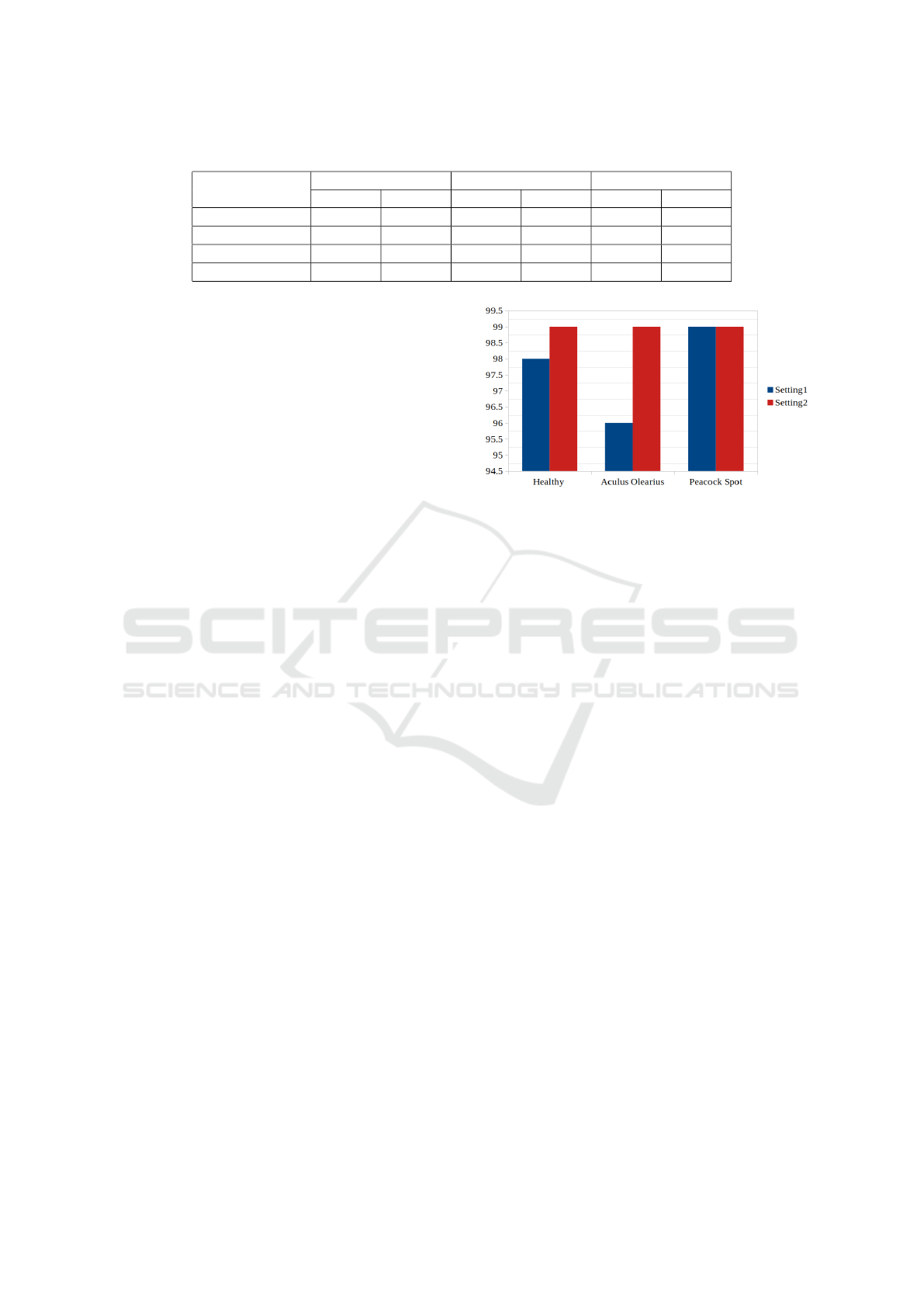
Table 1: Performance of YOLOv5-m: Setting 1 vs Setting 2.
Precision Recall mAP50-95
Setting1 Setting2 Setting1 Setting2 Setting1 Setting2
All 0.978 0.986 0.984 0.991 0.961 0.979
Healthy 0.988 0.987 0.989 0.994 0.955 0.972
Aculus Olearius 1 0.985 0.975 0.987 0.968 0.982
Peacock Spot 0.946 0.987 0.987 0.993 0.961 0.982
annotating them accordingly.
4 IMPLEMENTATION ISSUES
We implemented LIDL4Oliv using the SGD opti-
mizer with a momentum value of 0.9. The initial
learning rate, weight decay, and batch size were set
to 0.01, 0.0005, and 16, respectively. The training
and testing phases were conducted on an NVIDIA
RTX3080 GPU. For a fair comparison, all models un-
derwent training for 100 epochs.
5 EXPERIMENTAL EVALUATION
5.1 Experimental Settings
To evaluate our approach, we conducted a series of
preliminary experiments allowing us to identify our
experimental setting. These preliminary experiments
consist of training and evaluating the performance
of our teacher (i.e., YOLOv5-m) on the white back-
ground dataset, in two settings. Setting 1 involves
training the model from scratch, while in Setting 2 we
use pre-trained-model’s weights as a starting point.
These weights correspond to the YOLOv5-m model
after being trained on the COCO dataset (Lin et al.,
2014).
The main objective of this experiment is to iden-
tify the best setting to be used for the rest of the ex-
periments.
Table 1 and Figure 8 show the obtained results
when training YOLOv5-m in Setting 1 and Setting
2. Table 1 demonstrates that the overall precision, re-
call, and average mean precision (i.e., mAP50-90) are
higher when using Setting 2. These results also re-
veal that the recall and the mAP50-95 values of each
class individually exhibit higher values in compari-
son to Setting 1. However, the precision values of
the “Healthy” and the “Aculus Olearius” classes are
higher when using Setting 1. Figure 8 presents the
accuracy of predicting each class correctly. It shows
that Setting 2 shows a better classification accuracy
for the “Healthy” and the “Aculus Olearius” classes,
Figure 8: Classification accuracy of YOLOv5-m: Setting 1
vs Setting 2.
while, the accuracy of predicting the class “Peacock
spot” is the same for both settings. Thus, we can con-
clude that Setting 2 enables the model to achieve bet-
ter results in most of the evaluation metrics.
As a result, we work with Setting 2 in the rest of
this paper.
5.2 Evaluation of LIDL4Oliv
To evaluate the performance of LIDL4Oliv, we fol-
lowed three steps. First, we assessed the performance
of the pre-trained teacher model (i.e., trained on the
white background dataset) on images with complex
backgrounds. Second, we trained and evaluated the
performance of a teacher model trained on our com-
plex background dataset. Third, we evaluated the
performance of the student model with and without
knowledge distillation.
5.2.1 White Background Dataset
The results of the experiments conducted in the previ-
ous section show that YOLOv5-m, our teacher model,
achieves good results when evaluated on white back-
ground images. However, when we evaluate its per-
formance on images that have complex backgrounds,
we notice that its performance decreases, as illus-
trated in Table 2. YOLOv5-m achieves an overall
average mean precision of 0.298, demonstrating how
inefficient the pre-trained model can be when faced
with images containing complex backgrounds. Fur-
thermore, we identify the limitation of using a white
background dataset for training and also justify the
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
590
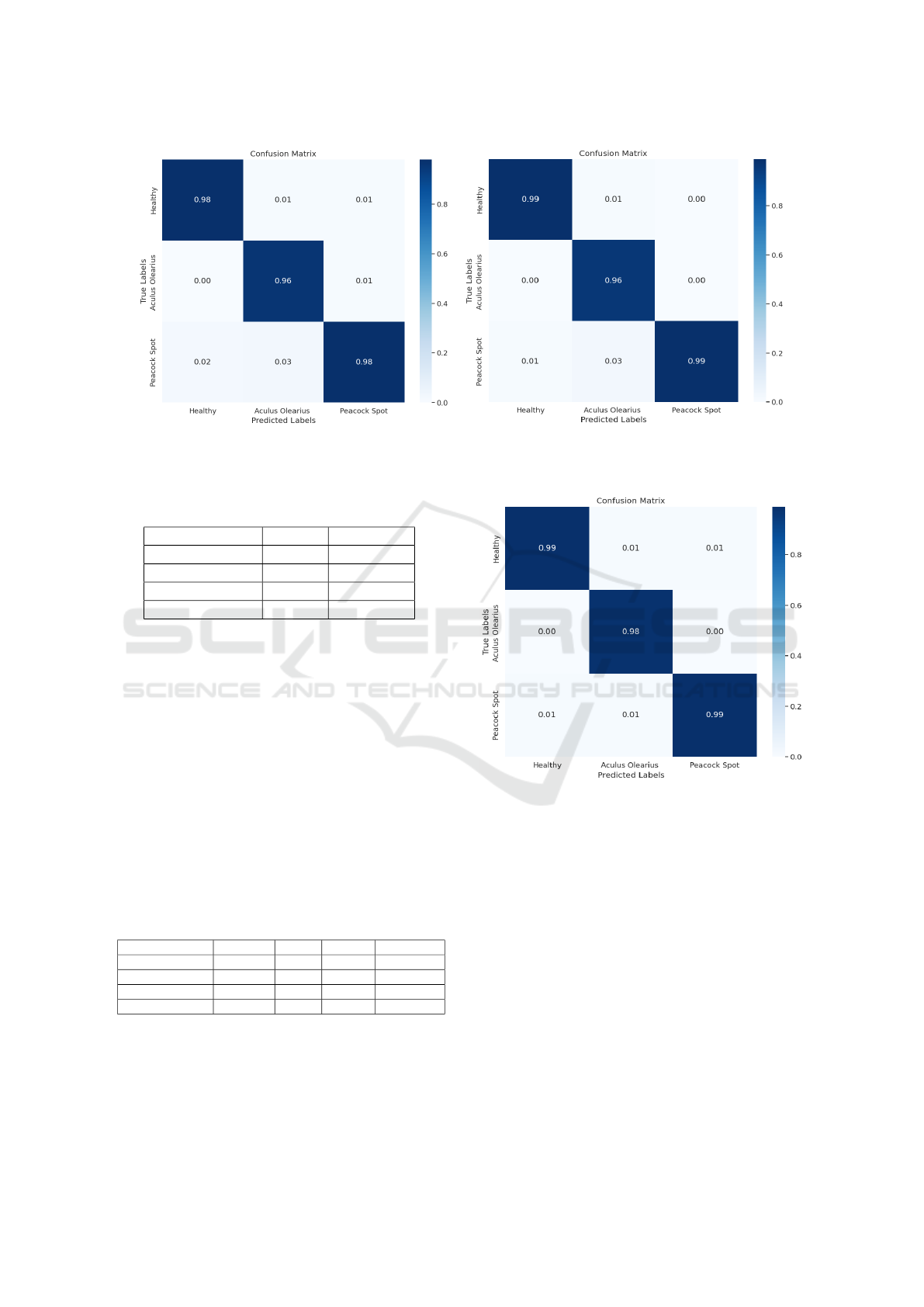
(a) Confusion matrix of YOLOv5-n (w/o) KD (b) Confusion matrix of YOLOv5-n with KD
Figure 9: Confusion matrix of YOLOv5-n with and (w/o) KD.
Table 2: Results of YOLOv5-m trained on a white back-
ground dataset and tested on a complex background images.
mAP50 mAP50-95
All 0.394 0.298
Healthy 0.347 0.266
Aculus Olearius 0.609 0.465
Peacock Spot 0.225 0.163
need to use a complex background.
5.2.2 Complex Background Dataset
Training on a dataset with a complex background is
of high importance. This is because a real-life im-
age usually includes a complex background. Thus,
to ensure the performance of our deep learning net-
work in the field, it is essential to train it on a similar
dataset. We use the same images when testing both
the teacher and the student. Samples of these images
are presented in Figure 11. Table 3 presents the ob-
tained results when training YOLOv5-m on our com-
plex background dataset.
Table 3: Results of YOLOv5-m trained and tested on com-
plex background dataset.
Class Precision Recall mAP50 mAP50-95
All 0.992 0.988 0.993 0.969
Healthy 0.989 0.989 0.994 0.959
Aculus Olearius 1 0.983 0.995 0.974
Peacock Spot 0.987 0.993 0.991 0.975
It is plausible that the achieved outcomes exhibit
remarkable performance in terms of precision, recall,
and mAP50-90. Indeed, its precision and recall val-
ues are 0.992 and 0.988, respectively. Also, its mean
average precision, mAP50-90, is equal to 0.969. Fig-
Figure 10: Confusion matrix of YOLOv5-m.
ure 10 presents the confusion matrix of YOLOv5-
m. It demonstrates that our teacher model achieves
reasonable classification results across all designated
classes.
5.2.3 Comparative Performance Analysis of
YOLOv5-n With and Without Knowledge
Distillation
To evaluate the performance of our student model
(YOLOv5-n) in detecting and classifying olive dis-
eases, we compare its performance with and without
using knowledge distillation.
We also present the performance of the teacher
model (YOLOv5-m) for a meaningful comparison
with the performance of the student model. Figure 9
presents the confusion matrix of our student model
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images
591

Figure 11: Detection and classification results of YOLOv5-n trained with KD.
Table 4: Performance of YOLOv5-n: With KD vs. without
KD.
YOLOv5-m
YOLOv5-n
(w/o KD)
YOLOv5-n
(with KD)
Precision 0.992 0.987 0.989
Recall 0.988 0.976 0.98
mAP50 0.993 0.994 0.994
mAP50-95 0.969 0.954 0.951
with and without KD. It is clear that knowledge dis-
tillation has improved the performance of the student
in classifying leaf diseases. Using KD enhanced the
student’s ability in classifying the ”Healthy“and the
”Peacock Spot“ state by an additional 1% for each.
The experimental results shown in Table 4 present
the precision, the recall, and the mean average pre-
cision of the teacher (YOLOv5-m), and the student
(YOLOv5-n trained with and without KD). It demon-
strates that YOLOv5-n trained using KD outperforms
YOLOv5-n trained without KD, in terms of precision
and recall. Specifically, YOLOv5-n trained using KD
obtains a precision value of 0.989 and a recall value of
0.980, while YOLOv5-n trained without KD achieves
a precision value of 0.987 and a recall value of 0.976.
We can also observe that the precision and the recall
of YOLOv5-n trained using KD achieves comparable
results when comparing it with the precision and the
recall values of the teacher YOLOv5-m, showing the
impact of fine-grained feature imitation knowledge
distillation approach in enhancing the performance
of the student’s performance. However, we notice
that the mean average precision, mAP50-90, achieved
by YOLOv5-m trained without KD is slightly higher
than that achieved by YOLOv5-m trained with KD.
This is quite plausible and acceptable. Examples
of detection and classification results of YOLOv5-n
trained with KD are shown in Figure 11. The test im-
ages are part of our generated dataset.
6 CONCLUSION
In this paper, we presented a novel lightweight incre-
mental deep learning model, called LIDL4Oliv, to de-
tect and classify olive leaf diseases in images. It is
based on YOLOv5-n for high-quality classification.
We used knowledge distillation to achieve very good
results and deployed our model as a cross-platform
mobile application. This mobile application supports
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
592

automatic olive leaf detection and serves as a farm
management system. Our mobile application enables
a farmer to ask for an expert opinion if (s)he wants
further evaluation of the leaf status. We established a
sustainable data collection process by gathering pro-
cessed images captured through the mobile applica-
tion. We also introduced an incremental update pro-
cess that ensures the adaptability of YOLOv5-n to
real-life images collected through the mobile appli-
cation. Our experimental results showed the perfor-
mance of our deployed model in terms of classifica-
tion accuracy, precision, recall, and mean average pre-
cision.
In the future, we intend to release our mobile ap-
plication on the Play Store and make it available for
free. This will allow us to collect annotated real-life
data. The collected data will be used to improve the
performance of our mobile application and effectively
assess and validate our incremental update process.
Moreover, the weights of our pre-trained YOLOv5-
n model can serve as initial weights when we extend
our task to include the detection of multiple leaves per
image and identifying their status.
ACKNOWLEDGEMENTS
This work is supported by the German Academic Ex-
change Service (DAAD) in the Ta’ziz Science Coop-
erations Program (AirFit Project; 57682841).
REFERENCES
Ahmed, A. A. and Reddy, G. H. (2021). A mobile-based
system for detecting plant leaf diseases using deep
learning. AgriEngineering, 3:478–493.
Aishwarya, N., Praveena, N., Priyanka, S., and Pramod, J.
(2023). Smart farming for detection and identifica-
tion of tomato plant diseases using light weight deep
neural network. Multimedia Tools and Applications,
82:18799–18810.
Alshammari, H. H. and Alkhiri, H. (2023). Optimized re-
current neural network mechanism for olive leaf dis-
ease diagnosis based on wavelet transform. Alexan-
dria Engineering Journal, 78:149–161.
Alshammari, H. H., Taloba, A. I., and Shahin, O. R.
(2023). Identification of olive leaf disease through
optimized deep learning approach. Alexandria Engi-
neering Journal, 72:213–224.
Canny, J. (1986). A computational approach to edge de-
tection. IEEE Transactions on Pattern Analysis and
Machine Intelligence, PAMI-8:679–698.
Chandan, J., Latha, D., Manisha, R., and Kishore, G.
(2022). Monitoring plant health and detection of plant
disease using iot and ml. International Journal of Re-
search in Engineering and Science, 10:1580–1588.
Chen, G., Choi, W., Yu, X., Han, T., and Chandraker, M.
(2017). Learning efficient object detection models
with knowledge distillation. In Proceedings of Neural
Information Processing Systems (NIPS), pages 742–
751, Long Beach, CA, USA. Curran Associates, Inc.
Gou, J., Sun, L., Yu, B., Wan, S., and Tao, D. (2023). Hier-
archical multi-attention transfer for knowledge distil-
lation. ACM Transactions on Multimedia Computing,
Communications and Applications, 20:1–20.
Gurav, S. S., Patil, S. A., Patil, S. L., Desai, P. P., and Bad-
kar, P. B. (2022). Android application on e-commerce
& plant disease identification using tensor flow. Inter-
national Journal of Innovative Science and Research
Technology, 7:2456–2165.
Hidayat, A., Darusalam, U., and Irmawati, I. (2019). De-
tection of disease on corn plants using convolutional
neural network methods. Jurnal Ilmu Komputer dan
Informasi, 12:51–56.
Hinton, G., Vinyals, O., Dean, J., et al. (2015). Distill-
ing the knowledge in a neural network. In NIPS
Deep Learning and Representation Learning Work-
shop, Montreal, Canada. Curran Associates, Inc.
Kruthiventi, S. S., Sahay, P., and Biswal, R. (2017).
Low-light pedestrian detection from rgb images using
multi-modal knowledge distillation. In Proceedings
of the International Conference on Image Processing
(ICIP), pages 4207–4211, Beijing, China. IEEE.
Ksibi, A., Ayadi, M., Soufiene, B. O., Jamjoom, M. M., and
Ullah, Z. (2022). Mobires-net: a hybrid deep learning
model for detecting and classifying olive leaf diseases.
Applied Sciences, 12:10278.
Li, Z., Xu, P., Chang, X., Yang, L., Zhang, Y., Yao, L., and
Chen, X. (2023). When object detection meets knowl-
edge distillation: A survey. IEEE Transactions on Pat-
tern Analysis and Machine Intelligence, 45:10555–
10579.
Lin, T.-Y., Maire, M., Belongie, S., Hays, J., Perona, P.,
Ramanan, D., Doll
´
ar, P., and Zitnick, C. L. (2014).
Microsoft coco: Common objects in context. In Pro-
ceedings of the 13th European Conference (ECCV),
pages 740–755, Zurich, Switzerland. Springer.
Mamdouh, N. and Khattab, A. (2022). Olive leaf dis-
ease identification framework using inception v3 deep
learning. In Proceedings of the IEEE International
Conference on Design & Test of Integrated Micro &
Nano-Systems (DTS), pages 1–6, Cairo, Egypt. IEEE.
Ney, B., Bancal, M., Bancal, P., Bingham, I., Foulkes, J.,
Gouache, D., Paveley, N., and Smith, J. (2013). Crop
architecture and crop tolerance to fungal diseases and
insect herbivory. mechanisms to limit crop losses. Eu-
ropean Journal of Plant Pathology, 135:561–580.
Osco-Mamani, E. F. and Chaparro-Cruz, I. N. (2023).
Highly accurate deep learning model for olive leaf
disease classification: A study in tacna-per
´
u. Inter-
national Journal of Advanced Computer Science and
Applications, 14:851–860.
Qin, X., Zhang, Z., Huang, C., Dehghan, M., Zaiane, O. R.,
and Jagersand, M. (2020). U2-net: Going deeper with
LIDL4Oliv: A Lightweight Incremental Deep Learning Model for Classifying Olive Diseases in Images
593

nested u-structure for salient object detection. Pattern
Recognition, 106:107404.
Savary, S., Ficke, A., Aubertot, J.-N., and Hollier, C.
(2012). Crop losses due to diseases and their impli-
cations for global food production losses and food se-
curity. Food Security, 4:519–537.
Soyyigit, S. and Yavuzaslan, K. (2018). Complex net-
work analysis of international olive oil market. Tarim
Ekonomisi Dergisi, 24:117–129.
Steen Lumholt and Guido van Rossum (2009). https://docs.
python.org/3/library/tkinter.html. Accessed: 2023-11-
19.
Tian, Y., Krishnan, D., and Isola, P. (2020). Contrastive
representation distillation. In Proceedings of the
International Conference on Learning Representa-
tions(ICLR), Addis Ababa, Ethiopia.
Tzutalin (2015). https://github.com/tzutalin/labelImg. Ac-
cessed: 2023-11-19.
Uguz,S. (2020). https://github.com/sinanuguz/CNN olive
dataset. Accessed: 2023-11-19.
Valdoria, J. C., Caballeo, A. R., Fernandez, B. I. D., and
Condino, J. M. M. (2019). idahon: An android based
terrestrial plant disease detection mobile application
through digital image processing using deep learning
neural network algorithm. In Proceedings of the 4th
International Conference on Information Technology
(InCIT), pages 94–98, Bangkok, Thailand. IEEE.
Wang, L., Lu, H., Wang, Y., Feng, M., Wang, D., Yin, B.,
and Ruan, X. (2017). Learning to detect salient ob-
jects with image-level supervision. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition (CVPR), pages 136–145, Honolulu, HI,
USA. IEEE.
Wang, T., Yuan, L., Zhang, X., and Feng, J. (2019). Dis-
tilling object detectors with fine-grained feature imi-
tation. In Proceedings of the IEEE/CVF Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 4933–4942, California, USA. IEEE.
Yadikar, N., Ubul, K., et al. (2023). A review of knowledge
distillation in object detection. IEEE Access, 11.
Ye, Y., Zhu, X., Chen, H., Wang, F., and Wang, C. (2023).
Identification of plant diseases based on yolov5. In In-
ternational Conference on Intelligent Computing and
Signal Processing (ICSP), pages 1731–1734, Xi’an,
China. IEEE.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
594
