
Formal Analysis of Uncertain Continuous Markov Chains in Systems
Biology
Krishnendu Ghosh
a
and Caroline Goodman
b
Department of Computer Science, College of Charleston, SC, U.S.A.
Keywords:
Probabilistic Model Checking, Uncertain Continuous-Time Markov Chain, Computational Feasibility,
Tractable, Phylogenetic Tree.
Abstract:
Data dependent abstraction for continuous-time Markov chain is often challenging given the incompleteness
and imprecision of data. Uncertainty in the environment is modeled in the form of uncertain continuous-time
Markov chain. In this work, a tractable model checking methodology, stochastic partial model set checking is
formalized by approximation of the uncertain continuous-time Markov chain. The methodology was applied in
querying to infer on a phylogenetic tree, constructed under uncertainty. Queries were posed on the formalism
using continuous stochastic logic formula. Experimental results demonstrate the computational feasibility of
the model.
1 INTRODUCTION
Modeling of biological processes is an important re-
search direction in systems biology. One of the prob-
lems in systems biology is the correct construction
of phylogenetic trees from biological data. Phylo-
genetic studies are key in understanding the evolu-
tion of species. Often the construction of phylo-
genetic tree is not precise because of imprecision
and uncertainty in the data. Stochastic models such
as discrete-time Markov chains and continuous-time
Markov chain have been used as modeling tools in
problems in systems biology and formal verifica-
tion methods such as model checking. Discrete-time
Markov chain (DTMC) have been used as stochas-
tic model in systems biology. The order of the pro-
cesses in biology is abstracted in the order of the states
in the dtmc. However biological applications, time
runs continuously, and so discrete-time mathematical
models are not always appropriate models. This is es-
pecially true in biology where organisms reproduce,
infect each other, etc. in continuous time. There-
fore, continuous-time Markov chains (CTMC) are of-
ten more suited to model abstraction. Continuous-
time Markov chains (CTMC) are one of the founda-
tional structure to model continuous probabilities in
the modeling processes such as in- rates of execution
a
https://orcid.org/0000-0002-8471-6537
b
https://orcid.org/0009-0009-1592-6029
of chemical reaction and rates of epidemiology as in
spread of diseases. However, rates are imprecise and
are uncertain because it is challenging to get precise
data given the experimental conditions are not always
possible to replicate exactly and there is presence of
noise. Parameter synthesis in formal modeling have
been applied to address imprecise data. However, of-
ten times the modeler would introduce imprecision to
quantify error and evaluate the constructed model ab-
straction in comparison with the real phenomena that
is modeled. In this work, we create a tractable for-
malism for model checking on uncertain continuous-
time Markov chain and evaluate the computational
feasibility of the application of the model in infer-
ence of phylogenetic tree, by posing temporal logic
queries.
1. A novel inference formalism for temporal logic
reasoning on uncertain continuous-time Markov
chain is described. A stochastic partial model set
is defined that provides the modeler the perfor-
mance of the models. The methodology is an ap-
proximation to perform model checking on uncer-
tain continuous-time Markov chain.
2. The formalism has been applied to model phylo-
genetic tree and computational feasibility of the
abstraction on a published dataset of phylogenetic
tree is evaluated.
Ghosh, K. and Goodman, C.
Formal Analysis of Uncertain Continuous Markov Chains in Systems Biology.
DOI: 10.5220/0012466000003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 519-526
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
519

2 BACKGROUND
The foundation of this work is the intersection of
probabilistic model checking for uncertain stochastic
structures, formal modeling and inference of phylo-
genetic tree under uncertainty.
2.1 Formal Verification of Stochastic
Models
Computation of reachability probabilities (Delahaye
et al., 2016) for parametric Markov model has been
studied by a closed form function (Hahn et al., 2011).
Given the imprecision in data, models require param-
eters for effective inference. Computation of param-
eters and its estimation is an active area of research.
The modeler executes temporal logic queries on the
model and computes the parameter (Han et al., 2008).
There are synthesis based approaches that have ad-
dressed uncertainty in data (Shavit et al., 2016). Ap-
proximations for analysis of biological systems us-
ing probabilistic model checking by removing the un-
likely states, thereby reducing the state space has been
studied (Neupane et al., 2019). Interval form of rep-
resentation of probabilities on Markov chains to in-
corporate uncertainty has been formalized (Sproston,
2018). Here the intervals were open intervals. In our
work, we focus on closed intervals of uncertain ki-
netic rates. Sampling techniques have been used to
learn CTMC (Sen et al., 2004). In our formalism
we use sampling on the subintervals after discretiza-
tion of the interval representing the rates. Sampling
having been applied to uncertain parametric CTMCs
with parametric transition rates and scalability of the
model is evaluated (Badings et al., 2022).
2.2 Formal Analysis in Systems Biology
There is a body of literature that described formal
analysis of stochastic models of biochemical path-
ways using continuous-time Markov chains using
probabilistic model checking (Calder et al., 2010;
Calder et al., 2006; Heath et al., 2008) and different
formalisms to address uncertainty in the environment.
Quantitative analysis using continuous-time stochas-
tic logic and have been compared with ordinary dif-
ferential equation based model(Calder et al., 2006).
However, the models do not take into account envi-
ronmental variations in which the rate of reactions,
i.e the speeds at which pathway (biochemical reac-
tion) will execute is assumed to be exact. Recent work
incorporating imprecision in CTMC is the formaliza-
tion of uncertain CTMCs (Cardelli et al., 2023). The
rates of the CTMCs have been assumed to vary non-
deterministically with time represented by continuous
intervals. The states of the CTMC are partitioned and
the states are lifted in a standard CTMC setting but
under uncertainty. The refinements were based on
bisimulation of stochastic models (Baier et al., 2003;
Neuhäußer and Katoen, 2007). Uncertain kinetic
rates using probabilistic model checking has been de-
scribed by creating a novel semantics (Barbuti et al.,
2012). Model checking of uncertain continuous-time
Markov chains have been reported(Bortolussi et al.,
2016). Chemical reaction network has been modeled
as parametric CTMC integrating with learning mod-
els with the goal of applying statistical model check-
ing (Molyneux and Abate, 2020).
2.3 Construction of Phylogenetic Tree
Under Uncertainty
Monte Carlo based method was studied as an in-
ference method on phylogenetic tree by represent-
ing CTMC and approximate transition probabilities
and compute the parameters (Hajiaghayi et al., 2014).
Bayesian phylogenetic inference using annealed se-
quential Monte Carlo method have been proposed
(Wang et al., 2020). This direction of research is on-
going and there is no substantial approximation that
has been accepted as the standard in the community.
3 PRELIMINARIES
In this section, we state the mathematical preliminar-
ies that are foundations of our formalism. A contin-
uous time Markov chain is a stochastic model with
a discrete states with a continuous time as a parame-
ter. In formal modeling, the continuous time Markov
chain is described in the form of a state based sys-
tem, called a labeled continuous-time Markov chain.
For consistency throughout the paper, we will denote
a labeled continuous-time Markov chain as CTMC.
Definition 1. A labeled CTMC (Baier et al., 2000) is
a tuple,M = ⟨S, S
0
, R,AP,L⟩ where:
1. S is a set of states.
2. S
0
⊂ S is the set of initial states.
3. R : S × S → R
≥0
as the rate matrix.
4. L : S ← 2
AP
is the labeling function.
The labeled CTMC described in Definition 2 elim-
inates the requirement R(s, s) =
∑
s̸=s
′
R(s, s
′
), unlike
non-state based definition of CTMCs. Self-loops are
modeled by R(s, s
′
) > 0.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
520

Definition 2. (Labeled Uncertain Continuous-time
Markov Chain) A labeled uncertain CTMC (UCTMC)
is a tuple, K = ⟨S, S
0
, R, A, AP, L⟩ where:
1. S is a set of states.
2. S
0
⊂ S is the set of initial states.
3. A is the the set of actions.
4. R : S × A × S → R
≥0
as the rate matrix and
each,R
i, j
is an interval,(m, n), m ≤ n and m, n ∈ Q.
5. L : S → 2
AP
is a state labeling function.
A path in a UCTMC is a non-empty sequence,
s
0
a
0
t
0
s
1
a
1
t
1
s
2
... where R(s
i
, a
i
, s
i+1
) > 0 and t
i
∈ R
>0
for all i ≤ 0. The value of t ∈ (
ˇ
t
i
,
ˆ
t
i
). The open interval
is a way to add imprecision.
3.1 Probabilistic Model Checking
Model checking is performed on labeled finite state
machine in the form of Kripke structure, M . For-
mally,
Definition 3. (Model Checking (Clarke, 1997)) Given
a model, M and formula,φ , model checking is the
process of deciding whether a formula φ is true in the
model, written M |= φ.
Here, φ represents a temporal logic formula that rep-
resents the properties of a system. The model, M
represents the description of the system. The system
description when represented by a stochastic struc-
ture, M
s
, then the model checking is performed on
a stochastic structure and the validity of the formula
is quantified by probability.
Definition 4. (Probabilistic Model Checking) Given
a model, M
s
and formula,φ , model checking is the
process of deciding whether a formula φ is true in the
model, written M |= φ and quantified by a probability.
Details on probabilistic computational tree logics can
be found in the literature (Baier et al., 2008; Aziz
et al., 1996) Probabilistic model checking on un-
certain stochastic structures such as interval discrete
Markov chains have been shown to be in PSPACE and
NP-hard (Benedikt et al., 2013; Sen et al., 2006).
3.1.1 Continuous Stochastic Logic
Stochastic Model Checking have been described
(Kwiatkowska et al., 2007). Model checking on
CTMC is performed by the logics known as contin-
uous stochastic logic (CSL) (Aziz et al., 1996; Baier
et al., 1999) The syntax of CSL (Aziz et al., 1996)
Syntax of CSL is:
φ ::= true | a | φ ∧ φ | ¬φ | P
⊕p
[ψ] | S
⊕p
[φ]
ψ ::= X φ | φU
≤k
φ |φUφ
where a is an atomic proposition,⊕ ∈ {≤, <, ≥, >
}, p ∈ [0, 1] and k ∈ R
≥0
. φ, ψ are state and path for-
mula respectively. P
⊕p
ψ represents the probability of
φ satisfied from a given state satisfies the bound ⊕p.
The bounded until operator φ
1
U
≤
kφ
2
is valid if φ
2
for
a time instant in the interval [0, k] and φ
1
is valid at all
preceding time instants. The path formula,Xφ is true
if φ is satisfied in the next state. The formula φ
1
U
≤k
φ
2
is true if φ
2
is satisfied within k time-steps and φ
1
is
true at that point. Similar is the description of φ
1
Uφ
2
where φ
2
is true some point in future till then φ
1
is
true. φ
2
is satisfied at some time instant in the interval
[0, k] and φ
1
holds at all preceding time steps. The
S operator describes the steady-state behaviour of the
CTMC. S
⊕
p[φ] asserts that the steady-state probabil-
ity of being in a state satisfying φ meets the bound
S
⊕p
The semantics of CSL are defined on CTMC and
can be found (Aziz et al., 1996; Baier et al., 1999) for
further reading.
4 STOCHASTIC PARTIAL
MODEL SET CHECKING
Probabilistic model checking is performed on
stochastic structures such as DTMC or CTMC. Model
checking on UCTMC is challenging because the in-
tervals for rates can generate infinite number of rates.
The following are keys to approximate the UCTMC
denoted by M
s
and construct a tractable computa-
tional model. A finite set of stochastic partial model,
M
pp
= {M
1
, M
2
, . . . M
n
} where n ∈ N. Each stochas-
tic partial model M ∈ M
pp
has identical state space
that of M
s
with the intervals for rates of M are the
subintervals for rates in M
s
.
The model checking problem is phrased as given
a model M
i
and a formula, φ, model checking is the
process of deciding whether a formula φ is true in the
model and represented by M
i
|= φ. The generalization
of the model checking on probabilistic partial models
will be denoted by probabilistic partial model check-
ing problem. Formally, given a set of probabilistic
partial models, M and a formula, φ model checking is
the process of deciding whether a formula is true for
each M ∈ M
pp
. and denoted by M |= φ.
4.1 Model Abstraction
The model abstraction is two step process: Initially, a
UCTMC is constructed from the data. The next step
is to construct set of stochastic partial models from
the UCTMC is constructed.
Construction of UCTMC Representing Phylogenetic
Tree: Given a set of species , G and set of rate of evo-
lution (transitions), R = {I
1
, ..., I
n
}where n ∈ N and
Formal Analysis of Uncertain Continuous Markov Chains in Systems Biology
521

Input: K
s
⟨S
0
, S, R, A, AP, L⟩ Number of
subintervals, k
Output: Set of Stochastic Partial Models, K
1: S = {s
o
},
ˆ
S =
/
0,
ˆ
I ; {S
O
is the initial set and
ˇ
T is
the set of transitions.}
2: IntervalSet ←
/
0
3: OrderedSetRate ←
/
0
4: for each s
i
r
i j
−→ s
j
where s
i
, s
j
∈ S do
5: Construct k subintervals for each r
i j
.
6: IntervalSet ← IntervalSet ∪ (r
i j
1
, r
i j
2
, . . . , r
i j
k
)
where r
i j
is a subinterval.
7: end for
8: OrderdedTuples ←
|IntervalSet|
∏
1
IntervalSet
9: K ←
/
0
10: for each T ∈ OrderedTuples do
11: R ← T
12: K
s
⟨S
0
, S, T, A, AP, L⟩
13: K ← K ∪ K
s
14: end for
15: K
Algorithm 1: StochasticPartialModel.
I
am
is the interval representing mth. clade. UCTMC
is constructed in the following way: Each state,s is
labeled as specie and a transition to another state,s
′
implies species A evolves into species B. The label
on the transition, I
m
is the interval that represents the
range of values the rate of evolution from species A
to species B. The state, s
′
is labeled with species.
Construction of Stochastic Partial Model Set: In the
construction of a set of stochastic partial models, each
intervals for rates on the transition in UCTMC is par-
titioned into subintervals. The partitions of the inter-
vals are user defined value, k amd k ∈ N. For exam-
ple, an interval in K
s
between a transition s → s
′
is
given by I = (a, b) where a, b ∈ (N) and a < b, the
intervals where s, s
′
∈ K
s
. For k = 2, the subinter-
vals are: (a,
a+b
2
], (
a+b
2
, b). Similarly, for n intervals
in K
s
, there will be nk subintervals and each stochastic
partial model will represent a UCTMC with a series
of subinterval from a series of intervals of transition
rates in K
s
. There are k
n
stochastic partial models. k
can be varied. Algorithm 1 shows the construction of
set of stochastic partial model set. The fixed point al-
gorithm constructs the set of stochastic partial models
with subintervals.
4.2 Construction of Simulation Model
The transition rates for probabilistic partial models
are represented in the form of intervals. In order to
make the models tractable, sampling on the intervals
are performed. Notation for upper bound and lower
bound of an interval in a stochastic partial model is
given by ub(I
p
) and lb(I
p
), respectively and I
p
repre-
sents an interval in a stochastic partial model and the
set of intervals is
ˆ
I
p
. Algorithm 2 demonstrates the
sampling of the rates from each intervals in a stochas-
tic partial model. The models in the set,K form the
basis of probabilistic model checking. If the values
of the probabilities for consecutive subintervals are
close to each other, then the intervals can be merged
and hence, the number of models get reduced. The
algorithm terminates because the number of intervals
are finite and there are finite states in each stochastic
partial model in the finite set, K. Each K
sim
∈ K
sim
is a CTMC, an approximation of K
s
∈ K and hence,
CSL logic queries can be executed. The computa-
tional complexity of model checking is polynomial in
the size of the model. Therefore, for a partial model
set of size n, the model checking complexity is poly-
nomial as the model checking is performed on each
of the partial model.
Input: K
Output: Set of Sampled Stochastic Partial Models,
K
sim
.
1: K
sim
← 0
2: for each K ∈ K do
3: for each r ∈ R do
4: if r is partly closed interval then
5: Generate random value, r such that
lb(I
p
) < r ≤ up(I
p
)
6: else
7: Generate random value, r such that
lb(I
p
) ≤ r ≤ up(I
p
)
8: SRate ← SRate ∪ r where i =|
ˆ
I
p
|.
9: end if
10: K
sim
⟨S
0
, S, srate, A, AP, L⟩.srate ∈ SRate.
11: end for
12: K
sim
← K
sim
∪ K
sim
13: end for
14: K
sim
Algorithm 2: Sampled Stochastic Partial Models.
5 APPLICATION OF UCTMC IN
PHYLOGENETIC TREE
The steps involved in reasoning for an abstraction of
phylogenetic tree under uncertainty is given by :
1. Construct an UCTMC, K
t
representing a phyloge-
netic tree,T .
2. Construct the set of stochastic partial models
from the UTMC, K
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
522
3. Perform probabilistic model checking by posing
queries using continuous stochastic logic on the
set of sampled stochastic partial models.
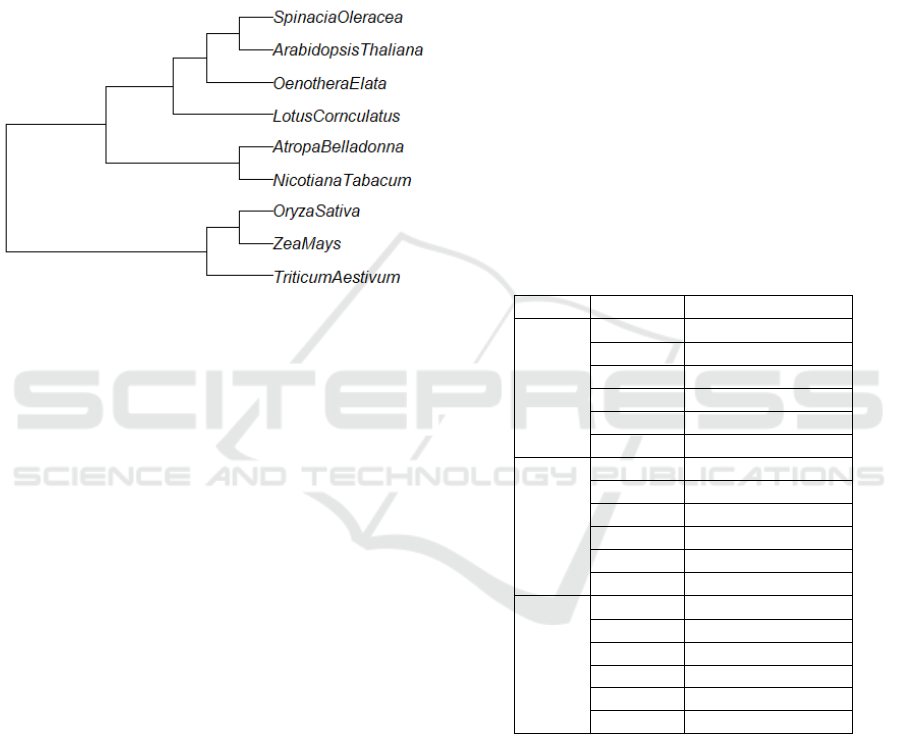
The data used as the phylogenetic tree model was
from published data by Baele et al. (Baele et al.,
2021), as shown in Figure 1. The models were con-
structed and evaluated in PRISM (Kwiatkowska et al.,
2011) The states in the model are labeled symboli-
cally with species.
Figure 1: Fragment of Phylogenetic tree representing the
speciation of eudicots, angiosperms, and seed plants (Baele
et al., 2021).
Notation: The various species of plants are
Arabidopsis Thaliana(at), Spinacia Oleracea(so),
Oenothera Elata(oe), Lotus Corniculatus(lc), Nico-
tiana Tabacum(nt), and Atropa Belladonna(ab) in the
model.
In the Figure 2, the boxplots show the variation
amongst each model variation. The data is based
on the randomly generated rates based on the ranges
specified. The figure is provided for the sole purpose
of conveying the variations among a select few mod-
els involved in the total calculations.
5.1 Evaluation of Temporal Logic
Properties
The evaluation of specifications, represented by
probabilistic temporal logics is performed using
PRISM (https://www.prismmodelchecker.org/)
model checker. The simulation was performed in a
Mac with 2.3 GHz with quad core Intel i5 proces-
sor and 8GB RAM. In this model, we are focused
on reachability properties and temporal logic proper-
ties. A sample of reachability biological queries rep-
resented as continuous stochastic logic formula are as
follows:
Property 1: P=? (ab & nt & oe). The meaning of
the statement is: Is there a state where all three
species ab, nt and oe are present? Note: This
property’s result will always evaluate to 0.0 be-
cause our model checks to see if a single species
formed, but not multiple species.
Property 2: P=? ( nt = true) The meaning of the
logic formula- is it possible to reach a state where
species nt is present in the model.
Property 3: P=? ( ab = true ) The meaning of the
logic formula- is it possible to reach a state where
species ab is present in the model.
Property 4: P=? ( at = true ). The meaning of the
logic formula- is it possible to reach a state where
species at is present in the model.
Property 5: P=? ( so = true ) The meaning of the
logic formula- is it possible to reach a state where
species so is present in the model.
Property 6: P=? ( oe = true ) This is a reachability
property asking if it is possible to reach a state
where species oe is present in the model.
Table 1: Execution Time of CSL Queries.
Model Property Time (in seconds)
n=6 1 0.088
2 0.009
3 0.001
4 0.001
5 0.001
6 0
n=10 1 0.018
2 0.202
3 0.202
4 0.245
5 0.23
6 0.227
n=13 1 0.453
2 41.65
3 40.76
4 53.945
5 52.851
6 45.922
Table 1 shows of the results obtained for model
checking for different problem sizes input sizes and
properties. The data consists of results obtained for
seven different input sizes: n=6, n=10, and n=13, with
varying properties. There are over 40 models, which
refer to the varied rates specified earlier. Each vari-
ation has been checked for the same set of proper-
ties. For n=6, the results are further broken down into
seven different variations of the input. For each input
size and property combination, the "Time" for exe-
cution of the queries are presented. The properties
checked are numbered from 1 to 6. The time taken
Formal Analysis of Uncertain Continuous Markov Chains in Systems Biology
523
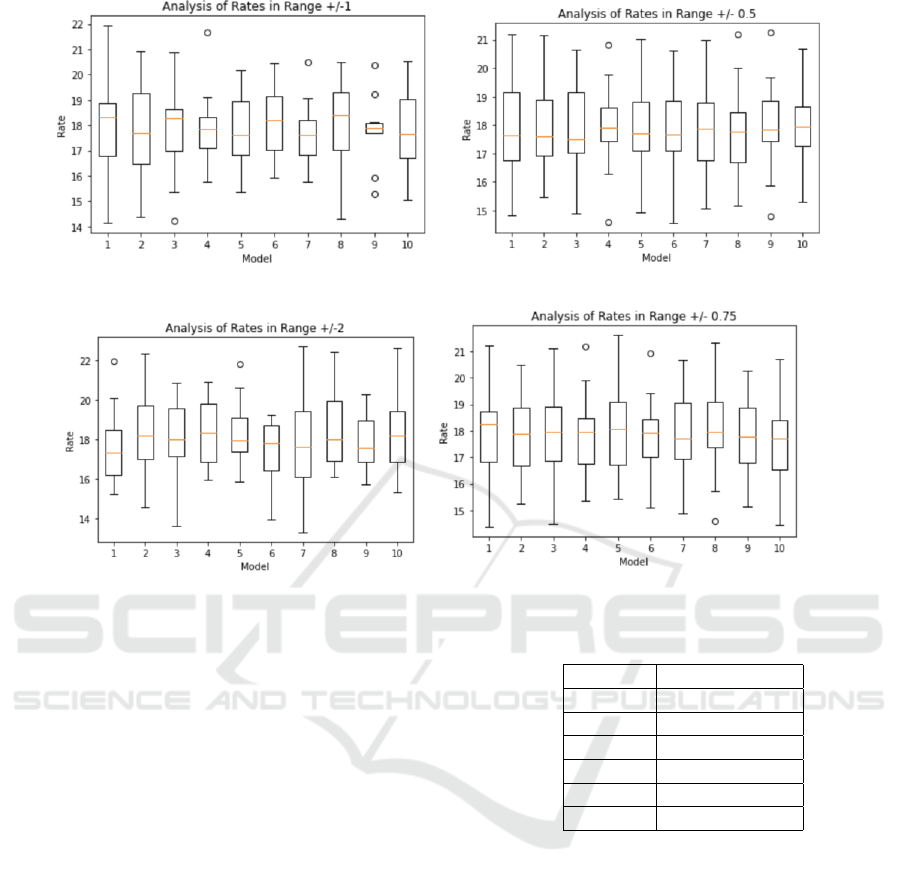
Figure 2: Random Rates of Phylogenetic Tree.
for model checking is recorded in seconds, and the
result indicates whether the model satisfies the prop-
erty checked. For example, for n=6 and property 2,
the time taken for model checking was 0.009 seconds,
and the result was 0.212459016, indicating that the
model satisfied the property. The times of the ex-
ecution of the logic queries demonstrate a correla-
tion with the increase in the input sizes. For exam-
ple, the time taken for model checking n=6 is signif-
icantly less than the time taken for n=13. However,
there are some variations in time taken for the same
input size, indicating that the properties of the input
can also have an impact on the time taken for model
checking.
5.2 Analysis of Properties
Table 2 shows some properties are satisfied for all
values of n, while others are not. Property 1 seems
to be satisfied for all values of n, as indicated by the
result of 0 for all entries. Here is the table with the
average result for each individual property (averages
are rounded to four decimal places):
The average result of each property reveals that
some properties are more likely to be present in
some models than others. Property 7 has the highest
average probability of presence, with a 34.9% chance
Table 2: Computed Probabilities from Properties.
Property Average Result
1 0.0
2 0.227723931
3 0.160131871
4 0.077751529
5 0.078001573
6 0.160862186
of eventually being present at some point in the
model checking process. This could be due to various
reasons, including but not limited to, the particular
organism specified in the property and the values of
the rates of transition associated with the property.
Analysis Results of Temporal Logic Queries: Table
3 shows the comparison of a property across all the
models. Property 2 is defined as follows: P=? [F nt =
true]. This property checks whether the state with nt
is eventually reached during the execution of the sys-
tem. The result of the property verification for differ-
ent problem sizes. For the model with n=6, the model
checker took 0.009 seconds to verify the property and
returned a result of 0.212459. This means that there
is a 21.25% chance that the system will eventually
reach the state labeled with nt during execution. For
the model with n=10, the model checker took 0.202
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
524

Table 3: Comparison of time of execution (in seconds) across different models.
Model Property Time (seconds) Result
n =6 2 0.009 0.212459
n =10 2 0.202 0.310325
n =13 2 40.76 0.220842
n =6 (variation 1) 2 0.001 0.187863
seconds to verify the property and returned a result of
0.310325. This means that there is a 31.03% chance
that the system will eventually reach the state nt dur-
ing execution. For the model with n=13, the model
checker took 40.76 seconds to verify the property and
returned a result of 0.220842649. This means that
there is a 22.08% chance that the system will even-
tually reach the state nt during execution. The model
with n=6 (variation 1) represents a larger size (size
of ± 2 ) of the interval for rates. The model checker
took 0.001 seconds to verify the property and returned
a result of 0.18786327, implying there is a 18.79%
chance that the system will eventually reach the state
nt during execution. The change in the size of the in-
terval of the rates did not influence the time for verifi-
cation of the specification. These results remain simi-
lar for each property, as the input size and model vari-
ables are altered. Based on these results, we can con-
clude that the larger the size of the model, the longer
it takes for the model checker to verify the property.
Additionally, we can see that the probability of reach-
ing the nt state during system execution is higher in
the model with n=10 compared to the other models.
This information can be useful in determining the fea-
sibility of the system and making design decisions to
optimize the system performance.
6 CONCLUSION
In this paper, we describe an abstraction of feasible
models, an approximation of a given uncertain
continuous-time Markov chain. The approximated
set of CTMC constructed is used for stochastic partial
model set checking. The computational feasibility
is evaluated on continuous stochastic logic queries,
on a prototype of a phylogenetic tree from published
data. The modeling technique can be improved
by quantifying error for model checking of partial
models and identifiability of the partial models
(Browning et al., 2020). The formalism is promising
to model other problems in systems biology where
there data is imprecise and provide the modeler a tool
to provide insights to the biological questions.
ACKNOWLEDGEMENTS
The first author, KG was supported by NSF CCF
2227898 during the research. The second author,
CG was supported by SC-INBRE funds in summer
of 2022. Research reported in this publication was
supported by the National Institute of General Medi-
cal Sciences of the National Institutes of Health under
Award Number P20GM103499. The content is solely
the responsibility of the authors and does not neces-
sarily represent the official views of the National In-
stitutes of Health.
REFERENCES
Aziz, A., Sanwal, K., Singhal, V., and Brayton, R. (1996).
Verifying continuous time markov chains. In Inter-
national Conference on Computer Aided Verification,
pages 269–276. Springer.
Badings, T. S., Jansen, N., Junges, S., Stoelinga, M.,
and Volk, M. (2022). Sampling-based verification of
ctmcs with uncertain rates. In Computer Aided Ver-
ification: 34th International Conference, CAV 2022,
Haifa, Israel, August 7–10, 2022, Proceedings, Part
II, pages 26–47. Springer.
Baele, G., Gill, M. S., Bastide, P., Lemey, P., and Suchard,
M. A. (2021). Markov-modulated continuous-time
markov chains to identify site-and branch-specific
evolutionary variation in beast. Systematic biology,
70(1):181–189.
Baier, C., Haverkort, B., Hermanns, H., and Katoen, J.-
P. (2000). Model checking continuous-time markov
chains by transient analysis. In International Confer-
ence on Computer Aided Verification, pages 358–372.
Springer.
Baier, C., Haverkort, B., Hermanns, H., and Katoen, J.-P.
(2003). Model-checking algorithms for continuous-
time markov chains. IEEE Transactions on software
engineering, 29(6):524–541.
Baier, C., Katoen, J.-P., and Hermanns, H. (1999). Approx-
imative symbolic model checking of continuous-time
markov chains. In International Conference on Con-
currency Theory, pages 146–161. Springer.
Baier, C., Katoen, J.-P., and Larsen, K. G. (2008). Princi-
ples of model checking. MIT press.
Barbuti, R., Caravagna, G., Maggiolo-Schettini, A., Mi-
lazzo, P., and Tini, S. (2012). Foundational aspects of
Formal Analysis of Uncertain Continuous Markov Chains in Systems Biology
525

multiscale modeling of biological systems with pro-
cess algebras. Theoretical Computer Science, 431:96–
116.
Benedikt, M., Lenhardt, R., and Worrell, J. (2013). Ltl
model checking of interval markov chains. In Inter-
national Conference on Tools and Algorithms for the
Construction and Analysis of Systems, pages 32–46.
Springer.
Bortolussi, L., Milios, D., and Sanguinetti, G. (2016).
Smoothed model checking for uncertain continuous-
time markov chains. Information and Computation,
247:235–253.
Browning, A. P., Warne, D. J., Burrage, K., Baker, R. E.,
and Simpson, M. J. (2020). Identifiability analy-
sis for stochastic differential equation models in sys-
tems biology. Journal of the Royal Society Interface,
17(173):20200652.
Calder, M., Gilmore, S., Hillston, J., and Vyshemirsky, V.
(2010). Formal methods for biochemical signalling
pathways. In Formal Methods: State of the Art and
New Directions, pages 185–215. Springer.
Calder, M., Vyshemirsky, V., Gilbert, D., and Orton, R.
(2006). Analysis of signalling pathways using con-
tinuous time markov chains. In Transactions on Com-
putational Systems Biology VI, pages 44–67. Springer.
Cardelli, L., Grosu, R., Larsen, K. G., Tribastone, M.,
Tschaikowski, M., and Vandin, A. (2023). Algo-
rithmic minimization of uncertain continuous-time
markov chains. IEEE Transactions on Automatic Con-
trol.
Clarke, E. M. (1997). Model checking. In Foundations of
Software Technology and Theoretical Computer Sci-
ence: 17th Conference Kharagpur, India, December
18–20, 1997 Proceedings 17, pages 54–56. Springer.
Delahaye, B., Lime, D., and Petrucci, L. (2016). Param-
eter synthesis for parametric interval markov chains.
In International Conference on Verification, Model
Checking, and Abstract Interpretation, pages 372–
390. Springer.
Hahn, E. M., Hermanns, H., and Zhang, L. (2011). Proba-
bilistic reachability for parametric markov models. In-
ternational Journal on Software Tools for Technology
Transfer, 13(1):3–19.
Hajiaghayi, M., Kirkpatrick, B., Wang, L., and Bouchard-
Côté, A. (2014). Efficient continuous-time markov
chain estimation. In International Conference on Ma-
chine Learning, pages 638–646. PMLR.
Han, T., Katoen, J.-P., and Mereacre, A. (2008). Ap-
proximate parameter synthesis for probabilistic time-
bounded reachability. In 2008 Real-Time Systems
Symposium, pages 173–182. IEEE.
Heath, J., Kwiatkowska, M., Norman, G., Parker, D., and
Tymchyshyn, O. (2008). Probabilistic model checking
of complex biological pathways. Theoretical Com-
puter Science, 391(3):239–257.
Kwiatkowska, M., Norman, G., and Parker, D. (2007).
Stochastic model checking. In International School
on Formal Methods for the Design of Computer, Com-
munication and Software Systems, pages 220–270.
Springer.
Kwiatkowska, M., Norman, G., and Parker, D. (2011).
PRISM 4.0: Verification of probabilistic real-time sys-
tems. In Gopalakrishnan, G. and Qadeer, S., edi-
tors, Proc. 23rd International Conference on Com-
puter Aided Verification (CAV’11), volume 6806 of
LNCS, pages 585–591. Springer.
Molyneux, G. W. and Abate, A. (2020). Abc (smc): Si-
multaneous inference and model checking of chem-
ical reaction networks. In International Conference
on Computational Methods in Systems Biology, pages
255–279. Springer.
Neuhäußer, M. R. and Katoen, J.-P. (2007). Bisimulation
and logical preservation for continuous-time markov
decision processes. In International Conference on
Concurrency Theory, pages 412–427. Springer.
Neupane, T., Zhang, Z., Madsen, C., Zheng, H., and Myers,
C. J. (2019). Approximation techniques for stochas-
tic analysis of biological systems. In Automated Rea-
soning for Systems Biology and Medicine, pages 327–
348. Springer.
Sen, K., Viswanathan, M., and Agha, G. (2004). Learning
continuous time markov chains from sample execu-
tions. In First International Conference on the Quan-
titative Evaluation of Systems, 2004. QEST 2004. Pro-
ceedings., pages 146–155. IEEE.
Sen, K., Viswanathan, M., and Agha, G. (2006). Model-
checking markov chains in the presence of uncertain-
ties. In International Conference on Tools and Algo-
rithms for the Construction and Analysis of Systems,
pages 394–410. Springer.
Shavit, Y., Yordanov, B., Dunn, S.-J., Wintersteiger, C. M.,
Otani, T., Hamadi, Y., Livesey, F. J., and Kugler, H.
(2016). Automated synthesis and analysis of switch-
ing gene regulatory networks. Biosystems, 146:26–34.
Sproston, J. (2018). Qualitative reachability for open in-
terval markov chains. In International Conference on
Reachability Problems, pages 146–160. Springer.
Wang, L., Wang, S., and Bouchard-Côté, A. (2020). An
annealed sequential monte carlo method for bayesian
phylogenetics. Systematic biology, 69(1):155–183.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
526
