
GENERATION: An Efficient Denoising Autoencoders-Based Approach for
Amputated Image Reconstruction
Leila Ben Othman
1 a
, Parisa Niloofar
2 b
and Sadok Ben Yahia
2 c
1
Faculty of Sciences of Tunis, University of Tunis, El Manar, Tunisia
2
Mærsk Mc-Kinney Møller Institute, University of Southern Denmark, Odense, Denmark
Keywords:
Missing Data Mechanism, Amputation, Data Quality, Imputation, Denoising Autoencoder, Image
Reconstruction.
Abstract:
Missing values in datasets pose a significant challenge, often leading to biased analyses and suboptimal model
performance. This study shows a way to fill in missing values using Denoising AutoEncoders (DAE), a type
of artificial neural network that is known for being able to learn stable ways to represent data. The observed
data are used to train the DAE, and then they are used to fill in missing values. Extensive tests on different
image datasets, taking into account different mechanisms of missing data and percentages of missingness, are
used to see how well this method works. The results of the experiments show that the DAE-based imputation
works better than other imputation methods, especially when it comes to handling informative missingness
mechanisms.
1 INTRODUCTION
In the realm of image processing and analysis, the ac-
curate reconstruction of images plays an important
role in numerous applications, ranging from medi-
cal imaging to computer vision. However, missing
data is a common problem in real-world datasets,
which presents a challenge to researchers and prac-
titioners. Imputing missing values into datasets is a
way to obtain a filled-in dataset that can be used for
further analysis. Many methods have been created
to help with imputation, but this difficult job needs
someone who knows how missing data causes incom-
plete datasets, such as missing completely at random
(MCAR), missing at random (MAR), or missing not
at random (MNAR). MCAR and MAR are considered
ignorable missingness mechanisms, while MNAR is
nonignorable. Nonignorable missing data, in which
the chance of missingness depends on data that has
not been observed, presents a unique set of problems
that need advanced imputation methods.
While most of the studies assume the missingness
mechanism to be ignorable, in practice there is often a
reason why a value is not observed, indicating a non-
a
https://orcid.org/0000-0002-0075-9848
b
https://orcid.org/0000-0002-3147-0078
c
https://orcid.org/0000-0001-8939-8948
ignorable mechanism. For example, in a cancer clini-
cal trial, suppose the outcome variable is a biomarker
reflecting the treatment effect and is only observed at
the end of the study. Subjects may decide to drop
out before the endpoint because the treatment seems
ineffective, which leads to a violation of the ignora-
bility assumption (Zhou et al., 2014). Moreover, the
randomness assumption is too restrictive and does not
take into account the specific reasons behind missing-
ness (Ben Othman et al., 2009).
Autoencoders, a class of neural networks well-
suited for learning representation, have demonstrated
remarkable capabilities in capturing complex patterns
and features within data. Autoencoders have also
proven to be promising in impuation tasks (Pereira
et al., 2020b). The primary goal of an autoencoder is
to encode input data into a lower-dimensional repre-
sentation and then decode it back to its original form.
Denoising AutoEncoders (DAE) add a denoising
aspect to the original autoencoder and hence they are
able to reconstruct clean or denoised versions of input
data, even when the input is corrupted by noise. This
ability makes them particularly useful for tasks such
as data imputation, where missing or noisy values in
the input need to be filled in or corrected.
This paper delves into the application of DAE
to imputing missing data for image reconstruction.
However, an important but unaddressed topic for au-
Ben Othman, L., Niloofar, P. and Ben Yahia, S.
GENERATION: An Efficient Denoising Autoencoders-Based Approach for Amputated Image Reconstruction.
DOI: 10.5220/0012460700003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 3, pages 1237-1244
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
1237

toencoders is the missing data mechanism. Consider-
ing the missingness mechanism when handling miss-
ing data has always been a topic not explicitly ad-
dressed by the majority of works in the literature. This
lack of thought was one of the challenges mentioned
in a study that reviewed autoencoders for filling in
missing data (Pereira et al., 2020b). Some researchers
have looked at how the missingness mechanism af-
fects the performance of statistical or machine learn-
ing models (Niloofar et al., 2013; Niloofar and Gan-
jali, 2014). However, as far as we know, no one has
looked into how missing data mechanisms with au-
toencoders affect missing data imputation. To fill this
gap, we introduce a new approach relying on a variant
of a Densoing Autoencoder, called GENERATION,
for denoisinG autoENcodEr foR AmpuTed Image re-
cOnstructioN, which has the ability to impute missing
values for reconstructing data
We provide an in-depth analysis of how well DAE
fills in missing data using a range of ignorable and
nonignorable missingness mechanisms. The evalu-
ation encompasses a range of metrics, including er-
ror reconstruction and the accuracy of a classification
technique. Our approach is also compared to other
imputation methods and the results show clear im-
provements on classification accuracy
The remainder of the paper is organized as fol-
lows: In Section 2, we present the problem of missing
data, the missing mechanisms and a scrutiny of im-
putation methodologies. A brief overview of autoen-
coders and denoising autoencoders is also presented.
In Section 3, we thoroughly describe the GENERA-
TION approach and discuss its ability to impute miss-
ing values for the reconstruction of data. Section 4 de-
scribes the dataset, the experimental study, the results,
and the discussion of the results. Section 5 wraps this
paper up and sketches future work issues.
2 PRELIMINARIES
Data analytics researchers are aware that handling
missing data, also referred to as missing values, is
a difficult problem that requires intensive considera-
tion. This well-recognized problem arises in a real-
world context. Dealing with missing values is still a
major challenge in data analytics since they can bias
the results of statistical inferences and machine learn-
ing models. Missing values can occur for a variety
of reasons, such as human error or machine failure.
The missing data mechanisms are the procedures that
control the probabilities of missing data, according
to Little and Rubin (Little and Rubin, 2002). These
mechanisms can be categorized into three types:
• Missing Completely at Random (MCAR): when
the missing value is unrelated to any observed or
unobserved data.
• Missing at Random (MAR): when the cause of the
missing value is related to observed data.
• Missing Not at Random (MNAR): when the value
being missing is related to unobserved data (the
value itself and/or other unobserved data).
It is very important to pick the right imputation
method for the missing data mechanism, as using the
wrong method can change how well the classification
works (Twala, 2009).
2.1 Denoising Autoencoders (DAE)
An autoencoder (Michelucci, 2022) has a structure
very similar to a feedforward neural network, where
the number of neurons in the output layer is equal to
the number of inputs (just a specific version of ar-
tificial neural networks). They consist of learning a
compact or compressed representation from unlabled
training data. This compact representation consists
of reducing the data dimensionality while preserving
the important features, which can improve the perfor-
mance of prediction and classification techniques. It
is also used to reconstruct the original data with high
accuracy. An autoencoder is composed of at least
three layers (input, hidden, and output layers) and it
is composed of two parts:
• the encoder part, which goes from the input layer
to the output of the hidden layer. It maps the input
data into a compressed representation in a lower-
dimensional space, called the latent space.
• the decoder part, which goes from the hidden
layer to the output layer. It reconstructs the origi-
nal input data from the latent space representation.
The autoencoder is trained to minimize the re-
construction error (Michelucci, 2022). It means that
it has to minimize a loss function, which measures
the difference between the reconstructed data and the
original data. Common loss functions include Mean
Squared Error (MSE) for continuous data or binary
cross-entropy for binary data. The minimization of
the loss function is made with an optimization method
such as Stochastic Gradient Descent.
The main objective of autoencoders is to make a
trade-off between learning a compact representation
while still providing an accurate reconstruction. For
example, if the dimension of the latent space is too
small, the autoencoder may not be able to capture all
the essential information and this leads to a high re-
construction error. Autoencoders have been success-
fully applied to a number of real-world applications
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1238

involving data compression (Sriram et al., 2022), fea-
ture extraction (Maggipinto et al., 2018), denoising
(Lee et al., 2021), data reconstruction (Liguori et al.,
2021), anomaly detection (Torabi et al., 2023), food
fraud detection on honey (Phillips and Abdulla, 2022)
or the classification of its botanical origins (Phillips
and Abdulla, 2019).
Denoising Autoencoders (DAE) are among the
most recent models to perform the imputation of
missing data (Costa et al., 2018), and present promis-
ing results in comparison to traditional methods.
A DAE intentionally introduces noise (for instance,
Gaussian noise) to the input data to prevent the net-
work from learning the identity function (Wang et al.,
2021). As a result, a DAE is trained to reconstruct
noisy data. It means that the model is forced to learn
the reconstruction of the input, given its noisy version.
2.2 Scrutiny of Imputation
Methodologies
There are many ways to handle missing values in data.
Simply deleting the rows and/or columns with miss-
ing values, also known as available case analysis (Lit-
tle and Rubin, 2002) is not feasible in practice. Im-
putation, a method of handling missing values, fre-
quently involves replacing them with plausible val-
ues. Imputation methods can be divided into two
categories: statistical and machine learning-based
methods. Statistical imputation techniques include
mean/mode/median imputation, which replaces miss-
ing values with the mean, the mode, or the median
of the non-missing values. But the resulting imputed
values may not be representative of the true distribu-
tion of the missing variable. However, we need accu-
rate imputations to avoid biasing the data. Therefore,
several advanced statistical imputation techniques ex-
ist, such as Expectation-Maximization (EM) (Demp-
ster et al., 1977) which is an iterative procedure that
proceeds in two steps where a complete dataset is
created by imputing one or more plausible values
for the missing data. Thus, this EM approach pro-
ceeds without constructing a predictive model (Costa
et al., 2018). Machine learning-based imputation
techniques build a predictive model based on the
available data to estimate those that are missing. The
k-Nearest Neighbors (KNN) imputation (Batista and
Monard, 2002) algorithm is to cite but a few. A num-
ber of different versions have been suggested in or-
der to make the original KNN imputation more accu-
rate (Keerin and Boongoen, 2022). In (Twala, 2009),
the authors made an empirical comparison of tech-
niques for handling incomplete data using decision
trees. Recently, a similar study was carried out in
(Gabr et al., 2023) and assessed the effect of incom-
plete datasets on the performance of five classifica-
tion models. The authors employed different ratios of
missing values in different datasets that vary in size,
type, and balance. In (Awan et al., 2022), the authors
introduced a reinforcement learning (RL) approach
for imputing missing data. The proposed approach
used an action-reward principle to learn a data impu-
tation policy, where the agent learns to take decisions
to make the best estimation of the missing values.
In recent years, deep learning-based methods have
proven to be increasingly popular for dealing with
missing values. Consequently, more advanced treat-
ment methods based on Deep learning have been pro-
posed (Phung et al., 2019; Pereira et al., 2020a; S. Li
and et al., 2022). In (Costa et al., 2018), a compar-
ison study between state-of-the-art imputation tech-
niques and a Denoising Autoencoders approach was
performed. Their research demonstrates that miss-
ing mechanisms have an impact on the imputation
methods. In (Venkataraman, 2022), Denoising Au-
toencoders have shown the ability to learn complex
patterns from data, which makes them convenient for
data reconstruction.
A classical approach to data imputation needs an
evaluation step. In the literature, imputation meth-
ods are usually evaluated by measuring the distance
between the reference data (the ground truth dataset
without missing values) and the imputed data (Ben
Othman and Ben Yahia, 2006). Another evaluation
technique consists of analyzing the impact of the im-
putation on the classification accuracy (Gabr et al.,
2023). In (Ben Othman and Ben Yahia, 2018), the au-
thors suggested using different criteria and considered
an evaluation technique addressing both of the fol-
lowing issues: the stability of a clustering technique
by preserving the characteristics of the original data
through the Rand (Rand, 1971) metric and the robust-
ness of association rules (Le Bras et al., 2010). Thus,
they considered the evaluation of missing value im-
putation as a multi-criteria decision-making problem
and used the TOPSIS method (Technique for Order
of Preference by Similarity to Ideal Solution) to rank
the different imputation methods according to differ-
ent criteria.
3 DENOISING AUTOENCODER
FOR DATA IMPUTATION
In the following, we thoroughly describe our ap-
proach called GENERATION. The broad strokes of our
approach are as follows:
• Integrating the different missing data mechanisms
GENERATION: An Efficient Denoising Autoencoders-Based Approach for Amputated Image Reconstruction
1239

Input
Data Complete
Data
Incomplete
Data
Latent space
Encoder
Decoder
Reconstructed
Data
Amputation
Imputation
Figure 1: Overall architecture of GENERATION.
(MCAR, MAR, and MNAR) by introducing miss-
ing values using an amputation technique;
• Training a Denoising Autoencoder to learn the un-
derlying data structure of missingness from the
different missing value mechanisms to reconstruct
the data; and
• Evaluating the impact of the different missing data
mechanisms on imputation quality in terms of the
reconstruction quality performed by the autoen-
coder and through the application of a classifica-
tion technique
Thus, we extend the denoising autoencoders by
replacing the denoising part with a data amputation.
In the following, we describe the methodology of our
proposed approach, GENERATION for reconstruct-
ing the amputed data.
3.1 The GENERATION Methodology
Description
Figure 1 shows the overall architecture of our ap-
proach, called GENERATION. It is composed of
two main steps: the amputation and the imputation
steps. The amputation step consists of artificially in-
troducing missing values with different mechanisms
(MCAR, MAR, and MNAR). The imputation or re-
construction step is made with an autoencoder to re-
construct the missing parts. By testing our suggested
method, we can see how well it handles the differ-
ent types of missing data and how it stacks up against
other imputation methods that are already out there.
The next few paragraphs talk about the different steps
of our approach.
1. Splitting Data. This step involves dividing the
dataset into two subsets: the training set and the test
set. The training set is used to train the autoencoder,
while the test set is used to assess the performance of
the model on unseen data. It is important to mention
that the autoencoder is not provided with labeled data;
it learns to encode and decode the input data in an
unsupervised manner.
2. Amputation. Amputation refers to the process
where missing values are generated artificially in
complete data for simulation purposes. Its main ob-
jective is to simulate different scenarios with differ-
ent missing data mechanisms to assess the imputation
quality. In this work, we employed a well-established
amputation technique proposed in (Schouten et al.,
2018), where the amputation is performed consider-
ing different missing patterns (MissingnessPattern).
A missing pattern defines a missingness mechanism
(MCAR, MAR, MNAR) and a proportion. When
this process is done, a linear regression equation with
user-specified coefficients gives a weighted sum of
scores that are used to figure out the probability of
missing data. Based on his weighted sum score, each
missing pattern receives a probability of being miss-
ing for a given set of data. Then a logistic distribu-
tion function is applied to transform the weighted sum
scores into probabilities indicating whether a value
becomes missing or not. Performing this procedure
on images involves replacing some pixels with miss-
ing values (missing pixels) and pre-imputing them
with 0. In this step, we created an amputed ver-
sion of both the train and the test sets that we call
x trainAmputed and x testAmputed.
3. Initialization of GENERATION. This step
consists of defining the Denoisy Autoencoder archi-
tecture. Taking into account the results and recom-
mendations of the literature, we adopted an architec-
ture consisting of an encoding network and a decod-
ing network. The characteristics of the chosen De-
noisy Autoencoder can be described as follows:
• The architecture is symmetrical.
• Both the encoder and the decoder have several
hidden layers.
• The number of neurons per layer decreases in the
encoder and increases in the decoder.
• The number of outputs of the autoencoder is equal
to the number of inputs.
The initialization of the Denoisy Autoencoder
consists of specifying the hyper-parameters. These
hyper-parameters include the number of layers, the
number of neurons per layer, the number of dimen-
sions of the latent space and the activation functions
for the encoding and decoding parts. In the experi-
ments, we varied the autoencoder architecture and the
hyper-parameters to select the one that minimizes the
reconstruction error. Figure 2 shows the architecture
built on 28 × 28 pixel gray-scale images.
4. GENERATION Training. The training sample
is made up of the amputed data (x trainAmputed) as
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1240

Output layer
Hidden layer 3
Hidden layer 2
784 neurons
49 neurons
196 neurons
196 neurons
Hidden layer 1
Reconstructed Data
+
Input layer784 neurons
Input Data
Latent
space
Amputation
Figure 2: The GENERATION architecture at a glance.
input and the original data (x train) as target. This al-
lows the DAE to learn to reconstruct data from data
with missing values. The main goal is to get the DAE
to learn a hidden representation that shows the im-
portant parts of the data and works even when some
values are missing. Using the original data as a target,
GENERATION is pushed to generate a reconstruction
that is as close as possible to the original data. In
the training step, the target is the version of data that
we want the autoencoder to learn to reconstruct later.
Training the autoencoder requires the application of
an optimization method for minimizing a loss func-
tion. Here, the hyperparameters include the number
of epoch used to train the model, the reconstruction
error (loss function) and the optimization method.
5. Reconstruction (Imputation). During the train-
ing step, GENERATION learns a compact represen-
tation that lets the previously cut data be put back to-
gether again. Once the autoencoder is trained, we give
the amputed test sample (x testAmputed) to the en-
coder to obtain its encoding (encodedData) and then
pass that encoding through the decoder to reconstruct
the original data (ReconstructedData). The recon-
structed data should be closely related to the input
data.
4 EXPERIMENTAL RESULTS
We usher in this section by providing information
about the considered dataset.
For these experiments, we used the well-known
and publicly available MNIST dataset (https://www.
kaggle.com/datasets/zalando-research/fashionmnist).
The latter covers images of a variety of fashion items,
including t-shirts, trousers, dresses, etc. It is designed
to facilitate the development and evaluation of ma-
chine learning models for the classification of differ-
ent types of clothing items. The dataset consists of
60, 000 training examples and 10, 000 test examples.
A 28 × 28-pixel gray-scale image serves as the repre-
sentation for each example.
We carried out a series of experiments with the
aim of evaluating our proposed approach. All the
steps described in Section 3 were conducted in Python
using the TensorFlow (Martin. Abadi and et al., 2015)
framework, Pandas and scikit-learn libraries. For data
amputation, we used the pyampute https://riannesc
houten.github.io/pyampute/build/html/index.html
python library for the amputation technique. Af-
ter tuning and testing the autoencoder in terms of
the number of hidden layers and their neurons, we
achieved satisfactory training loss values for the spec-
ified autoencoder architecture depicted by figure 2.
We also used the Adam (Adaptive Moment Estima-
tion) (Kingma and Ba, 2014) optimizer to minimize
the error (loss function) with the mean squared error,
and 10 epochs proved to be suitable for the learning
process. In the following, we thoroughly describe our
experimental validation outcomes.
Serie 1 of Experiments: Gauging the Quality of the
Reconstruction Visually. In this first experiment,
we evaluated the quality of the reconstruction of GEN-
ERATION visually. Figure 3 shows this reconstruction
for the 10 first test images.with 50% of MAR values
missing. It is easy to see that the quality of the ampu-
tated images has improved after reconstruction. The
second series of experiments will validate this visu-
alization by studying the loss function during the re-
construction step.
Serie 2 of Experiments: Impact of the
Mechanisms of Missing Data. In this second
series of experiments, we study the impact of the
mechanisms of missing data: missing completely
at random data (MCAR), missing at random data
(MAR) and missing not at random data (MNAR).
We plot the loss function curves for each mechanism
with 60% of a proportion of missing values. We
present the result in Figure 4, from which we can
easily notice that:
• For the three different mechanisms (MCAR,
MAR, and MNAR), the loss function is decreas-
ing over the learning process. This indicates that
the model is learning and improving its ability to
reconstruct the missing values.
GENERATION: An Efficient Denoising Autoencoders-Based Approach for Amputated Image Reconstruction
1241
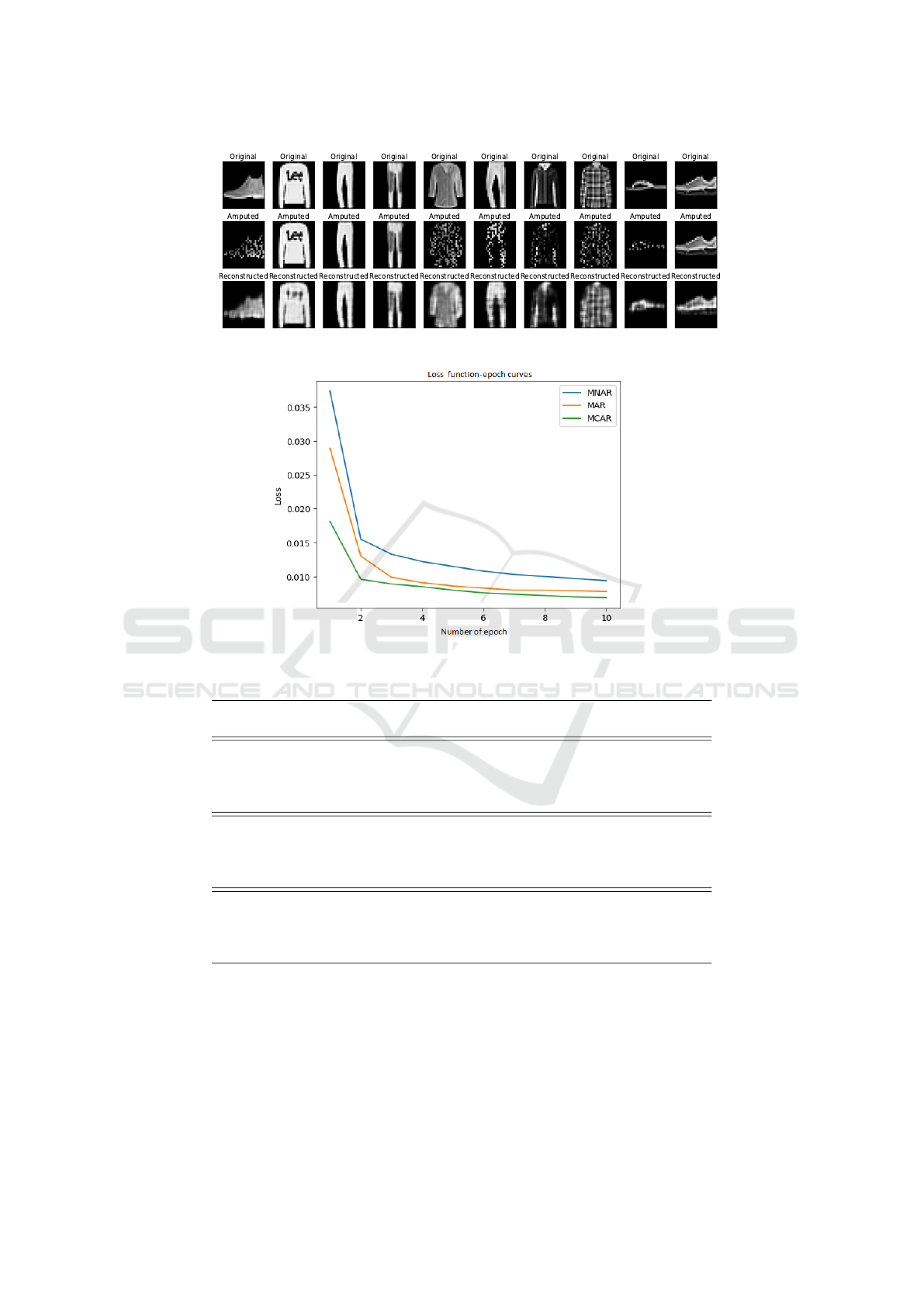
Figure 3: Visualization of original, amputed and reconstructed images with GENERATION.
Figure 4: The loss function curves for MCAR, MAR, and MNAR mechanisms: 60% of missing data.
Table 1: Accuracy using KNNeighborsClassifier (k = 5) for different missingness mechanisms.
Missingness Mechanism #MV (%) Before imputation After imputation
MCAR 30 0.6379 0.8421
50 0.6361 0.8399
70 0.6321 0.8332
90 0.6401 0.8464
MAR 30% 0.6410 0.8422
50 0.5360 0.8399
70 0.5325 0.8432
90 0.5306 0.8405
MNAR 30 0.6175 0.8418
50 0.5165 0.8531
70 0.5018 0.8620
90 0.5001 0.8710
• We can notice that the MNAR and MAR mecha-
nisms have higher loss values than the MCAR. It
means that filling in missing values in the MNAR
and MAR scenarios is harder or more complicated
to do than in the MCAR scenario. Otherwise,
performance varies according to the mechanism;
MCAR is the easiest to process. The MAR mech-
anism requires observed variables to be taken into
account and the MNAR mechanism presents ad-
ditional difficulties due to the relationships be-
tween observed and unobserved variables. The
result highlights the need for specific approaches
and consideration of the underlying missing data
mechanisms in order to deal efficiently with miss-
ing values.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1242

Table 2: Obtained accuracy values for KNNeighborsClassifier (k = 5).
Mechanism
Imputation
GENERATION KNN Median Mean
Imputer
MCAR 0.8331 0.7336 0.7357 0.7340
MAR 0.8379 0.7328 0.7325 0.7361
MNAR 0.8397 0.7374 0.7303 0.7355
Serie 3 of Experiments: Assessing the Impact of
the Reconstruction on a Classification Technique
In order to figure out how well GENERATION did at
imputation, we looked at how the reconstruction af-
fected a classification method. This constitutes an
evaluation technique for missing data handling meth-
ods. To perform this evaluation, we used the KN-
NeighborsClassifier. This involves using the autoen-
coder to impute missing values and then using the
KNNeighborsClassifier to make predictions based on
the imputed (reconstructed) data on the one hand and
amputed data on the other hand. The idea is to study
the impact on accuracy before and after imputation.
Table 1 illustrates the results obtained when applying
the classifier before and after imputation. We can tell
how well GENERATION did the imputation by com-
paring the predicted labels to the actual labels. The
results show the importance of handling missing data
when applying a classification technique. Regard-
less of the missing data mechanism, the accuracy im-
proves after imputation. In addition, we observe once
again the impact of the missing data mechanism. In
fact, accuracy tends to deteriorate when missing val-
ues increase and are MAR or NMAR before imputa-
tion.
Serie 4 of Experiments: Comparaison of the
Imputation Technique with Baseline Imputation.
In the remainder, we compare our imputation based
on GENERATION with baseline imputation tech-
niques. We employed KNN imputation (KNNeigh-
borsImputer) with k = 3, Median imputation and
Mean imputation. We then measure the accuracy
of the imputed data using the KNNeighborsClassi-
fier with k = 5. We present the results in Table 2.
No matter how the missing data is handled, our sug-
gested method using GENERATION is more accurate
than the standard imputation methods that were used.
This confirms the effectiveness of autoencoders for
handling missing values. However, the classification-
based evaluation technique does not provide addi-
tional information in relation to the missing data
mechanism. In fact, our autoencoder obtained an ac-
curacy equal to 0.83 with the different mechanisms.
However, we can notice that this was not the case with
the evaluation using the loss function. This result en-
courages us to reconsider the suitability of this evalu-
ation technique for handling missing data.
5 CONCLUSION AND FUTURE
WORK
Addressing missing values in image datasets is a crit-
ical step in ensuring the reliability and accuracy of
analyses and model performance. It was shown in this
study that denoising autoencoders are a good way to
fill in missing data in images for reliability. First, the
image dataset is amputed with missing values follow-
ing different percentages of informative and noninfor-
mative missingness mechanisms. Second, the DAE is
trained on the training set and evaluated on the test-
ing set. Third, its performance is compared with that
of other well-known prediction methods. Although
our proposed methodology shows promising results
even for noninformative missingness, there are certain
other directions that should be investigated further: (i)
The major limitation of DAEs stands in the absence of
uncertainty quantification in the model parameter es-
timation. Indeed, this could be addressed by applying
a generative model like variational autoencoders, and
(ii) the way we artificially impose missing values in
the dataset has a prominent effect on the imputation
accuracy and its outcomes. Exploring other method-
ologies and algorithms employed for this purpose is
also deemed worth further investigation.
REFERENCES
Awan, S., Bennamoun, M., Sohel, F., Sanfilippo, F., and
Dwivedi, G. (2022). A reinforcement learning-based
approach for imputing missing data. Neural Comput-
ing and Applications, 34:1–16.
Batista, G. and Monard, M.-C. (2002). A study of k-nearest
neighbour as an imputation method. volume 30, pages
251–260.
Ben Othman, L. and Ben Yahia, S. (2006). Yet another ap-
proach for completing missing values. In Proceedings
of the 4th Intl. Conf. on Concept Lattices and their
Applications (CLA 2006), Hammamet, Tunisia.
GENERATION: An Efficient Denoising Autoencoders-Based Approach for Amputated Image Reconstruction
1243

Ben Othman, L. and Ben Yahia, S. (2018). A multiple cri-
teria evaluation technique for missing values imputa-
tion. In 12th Intl. Conf. on Research Challenges in In-
formation Science, RCIS 2018, Nantes, France, May
29-31, 2018, pages 1–12. IEEE.
Ben Othman, L., Rioult, F., Ben Yahia, S., and Cr
´
emilleux,
B. (2009). Missing values: Proposition of a typology
and characterization with an association rule-based
model. volume 5691, pages 441–452.
Costa, A. F., Santos, M. S., Soares, J. P., and Abreu, P. H.
(2018). Missing data imputation via denoising autoen-
coders: The untold story. In Duivesteijn, W., Siebes,
A., and Ukkonen, A., editors, Advances in Intelligent
Data Analysis XVII, pages 87–98, Cham. Springer
Intl. Publishing.
Dempster, A., Laird, N., and Rubin, D. (1977). Max-
imum likelihood from incomplete data via the EM
algorithm. Journal of the Royal Statistical Society,
39(1):1–38.
Gabr, M. I., Helmy, Y. M., and Elzanfaly, D. S. (2023). Ef-
fect of missing data types and imputation methods on
supervised classifiers: An evaluation study. Big Data
and Cognitive Computing, 7(1).
Keerin, P. and Boongoen, T. (2022). Improved knn imputa-
tion for missing values in gene expression data. Com-
puters, Materials and Continua, 70:4009–4025.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Le Bras, Y., Meyer, P., Lenca, P., and Lallich, S. (2010).
A robustness measure of association rules. In Proc.
of ECML PKDD’10: Part II, ECML PKDD’10, pages
227–242, Berlin, Heidelberg. Springer-Verlag.
Lee, W.-H., Ozger, M., Challita, U., and Sung, K. W.
(2021). Noise learning-based denoising autoencoder.
IEEE Communications Letters, 25(9):2983–2987.
Liguori, A., Markovic, R., Dam, T. T. H., Frisch, J., Treeck,
C., and Causone, F. (2021). Indoor environment data
time-series reconstruction using autoencoder neural
networks. Building and Environment, 191:107623.
Little, R. and Rubin, D. (2002). Statistical Analysis with
Missing Data, Second Edition. John Wiley, New York.
Maggipinto, M., Masiero, C., Beghi, A., and Susto, G. A.
(2018). A convolutional autoencoder approach for
feature extraction in virtual metrology. Procedia
Manufacturing, 17:126–133. 28th Intl. Conf. on
Flexible Automation and Intelligent Manufacturing
(FAIM2018), June 11-14, 2018, Columbus, OH, US-
AGlobal Integration of Intelligent Manufacturing and
Smart Industry for Good of Humanity.
Martin. Abadi, A. A. and et al. (2015). TensorFlow: Large-
scale machine learning on heterogeneous systems.
Software available from tensorflow.org.
Michelucci, U. (2022). An introduction to autoencoders.
CoRR, abs/2201.03898.
Niloofar, P. and Ganjali, M. (2014). A new multivariate im-
putation method based on bayesian networks. Journal
of applied statistics, 41(3):501–518.
Niloofar, P., Ganjali, M., and Rohani, M. F. (2013). Im-
proving the performance of bayesian networks in non-
ignorable missing data imputation. Kuwait Journal of
Science, 40(2).
Pereira, R. C., Santos, J. C., Amorim, J., Rodrigues, P., and
Henriques Abreu, P. (2020a). Missing image data im-
putation using variational autoencoders with weighted
loss.
Pereira, R. C., Santos, M. S., Rodrigues, P. P., and Abreu,
P. H. (2020b). Reviewing autoencoders for missing
data imputation: Technical trends, applications and
outcomes. Journal of Artificial Intelligence Research,
69:1255–1285.
Phillips, T. and Abdulla, W. (2019). Class embodiment au-
toencoder (ceae) for classifying the botanical origins
of honey. pages 1–5.
Phillips, T. and Abdulla, W. (2022). A new honey adulter-
ation detection approach using hyperspectral imaging
and machine learning. European Food Research and
Technology, 249.
Phung, S., Kumar, A., and Kim, J. (2019). A deep learning
technique for imputing missing healthcare data. vol-
ume 2019, pages 6513–6516.
Rand, W. M. (1971). Objective criteria for the evaluation of
clustering methods. Journal of the American Statisti-
cal association, 66(336):846–850.
S. Li, M. L. and et al. (2022). Handling missing values in
healthcare data: A systematic review of deep learning-
based imputation techniques.
Schouten, R., Lugtig, P., and Vink, G. (2018). Generating
missing values for simulation purposes: a multivariate
amputation procedure. Journal of Statistical Compu-
tation and Simulation, 88:1–22.
Sriram, S., Dwivedi, A., Chitra, P., Sankar, V., Abirami, S.,
Durai, S., Pandey, D., and Khare, M. (2022). Deep-
comp: A hybrid framework for data compression us-
ing attention coupled autoencoder. Arabian Journal
for Science and Engineering, 47.
Torabi, H., Mirtaheri, S., and Greco, S. (2023). Practical
autoencoder based anomaly detection by using vector
reconstruction error. Cybersecurity, 6.
Twala, B. (2009). An empirical comparison of techniques
for handling incomplete data using decision trees. Ap-
plied Artificial Intelligence, 23:373–405.
Venkataraman, P. (2022). Image denoising using convolu-
tional autoencoder.
Wang, Z., Akande, O., Poulos, J., and Li, F. (2021). Are
deep learning models superior for missing data impu-
tation in large surveys? evidence from an empirical
comparison. CoRR, abs/2103.09316.
Zhou, X.-H., Zhou, C., Lui, D., and Ding, X. (2014). Ap-
plied missing data analysis in the health sciences.
John Wiley & Sons.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1244
