
Microplankton Discrimination in FlowCAM Images Using Deep
Learning
∗
Francisco Bonin-Font
1 a
, Gorka Buenvaron
2
, Mary K. Kane
3
and Idan Tuval
3
1
Systems, Robotics and Vision Group (SRVG), University of the Balearic Islands,
ctra de Valldemossa km 7.5, Palma de Mallorca, Balearic Islands, Spain
2
Institute for Cross-Disciplinary Physics and Complex Systems (IFISC), University of the Balearic Islands,
ctra de Valldemossa km 7.5, Palma de Mallorca, Balearic Islands, Spain
3
Department of Marine Ecology, Mediterranean Institute of Advanced Studies (IMEDEA),
Miquel Marqu
´
es 21, Esporles, Balearic Islands, Spain
Keywords:
Phytoplankton, Zooplankton, Convolutional Neural Networks.
Abstract:
Marine plankton are omnipresent throughout the oceans, and the Mediterranean Sea is no exception. Inno-
vation on microscopy technology for observing marine plankton over the last several decades has enabled
scientist to obtain large quantities of images. While these new instruments permit generating and recording
large amounts of visual information about plankton, they have produced a bottleneck and overwhelmed our
abilities to provide meaningful taxonomic information quickly. The development of methods based on Artifi-
cial Intelligence or Deep Learning to process these images in efficient, cost-effective manners is an active area
of continued research. In this study, Convolutional Neural Networks (CNNs) were trained to analyze images
of natural assemblages of microplankton (< 100µm) and laboratory monocultures. The CNN configurations
and training were focused on differentiating phytoplankton, zooplankton, and zooplankton consuming phyto-
plankton. Experiments reveal high performance in the discrimination of these different varieties of plankton,
in terms of Accuracy, Precision, F1 scores and mean Average Precision.
1 INTRODUCTION
Marine phytoplankton are the base of most marine
ecosystems. In the Mediterranean, algal blooms
which cause harm to the environment, economy, and
human health, also known as Harmful Algal Blooms
(HABs), are increasing in frequency (Zingone et al.,
2021). Along the Spanish and Balearic coast, HABs
caused by dinoflagellates occur frequently in summer
driven by pervasive sea breezes that push plankton to-
a
https://orcid.org/0000-0003-1425-6907
∗
This work is partially supported by ”ERDF
A way of making Europe”, Grant PLEC2021-
007525/AEI/10.13039/501100011033 funded by the
Agencia Estatal de Investigaci
´
on, under Next Generation
EU/PRTR, Grant PID2020-115332RB-C33 funded by
MCIN/AEI/10.13039/501100011033, Grant JAEICU-
2021-IMEDEA-07, funded by AEI, and by European
Union’s Horizon 2020 research and innovation programme
funds under the Marie Skłodowska-Curie grant agreement
No. 896043. The present research was carried out within
the framework of the activities of the Spanish Government
through the ”Maria de Maetzu Center of Excellence” ac-
creditation to IMEDEA (CSIC-UIB) (CEX2021-001198).
wards the shore (Basterretxea et al., 2007) (Figueroa
et al., 2008). Submarine groundwater discharges also
provide increased nutrient concentrations nearshore,
creating optimal conditions for phytoplankton growth
(Tovar-S
´
anchez et al., 2014) (Rodellas et al., 2015).
Being able to sample local blooms and identify poten-
tially toxic species of dinoflagellates from other mi-
croplankton may enable us to create an early warning
system and avoid harm to human health (Buskey and
Hyatt, 2006) (Henrichs et al., 2021). Novel methods
to classify and quantify plankton populations created
in the last several decades have increased the speed
and accuracy of their identification. Traditional mi-
croscopy involves looking at samples under a micro-
scope and can be very time consuming, depending on
the quantity of data and the ability of the involved
technicians (Menden-Deuer et al., 2020). Flow cy-
tometry permits a fast resolution of the size particles
within a sample, but it does not always gives an accu-
rate identification of plankton (Dubelaar and Jonker,
2000). Instruments like the FlowCAM and FlowCy-
tobot generate images of plankton from water sam-
606
Bonin-Font, F., Buenvaron, G., Kane, M. and Tuval, I.
Microplankton Discrimination in FlowCAM Images Using Deep Learning.
DOI: 10.5220/0012460200003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 4: VISAPP, pages
606-613
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

ples, creating a digital record which can be opened
publicly to be used and referenced by multiple scien-
tists (Ullah et al., 2022) (Yamazaki, 2022). However,
having thousands of images of different plankton to
obtain meaningful data is usually infeasible for a sin-
gle person, creating a bottleneck. This problem is
difficult to solve without using innovative automatic
image classification algorithms based on Machine or
Deep Learning.
Computers have become more powerful, and
cloud-based servers or super-computers enable the
processing of ever-larger quantities of data. Improve-
ments in Artificial Intelligence (AI), Machine Learn-
ing and Neural Networks have produced impressive
advances in visual object recognition, including auto-
mated detection and identification of plankton (Kerr
et al., 2020) (Zhang et al., 2021) (Fuchs et al., 2022)
(Zhang et al., 2023) (Sosa-Trejo et al., 2023). Al-
though these advances have greatly enhanced our
ability to classify organisms from images of natural
populations, state of the art approaches and public im-
age databases are not always applicable for localized
studies and often need to be refined before use, leav-
ing a wide range of possibilities for innovation. The
work presented in this paper goes one step forward in
using CNNs to detect and classify plankton compared
to previous approaches with respect to the quality and
origin of images used; the software platform used to
implement, train, and validate the system; and the lo-
calized character of the problem to which the CNNs
have been applied. Training, Validation and Testing
datasets are partially formed from a time series of
microplankton photography taken with a FlowCAM
VS series (Yokogawa Fluid Imaging Technologies,
Inc.) (Yokogawa, 2023) and collected at Cala San-
tany
´
ı, Mallorca, (Spain). Unlike previous approaches,
our work was centered on, in a first phase, discrim-
inating autotrophic from heterotrophic microplank-
ton, including heterotrophic microplankton that had
ingested phytoplankton, and, in a second phase, dis-
tinguishing different types of phytoplankton. Images
from cultures of Phaeodactylum tricornutum, a di-
atom, Alexandrium minutum, a dinoflagellate, and
Chlamydomonas reinhardtii, a freshwater algae, were
also included in the datasets for the second phase, in
order to increase the efficiency and performance of
the second trained Network. Experiments showed ex-
cellent testing performances in the discrimination of
different types of plankton, evaluated using classical
metrics, such as Accuracy, Recall, Precision, F1 score
and Mean Average Precision (mAP).
2 MATERIALS AND PROCEDURE
2.1 Study Site and Sample Collection
Sampling was conducted at Cala Santany
´
ı, a beach
on the southeastern side of Mallorca (Spain) between
June 2021 and mid-September 2022, with two addi-
tional dates on 27 September 2022 and 26 October
2022. This beach frequently experiences HABs in
summer. The submarine groundwater discharge pro-
vides increased nutrient availability for phytoplank-
ton growth (Basterretxea et al., 2010). Additionally,
as the beach is very well protected, water renewal
is very weak, with shoreward currents generated by
the sea breezes pushing water and plankton towards
the shore. From June to October 2021 and again
from April until mid-September 2022, sampling oc-
curred every 5-10 days, except during phytoplank-
ton blooms, when sampling occurred every 2-3 days.
From November 2021 to March 2022, sampling oc-
curred every 10-20 days.
Water samples were collected at approximately
the same location. We collected 6 L of water from
the surface and 6 L of water from a depth of ∼ 1.4 m.
After returning to the laboratory, the collected water
was passed through a 100 µm filter to remove larger
plankton and debris.
2.2 Microplankton Community
Sampling and Incubations
Fresh samples were generally processed within 24
hours of collection. Up to 1 L of water from the
surface and depth were concentrated to 50 mL us-
ing ≤ 5 µm filters to retain the microplankton size
community of interest. Sub-samples were then pro-
cessed using a FlowCAM VS series (Yokogawa Fluid
Imaging Technologies, Inc) (Yokogawa, 2023). The
FlowCAM combines flow cytometry with image mi-
croscopy, creating a laminar flow and drawing sam-
ples through a flow cell. The camera microscope
views the sample through the flow cell and captures
an image of any particle it sees using background sub-
traction. We used the 20x objective setting, meaning
objects between 2 µm and 100 µm could be imaged;
we ran each sample through the FlowCAM at a rate
of 0.05 mL/min for 20 minutes.
From August 2021 to October 2022, 1 L of wa-
ter from both the surface and depth was incubated for
4–7 days on a 14:10 light:dark cycle. Samples were
then condensed and processed in the FlowCAM us-
ing the same methodology as with the fresh samples
described above.
Microplankton Discrimination in FlowCAM Images Using Deep Learning
607

2.3 Separation of Images
The FlowCAM saves image collages of the objects it
photographs. Therefore, it is necessary to crop and
separate the images from the collages to obtain and
save photographs of individual plankton. All images
used in this study were obtained from FlowCAM col-
lages of the fresh or incubated natural samples from
Cala Santany
´
ı, and from laboratory monocultures of
P. tricornutum, A. minutum, or C. reinhardtii. Flow-
CAM images of laboratory cultures were obtained on
the same FlowCAM VS series using the 20x objective
from previous experiments.
2.4 Convolutional Neural Networks
(CNNs)
A Convolutional Neural Network is a network used
for Deep Learning composed of a set of intercon-
nected layers that consist of several neurons (matri-
ces) which perform successive convolution operations
guided by weights and biases (Alzubaidi et al., 2021).
Each layer processes grid-like topological data to out-
put also a matrix-type data structure. Weights and bi-
ases of neurons are learned and continuously updated
as new images get into the training process. CNN
models are optimized to minimize the so-called loss
function, which is the difference between each predic-
tion output by the CNN model and the corresponding
ground-truth. In addition to convolutions, CNN lay-
ers can include other operations, such as Linear Unit
Rectifications (ReLUs) or Poolings. ReLUs map neg-
ative values to 0 and maintain positive values, while
Poolings downsample the data packages transmitted
to the following layer (Alzubaidi et al., 2021). A CNN
can be trained from scratch, but it needs significant
computational resources and a huge number of train-
ing images. An alternative consists of initializing, re-
training, and reconfiguring pre-trained models, a tech-
nique commonly known as Transfer Learning (Hus-
sain et al., 2019). This method requires less data and
fewer computational resources and is the one used to
approach our plankton discrimination problem.
2.5 Differentiation Between
Phytoplankton, Zooplankton, and
Zooplankton Consuming
Phytoplankton
A first dataset composed of 2188 color images from
natural sampling contained different types of phyto-
plankton (PHY), zooplankton (ZOO), and zooplank-
ton consuming phytoplankton (ZCP). Each image
showed just one individual, so this first stage solved
an image classification problem. Figure 1 shows some
examples of zooplankton, phytoplankton and zoo-
plankton consuming phytoplankton. Since each im-
age of the dataset must contain a single organism and
were obtained cropping the collates formed by the mi-
croscopy, their resolution is not homogeneous, vary-
ing with the type, size and form of each individual.
For instance, resolutions of images of Figure 1-(a) to
(d) were, respectively, 138 × 160, 119 × 112, 82 ×
248, and 139 × 190 pixels, and resolutions of images
of Figure 1-(e) to (h) were, respectively, 295 × 388,
189 × 192, 134 × 215, and 168 × 138 pixels.
This dataset was used to re-train, validate and test
an EfficientNetv2 B3 (Tan and Le, 2019) pre-trained
model obtained from the Tensor Flow Hub (Tensor-
Flow-Org, 2023). Image resolution required a pre-
adjustment to 300 × 300 pixels. EfficientNet is one of
the most powerful Convolutional Neural Network ar-
chitectures available online, used worldwide to clas-
sify images in numerous relevant applications with
excellent results (de Zarz
`
a et al., 2022) (Huang and
Liao, 2023). Images containing either PHY, ZOO
or ZCP were clustered in 3 completely disjointed
groups: Training, Validation and Test, each account-
ing for 40, 10 and 50% of the whole image set, and
distributed as shown in Table 1. The Training and Val-
idation groups were used to re-configure and re-train
the EfficientNetv2 B3 model, and the Test group was
used to evaluate the performance of the resulting re-
trained structure with a set of images not involved in
the training phase. All aforementioned images were
manually classified to generate a ground truth needed
for both phases, training and testing: manual classifi-
cations are needed for the Neural Network to learn the
specific weights and biases of the model, and to quan-
tify the trained model performance comparing each
predicted clustering with the corresponding ground
truth. During the testing phase, the outputs of the
trained model were visually identified as True Pos-
itive (TP), True Negative (TN), False Positive (FP),
and False Negative (FN) to compute the following
metrics:
Precision =
T P
(T P + FP)
Recall =
T P
(T P + FN)
F1 − score =
(2 ∗ precision ∗ recall)
(precision + recall)
Accuracy =
(T P + T N)
(T P + T N + FP + FN)
.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
608
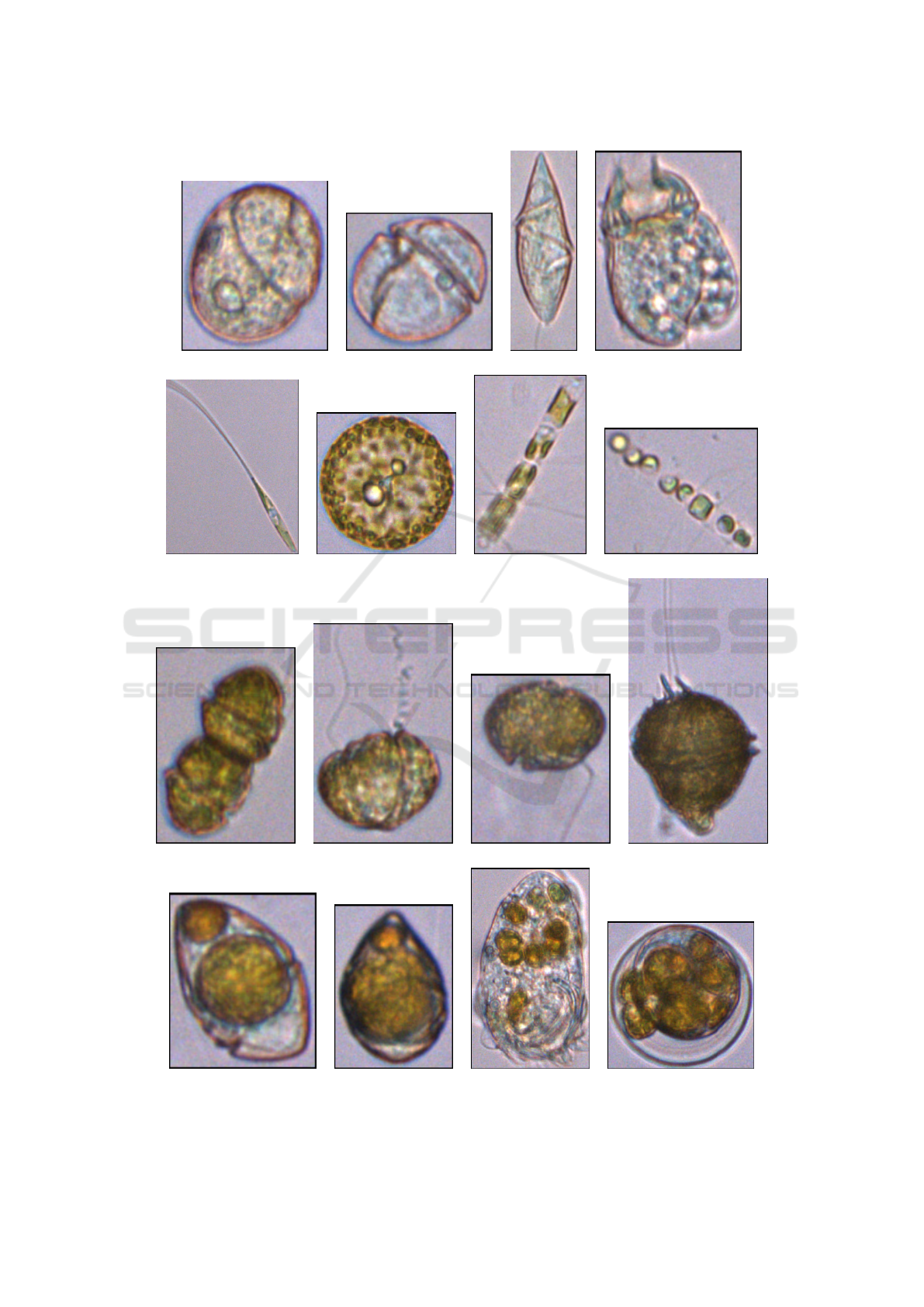
(a) (b) (c) (d)
(e) (f) (g) (h)
(i) (j) (k) (l)
(m) (n) (o) (p)
Figure 1: Samples of zooplankton ((a)-(d)), phytoplankton ((e)-(l)) and zooplankton consuming phytoplankton ((m)-(p)),
included in the first dataset. (e)-(h) are diatoms, singles ((e) and (f)) and forming chains ((g) and (h)). (i)-(l) are dinoflagellates.
Microplankton Discrimination in FlowCAM Images Using Deep Learning
609

Table 1: Number of images of each microplankton type
used to train, validate, and test the EfficientNetv2 B3 net-
work.
Training Validation Test Total
ZCP 180 44 224 448
ZOO 112 28 139 279
PHY 585 146 730 1461
Total 877 218 1093 2188
2.6 Object Detection: Identifying
Multiple Cells in an Image
One of the challenges with identifying and deter-
mining phytoplankton abundance is the ability of di-
noflagellates and diatoms, two distinct types of phyto-
plankton, to form chains. This complicates the identi-
fication of different species and also makes it difficult
to count the total number of individuals using just im-
age classification. Figures 1-(g)-(h) shows some sam-
ples of diatom chains from natural samples.
This second stage centers on phytoplankton and
approaches an object classification problem rather
than an image classification one. Now, the prob-
lem is solved using the fourth version of Ultralitics
You Only Look Once (YOLO) software infrastructure
(Bochkovskiy et al., 2020). YOLO is an open source
software package that implements deep learning and
neural networks, easy, efficient and proven to perform
excellently in object detection and image segmenta-
tion in multiple environments (Diwan et al., 2023)
(Lee and Hwang, 2022).
More than 5000 color images were manually la-
beled and clustered into 3 classes: Dinoflagellates,
Diatoms and Chlamydomonas. Dinoflagellates and
diatoms from natural sampling and used in the phy-
toplankton images of the previous dataset were sep-
arated. Monoculture images of P. tricornutum were
added to the images of diatoms from natural samples,
and monoculture images of A. minutum were added
to the images of dinoflagellates from natural samples.
As C. reinhardtii is a freshwater algae and not found
in marine environments, it was kept in its own class.
Labeled images were also split in 3 different groups:
Training, Validation and Testing. The ground truth
consists of bounding boxes framing each individual
of each class found in each image. These bounding
boxes found in the Training and Validation images are
the elements used by the network to learn the internal
weights and biases, instead of the whole images. The
resolution of all these images was fixed to 416 × 416
pixels.
Table 2 shows the number of images and individu-
als of different classes visually identified in the image
set. The last column shows the average number of in-
dividuals per class and per image. Table 3 shows the
distribution of the individuals of different classes in
the three subsets for training, validation and testing.
Original and hand-labeled Testing images were in-
put in the trained model to assess its performance,
using Precision, Recall, Accuracy, and F1-score, as
defined in section 2.5. In object detection, the In-
tersection over Union (IoU) is defined as the area of
the intersection of two bounding boxes over that of
their union. A common strategy to evaluate object
detectors, such as YOLO v4 in this case, is to select
the bounding boxes predicted by the trained model
whose detection score exceed a predefined threshold
α. Each bounding box output by the network is as-
sociated with the ground truth bounding box of the
same image that has a maximum IoU, if that IoU
exceeds a certain threshold β. The number of True
Positives (TP) are the predicted bounding boxes that
have been associated with a ground truth bounding
box, the False Positives (FP) are the predicted bound-
ing boxes that have not been associated to any ground
truth bounding box, and the False Negatives (FN) are
the ground truth bounding boxes not associated to any
prediction (Padilla et al., 2020). The Precision-Recall
Curve is built by computing these metrics for differ-
ent β values. Then, the pairs of obtained Precisions
and Recalls are sorted by the Recall value and plot-
ted. The area below this Precision-Recall Curve will
range from 0 to 1, and it is called the Average Pre-
cision (AP). A perfect object detector generates AP
close to 1, and random object detectors result in a AP
around 0.5. The mAP can be defined as the average of
AP, for the different classes. A common approach is
to use β = 0.5 (AP@0.5) as a good indicator of detec-
tion ability. According to several approaches widely
used in Deep Learning, AP and mAP correspond to
the same calculation (Wood and Chollet, 2022).
Trainings and Validations were performed under
a Google Colaboratory (Google, 2023) environment
using a GPU. Colaboratory is a hosted Jupyter Note-
book service with no setup required, and ready to use.
It provides free access to Machine and Deep learning
computing resources, including GPUs and TPUs.
3 EXPERIMENTAL RESULTS
This section gives the first results obtained in two
fronts: 1) the assessment of the image classification
network used to distinguish among different types of
plankton (PHY, ZOO and ZCP), and 2) the evaluation
of the object detector model to discriminate among
different types of phytoplankton.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
610
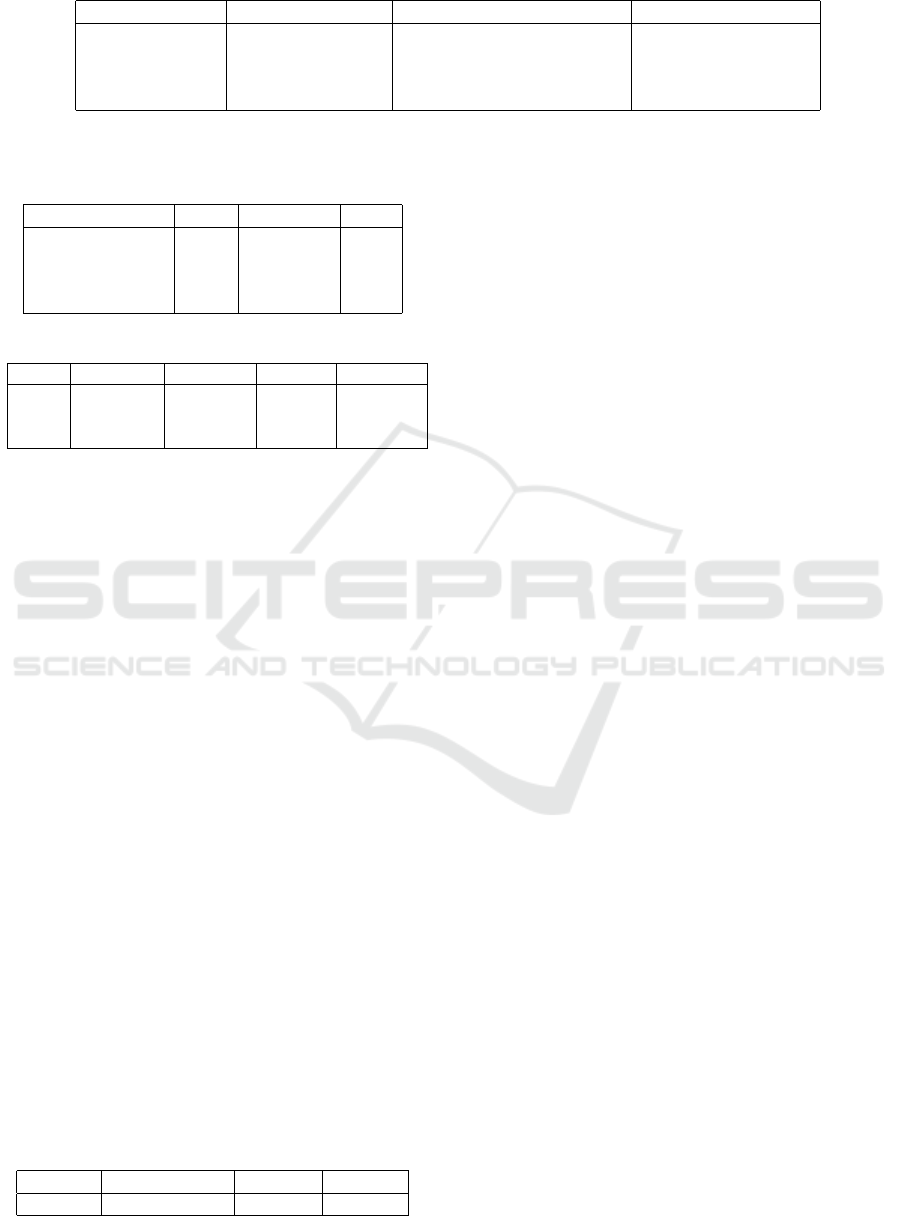
Table 2: Number of individuals identified in all the images used to train the object detection network.
Class Number of Images Total Number of Individuals Individuals per image
Chlamydomonas 1520 1666 1.10
Dinoflagellates 2940 3148 1.07
Diatoms 785 2302 2.93
Total 5245 7116 1.35
Table 3: Number of individuals of each class used to train,
validate, and test the YOLO network.
Class Train Validation Test
Chlamydomonas 595 458 467
Dinoflagellates 1179 880 881
Diatoms 324 235 226
Total 2098 1573 1574
Table 4: Evaluation metrics for image classification.
Class Accuracy Precision Recall F1-Score
PHY 97.43% 99.29% 96.84% 98.05%
ZCP 96.43% 87.14% 96.87% 91.75%
ZOO 98.07% 94.73% 90.00% 92.30%
3.1 Image Classification
Table 4 shows the scores of the different metrics
used to assess the performance of the image classi-
fier model. Accuracy, Precision and Recall surpass all
the 90%, except the Precision in the detection of ZCP,
which is greater than 87%. Accordingly, F1-Scores
are all > 90%. All these results were obtained from
the Testing dataset.
3.2 Object Detection
Table 5 shows the value of the Average Precision with
the IoU threshold β fixed in 0.5 (AP@0.5) for the
three different types of phytoplankton. The Mean Av-
erage Precision computed as the mean of all the AP
values included in table 5 is mAP(0.5)= 97.47%, a
reference value that indicates a high performance of
the trained object classifier.
Figure 2 shows several samples of inferences out-
put by the trained model.
Four diatoms at the top, in the middle, four di-
noflagellates and at the bottom, four images with C.
reinhardtii. Notice how, in the images of diatoms that
form chains, each element of the chain is marked sep-
arately as one inference of one diatom, although all
diatoms inferred in the image form a larger biological
structure.
Table 5: Average Precision per class with a β=0.5.
Class Dinoflagelates Diatoms Chlamy.
AP(0.5) 99.70% 99.60% 93.10%
4 CONCLUSIONS
This paper advances automatic classification of dif-
ferent species of plankton viewed in images recorded
using a FlowCAM. Our approach is based on train-
ing CNNs using an extensive and widely assorted im-
age set obtained from field samples collected at a lo-
cal beach in Mallorca and supplemented with images
from monocultures to identify zooplankton and phy-
toplankton, and to identify different individuals of
phytoplankton in a single image. The system, once
trained, is a potential solution to separate and taxo-
nomically identify plankton in the thousands of im-
ages which can be potentially recorded from micro-
scopic imaging methods, avoiding tedious, slow, and
sometimes imprecise manual identification and label-
ing.
The work has been divided in two parts: firstly, an
image classification network trained with a structure
based on EfficientNetv2 B3, focused on distinguish-
ing images that contain either phytoplankton or zoo-
plankton; and secondly, a YOLO v4 object classifica-
tion model trained to discriminate different types of
phytoplankton. In the first case, more than 2000 im-
ages were used to train the system, and in the second
case, more than 7000 individuals of different species
from more than 5000 images were input in the YOLO
v4 network to be trained, validated and tested. All
images involved in both parts were carefully hand-
labeled to build the ground truth. The preliminary re-
sults show how well the classification succeeds with
both models, reaching, in the first one, 90% in terms
of F1-Score, and > 93% of Average Precision with a
IoU of 0.5 in the second one.
These results encourage the authors of this pa-
per to extend the work in several directions. Ongo-
ing work is centered on the analysis of results with
a wider range of IoU thresholds. However, there
are other points that deserve our attention and could
also be included in the forthcoming work. Metrics
of Precision, Recall, Fall-out and Accuracy need to
be analyzed for the different classes of phytoplank-
ton, so that further and more solid conclusions about
the classifier performance can be inferred, and see
if the model needs to be re-trained with more or
different images. Additionally, the image database
Microplankton Discrimination in FlowCAM Images Using Deep Learning
611

Figure 2: Sample inferences output by the YOLO model.
used to build the different train, validation and test-
ing datasets can be expanded with other samples col-
lected in different areas of the Balearic shoreline and
globally to increase and expand the detection of other
individuals of other types of plankton organisms. Fur-
thermore, more advanced network infrastructures can
be tested, such as the 3 different versions of YOLO
v8, the small, the medium and the large; usually, the
greater is the size of the trained model, the finest is its
performance; however, larger models entail more pro-
cessing power. Testing different sizes of trained clas-
sifiers will permit us to find the compromise between
desired or needed detection quality and the available
computational resources. All in all, this work has
demonstrated the power of the Deep Learning infras-
tructures to ease and automate the analysis and pro-
cessing of thousands of images which are obtained
from Imaging Microscopy machines like the Flow-
CAM, and the absolute feasibility for this type of bi-
ological applications of CNNs.
ACKNOWLEDGEMENTS
The authors thank Raquel Guti
´
erez Cuenca, Paula Is-
abel Gonzalo Valmala, Alba G
´
omez Rubio, Benjam
´
ın
Casas, and both current and past members of the In-
FiBio group for their support collecting and process-
ing water samples from Cala Santany
´
ı.
REFERENCES
Alzubaidi, L., Zhang, J., Humaidi, A., Al-Dujaili, A., Duan,
Y., Al-Shamma, O., Santamar
´
ıa, J., Fadhel, M., Al-
Amidie, M., and Farhan., L. (2021). Review of Deep
Learning: Concepts, CNN Architectures, Challenges,
Applications, Future directions. Journal of Big Data,
8(53):12853–12884.
Basterretxea, G., Garc
´
e, E., Jordi, A., and Mas
´
o, M. (2007).
Modulation of Nearshore Harmful Algal Blooms by in
situ Grouth Rate and Water Renewal. Marine Ecology
Progress Series, 352:53–65.
Basterretxea, G., Tovar-S
´
anchez, A., Beck, A. J., Masqu
´
e,
P., Bokuniewicz, H. J., Coffey, R., Duarte, C. M.,
Garcia-Orellana, J., Garcia-Solsona, E., Martinez-
Ribes, L., and Vaquer-Sunyer, R. (2010). Submarine
Groudwater Discharge to the Coastal Environment of
a Mediterranean Island. Ecosystem and Biogeochemi-
cal Significance. Ecosystems, 13:629–643.
Bochkovskiy, A., Wang, C.-Y., and Liao, H.-Y. M.
(2020). Yolov4: Optimal Speed and Accuracy
of Object Detection. https://github.com/ultralytics/,
https://arxiv.org/abs/2004.10934.
Buskey, E. J. and Hyatt, C. J. (2006). Use of the FlowCAM
for Semi-automated Recognition and Enumeration of
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
612

Red Tide Cells (Karenia brevis) in Natural Plankton
Simples. Harmful Algae, 5(6):685–692.
de Zarz
`
a, I., de Curt
`
o, J., and Calafate, C. T. (2022). De-
tection of Glaucoma using Three-stage Training with
EfficientNet. Intelligent Systems with Applications,
16:200140.
Diwan, T., Anirudh, G., and Tembhurne, J. V. (2023). Ob-
ject Detection using YOLO: Challenges, Architectural
Successors, Datasets and Applications. Multimedia
Tools and Applications, 82(6):9243–9275.
Dubelaar, G. B. and Jonker, R. R. (2000). Flow Cytometry
as a Tool for the Study of Phytoplankton. Scientia
Marina, 64(2):135–156.
Figueroa, R., Garc
´
es, E., Massana, R., and Camp, J. (2008).
Description, Host-specificity and Strain Selectivity of
the Dinoflagellate Parasite Parvilucifera sinerae sp.
Nov. (Perkinsozoa). Protist, 159(4):563–578.
Fuchs, R., Thyssen, M., Creach, V., Dugenne, M., Izard,
L., Latimier, M., Louchart, A., Marrec, P., Rijkeboer,
M., Gr
´
egori, G., et al. (2022). Automatic Recognition
of Flow Cytometric Phytoplankton Functional Groups
using Convolutional Neural Networks. Limnology and
Oceanography: Methods, 20(7):387–399.
Google (2023). Google Colaboratory: a Hosted Jupyter
Notebook Service. https://colab.google/.
Henrichs, D., Angl
`
es, S., Gaonkar, C., and Campbell, L.
(2021). Application of a Convolutional Neural Net-
work to Improve Automated Early Warning of Harm-
ful Algal Blooms. Environmental Science and Pollu-
tion Research, 28:28544–28555.
Huang, M.-L. and Liao, Y.-C. (2023). Stacking Ensem-
ble and ECA-EfficientNetV2 Convolutional Neural
Networks on Classification of Multiple Chest Dis-
eases Including COVID-19. Academic Radiology,
30(9):1915–1935.
Hussain, M., Bird, J. J., and Faria, D. R. (2019). A Study
on CNN Transfer Learning for Image Classification.
In Advances in Computational Intelligence Systems,
pages 191–202.
Kerr, T., Clark, J. R., Fileman, E. S., Widdicombe, C. E.,
and Pugeault, N. (2020). Collaborative Deep Learn-
ing Models to Handle Class Imbalance in FlowCam
Plankton Imagery. IEEE Access, 8:170013–170032.
Lee, J. and Hwang, K.-i. (2022). YOLO with Adap-
tive Frame Control for Real-time Object Detection
Applications. Multimedia Tools and Applications,
81(25):36375–36396.
Menden-Deuer, S., Morison, F., Montalbano, A. L., Franz
`
e,
G., Strock, J., Rubin, E., McNair, H., Mouw, C.,
and Marrec, P. (2020). Multi-Instrument Assess-
ment of Phytoplankton Abundance and Cell Sizes in
Mono-Specific Laboratory Cultures and Whole Plank-
ton Community Composition in the North Atlantic.
Frontiers in Marine Sciences, 7(254).
Padilla, R., Netto, S. L., and da Silva, E. A. B. (2020). A
Survey on Performance Metrics for Object-Detection
Algorithms. In Proceedings of the International Con-
ference on Systems, Signals and Image Processing
(IWSSIP), pages 237–242.
Rodellas, V., Garcia-Orellana, J., Masqu
´
e, P., and Wein-
stein, Y. (2015). Submarine Groundwater Discharge
as a Major Source of Nutrients to the Mediterranean
Sea. The Proceedings of the National Academy of Sci-
encese PNAS, 112(13):3926–3930.
Sosa-Trejo, D., Bandera, A., Gonz
´
alez, M., and Hern
´
andez-
Le
´
on, S. (2023). Vision-based Techniques for Auto-
matic Marine Plankton Classification. Artificial Intel-
ligence Reviews, 56:12853–12884.
Tan, M. and Le, Q. (2019). EfficientNet: Rethinking Model
Scaling for Convolutional Neural Networks. In Pro-
ceedings of the 36th International Conference on Ma-
chine Learning, pages 6105–6114.
Tensor-Flow-Org (2023). Tensor Flow Hub, a Repos-
itory of Trained Machine Learning Models.
https://www.tensorflow.org/hub.
Tovar-S
´
anchez, A., Basterretxea, G., Rodellas, V., S
´
anchez-
Quiles, D., Garc
´
ıa-Orellana, J., Masqu
´
e, P., Jordi, A.,
L
´
opez, J. M., and Garcia-Solsona, E. (2014). Contri-
bution of Groundwater Discharge to the Coastal Dis-
solved Nutrients and Trace Metal Concentrations in
Majorca Island: Karstic vs. Detrital Systems. Environ
Sci. Technol, 48:11819–11827.
Ullah, I., Carri
´
on-Ojeda, D., Escalera, S., Guyon, I. M.,
Huisman, M., Mohr, F., van Rijn, J. N., Sun, H., Van-
schoren, J., and Vu, P. A. (2022). Meta-Album: Multi-
domain Meta-Dataset for Few-Shot Image Classifica-
tion. In Neural Information Processing Systems.
Wood, L. and Chollet, F. (2022). Efficient Graph-Friendly
COCO Metric Computation for Train-Time Model
Evaluation. https://arxiv.org/abs/2207.12120.
Yamazaki, H. (2022). Plankton Image Dataset from a Ca-
bled Observatory System (JEDI System/OCEANS)
Deployed at Coastal Area of Oshima Island, Tokyo,
Japan. https://doi.org/10.48518/00014.
Yokogawa (2023). FlowCam: Flow Imaging Microscopy.
https://www.yokogawa.com/solutions/products-
and-services/life-science/flowcam-flow-imaging-
microscopy/.
Zhang, J., Li, C., Yin, Y., Zhang, J., and Grzegorzek, M.
(2023). Applications of Artificial Neural Networks
in Microorganism Image Analisis: a Comprehensive
Review from Conventional Multilayer Perceptron to
Popular Convolutional Neural Network and Potential
Visual Transformer. Artificial Intelligence Review,
56:1013–1070.
Zhang, Y., Lu, Y., Wang, H., Chen, P., and Liang, R.
(2021). Automatic Classification of Marine Plankton
with Digital Holography Using Convolutional Neural
Network. Optics and Laser Technology, 139:106979.
Zingone, A., Escalera, L., Aligizaki, K., Fern
´
andez-Tejedor,
M., Ismael, A., Montresor, M., Mozeti
ˇ
c, P., Tas¸, S.,
and Totti, C. (2021). Toxic Marine Microalgae and
Noxious Blooms in the Mediterranean Sea: A Con-
tribution to the Global HAB Status Report. Harmful
Algae, 102:101843.
Microplankton Discrimination in FlowCAM Images Using Deep Learning
613
