
A Deep Analysis for Medical Emergency Missing Value Imputation
Md Faisal Kabir
a
and Sven Tomforde
b
Institute of Computer Science, Department of Intelligent Systems, University of Kiel,
Christian-Albrechts-Platz 4, Kiel, Germany
Keywords:
Medical Emergency System, Missing Value, Data Imputation, Autoencoder, Deep Learning.
Abstract:
The prevalence of missing data is a pervasive issue in the medical domain, necessitating the frequent deploy-
ment of various imputation techniques. Within the realm of emergency medical care, multiple challenges have
been addressed, and solutions have been explored. Notably, the development of an AI assistant for telenotary
service (TNA) encounters a significantly higher frequency of missing values compared to other medical appli-
cations, with these values missing completely at random. In response to this, we compare several traditional
machine learning algorithms with denoising autoencoder and denoising LSTM autoencoder strategies for im-
puting numerical (continuous) missing values. Our study employs a genuine medical emergency dataset, which
is not publicly accessible. This dataset exhibits a significant class imbalance and includes numerous outliers
representing rare occurrences. Our findings indicate that the denoising LSTM autoencoder outperforms the
conventional approach.
1 INTRODUCTION
Medical emergencies represent a paramount concern,
posing substantial challenges not only in addressing
patients’ life-threatening conditions but also in the
prompt assessment and treatment initiation faced by
healthcare professionals. This process is inherently
time-intensive. Therefore, an existing telenotary ser-
vice (TeleNotarzt, TNA) (Aachen, 2023) is being un-
dertaken to enhance support during medical emergen-
cies.
The successful categorization of diseases in future
emergency medical applications requires a substan-
tial dataset. Our dataset, sourced from a decade of
TNA operations in Aachen, Germany, includes mea-
surement data records for patients and their emer-
gency cases. However, numerous patient samples
lack essential parameters, potentially leading to inac-
curate classification results. Furthermore, the dataset
displays a notable class imbalance and incorporates
outliers reflective of genuine medical events, which
are crucial for understanding specific diseases. This
study aims to investigate missing value imputation
techniques that effectively handle minority class in-
stances while accurately identifying outliers linked to
rare disease events.
a
https://orcid.org/0009-0000-2354-2193
b
https://orcid.org/0000-0002-5825-8915
Within this research paper, we propose two de-
noising autoencoder models, namely the conventional
denoising autoencoder and the denoising LSTM au-
toencoder, with the primary objective of addressing
missing value imputation. Subsequently, we per-
form a comparative analysis of both models with an
emphasis on optimization. Additionally, our pro-
posed model is systematically evaluated against var-
ious machine learning models and statistical tech-
niques, including mean imputations, K-nearest neigh-
bors (KNN), Iterative Imputer (I.Imp), Decision
Tree (D.Tree), and Linear Regression (LR). This is
the first study to complete the missing imputation for
the emergency medical dataset, a corresponding real-
world-based. With this paper, we aim to answer the
question of which techniques are most suitable for
this kind of scenario and which challenges need to be
tackled.
The paper is organized as follows: Section 2 re-
views existing literature on missing data patterns,
techniques, and methodologies. Section 3 discusses
current state-of-the-art developments. Section 4 ex-
plains the formulation of proposed models, and Sec-
tion 5 details data description and preparation. Model
evaluation is covered in Section 6, while Section 7
analyzes experimental outcomes, presenting insights
and findings. Finally, Section 8 summarises the paper
and outlines future research directions.
Kabir, M. and Tomforde, S.
A Deep Analysis for Medical Emergency Missing Value Imputation.
DOI: 10.5220/0012457300003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 3, pages 1229-1236
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
1229

2 BACKGROUND
Missing values are a common problem in a dataset.
The causes of missing values are the data entry mech-
anism not working, updating the new system, human
error, system error, faulty measurements, etc. In this
case, the critical issue needs values for future product
improvement. So, it is necessary to know 1) types of
missing value, 2) strategies of missing value imputa-
tion and 3) imputation techniques.
2.1 Types of Missing Values
In situations where data are classified as Missing
Completely at Random (MCAR), there is an absence
of any discernible connection between missing and
non-missing values. In other words, missing and non-
missing variables contain no relationship. The miss-
ing value placed in a dataset is entirely random (Em-
manuel et al., 2021; Hameed and Ali, 2023). So, the
probability is represented as: P(X
j
|X
j
, X
i
) = P(X
j
).
This equation indicates that the probability of miss-
ingness (X
j
) is not influenced by the observed data
(X
i
) or the missing data (X
j
), as the conditional prob-
ability is equal to the unconditional probability (Em-
manuel et al., 2021; Hameed and Ali, 2023).
Missing at Random (MAR) considers if the miss-
ing data is connected to observed variables and has a
relationship between the subset of that variable. For
example, if we consider X
i
as an observed variable,
which is the person’s name, X
i
is age, X
m
is monthly
income, and other variables are different parameters.
Here X
j
is missing variables (Emmanuel et al., 2021;
Hameed and Ali, 2023). We can write that the proba-
bility of missing is P(X
m
|X
m
, X
i
) = P(X
m
|X
i
).
When the missing variable depends on itself and
the observed variable, we call it Missing Not at Ran-
dom (MNAR) (Emmanuel et al., 2021; Hameed and
Ali, 2023). This can be expressed as: P(X
m
|X
m
, X
i
) =
f (X
m
X
i
).
2.2 Missing Data Handling
AI missing imputation divides into two parts: ma-
chine learning and deep learning. Deep learning is
a subsection of machine learning, but deep learning is
one of the best approaches to complete missing val-
ues. The Deep learning approach mainly focuses on
model-based and predicts missing values to achieve
the target dataset. Deep learning uses a neural net-
work with multiple layers to discover the dataset’s
complex pattern. Conversely, deep learning is com-
plicated to construct a model and requires high com-
putational capacity (Emmanuel et al., 2021; Liu et al.,
2023a; Sun et al., 2023; Liu et al., 2023b).
2.3 Missing Imputation Techniques
Machine learning missing value imputation tech-
niques based on Classification and clustering mod-
els. Bayesian, neural network, decision tree, ran-
dom forest and support vector machine are con-
sidered classification-based models. On the other
hand, k-means, subspace, self-organising map, and
kernel are clustering-based models. To complete
missing imputation, autoencoder (AE), variational
autoencoder (VAE), generative adversarial networks
(GANs), Long short-term memory (LSTM) networks,
and transformers are used in deep learning (Adhikari
et al., 2022; Emmanuel et al., 2021; Liu et al., 2023a;
Sun et al., 2023).
3 RELATED WORK
Recently, many methods have been employed for
missing data imputations across diverse data types
spanning various sectors. This section mainly con-
centrates on the application of these methods in the
context of medical and healthcare datasets. The cur-
rent state of the art reveals the utilization of multi-
ple models based on deep learning techniques for ad-
dressing missing data across various data types.
In the medical domain, missing imputation tech-
niques are extensively utilized for tabular static data,
signals, temporal data, images, and genetic and ge-
nomic data. Deep learning models have demonstrated
superior accuracy compared to alternative algorithms.
Typically, missing data imputation serves as a critical
step in data preprocessing, with the ultimate objec-
tive often being prediction or classification analysis.
Concurrently, various statistical and machine learning
models are also employed to assess imputation accu-
racy for subsequent processes (Liu et al., 2023a).
Notably, multilayer perceptron (MLP) with gradi-
ent descent emerges as a successful method for imput-
ing missing values, showcasing superior performance
compared to other statistical methods such as mode,
random, hot-deck, KNN, decision tree, and random
forest (Pan et al., 2022). GapNet (Chang et al., 2022),
another approach rooted in the MLP model, exhibits
similar imputation performance as MLP with gradi-
ent descent. MLP models, known for their effective-
ness in handling static data, also demonstrate note-
worthy performance in genetic datasets, exemplified
by deepMC (Mongia et al., 2020).
For temporal data, models such as Long Short-
Term Memory (LSTM), Recurrent Neural Network
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1230

(RNN), and Gated Recurrent Unit (GRU) are specif-
ically designed and proficient in handling time se-
ries missing values. To predict hypertensive disor-
der in pregnancy (HDP) using pregnancy examination
data, a Bidirectional LSTM model outperforms cubic
spline interpolation, KNN filling, the LSTM model,
and the ST-MVL model (Lu et al., 2023). On the other
hand, a forward-to-backward bidirectional model em-
ploying missing imputation demonstrates enhanced
accuracy in predicting Alzheimer’s disease compared
to other methods (Ho et al., 2022).
Within the medical domain, autoencoders are ex-
tensively employed for missing value imputation. An
illustrative experiment utilized a sample autoencoder
to impute missing values using a dataset with ran-
domly masked values in the missing positions. This
approach was compared with mean imputation, re-
vealing superior results in favour of the autoencoder
method over mean imputation (Macias et al., 2021).
Additionally, employing a random mask as an auxil-
iary input proved beneficial in assisting the network
to discern between missing and zero features. In
this context, autoencoders demonstrated superior per-
formance across five distinct datasets, namely Breast
Cancer Wisconsin (WDBC), Parkinson’s disease, di-
abetic data, Indian Liver Patient Dataset (ILPD),
and National Surgical Quality Improvement Program
(NSQIP). This performance surpassed that of the
support vector machine (SVM), k-nearest neighbour
(KNN), artificial neural network (ANN), linear re-
gression (LR), and random forest (RF) (Kabir and
Farrokhvar, 2022).
A stacked denoising autoencoder (DAE) was im-
plemented with a low-dimensional representation.
The weight matrix of the last layer in the encoder
was utilized as the transpose weight matrix in the first
layer of the decoder (Abiri et al., 2019). In the Fram-
ingham Heart dataset, the process of missing impu-
tation was conducted using a DAE as the generator
within an Improved Generative Adversarial Network
(IGAN). Before inputting the data into the model,
missing values were filled using K-Nearest Neigh-
bors (KNN). Notably, IGAN demonstrated superior
performance when compared to Simple imputation,
KNN, MissForest, Neighborhood Aware Autoen-
coder (NAA), and Improved Neighborhood Aware
Autoencoder (INAA) (Psychogyios et al., 2022).
The current state of the art reveals numerous stud-
ies addressing missing imputation in various medical
datasets. However, no existing work has specifically
focused on addressing the challenge of emergency
medical missing data imputation. In response, we
study two autoencoder models—the Denoising Au-
toencoder and the Denoising LSTM Autoencoder for
the imputation of emergency medical data.
4 PROPOSED MISSING VALUE
IMPUTATION
4.1 Autoencoder
An autoencoder (AE) comprises an encoder and
a decoder. The encoder processes the input data
and generates an output that represents a reduced-
dimensional vector space, commonly referred to as
the latent space or information bottleneck. The math-
ematical functions governing the encoder and decoder
are denoted as f (x) = z and g(z) = x
′
. Let us pro-
vide a more detailed elaboration of these functions.
In the context of model construction for the encoder,
w1 and b1 represent the weight matrix and bias of the
first layer, respectively. Here, x and f denote the in-
put and activation functions, leading to the presenta-
tion of the output as z. (Wang et al., 2016; Chen and
Guo, 2023) The equation can be expressed as follows:
z = f (w × x + b). Same as for the decoder, w
′
, b
′
,g
and x
′
are weight, bias, activation function of decoder
output layer and reconstruction data presented. z is
the input of the decoder. So the equation presented
as: x
′
= g(w
′
× z + b
′
). Training an autoencoder loss
function is also an important issue. The loss func-
tion calculates how input data is reconstructed. To
define the loss function, the following expression is
used: L(x, g( f (x)))
4.2 Denoising Autoencoder
The denoising autoencoder (DAE) represents an ex-
tension of the standard autoencoder. While the AE is
designed to reconstruct input data, the DAE, in con-
trast, is tasked with reconstructing the original data
from noisy input. In essence, the encoder of the DAE
processes noisy data denoted as x
′′
, and the decoder
reconstructs the corresponding noise-free data, which
is presented as x. It is important to note that apart from
this distinction in their reconstruction objectives, both
the AE and DAE share an identical model construc-
tion. The input data of DAE adds some extra noise,
which could be random noise(Vincent et al., ; Vincent
et al., 2008).
Figure 1 illustrates our proposed DAE, delineat-
ing two distinct components: one for training the
DAE and the other one for imputation of missing val-
ues. Initially, the dataset was partitioned into two
sub-datasets: one is samples with missing values, and
the other contains samples with non-missing values.
A Deep Analysis for Medical Emergency Missing Value Imputation
1231

Figure 1: Denoising LSTM autoencoder imputation archi-
tecture.
The non-missing dataset adds some noise as mean
values in the first block. The mean value replaces
the non-missing dataset’s exact location where data
are missing in the missing value dataset. So, the
encoder, decoder and the loss function of the DAE
model are expressed as follows: z = f (w × x
′′
+ b),
x = g(w
′
× z + b
′
) and L(x, g( f (x
′′
))).
The second block of Figure 1 depicts the impu-
tation procedure. In this process, complete the miss-
ing value with the mean value as noise in the missing
dataset. After training the model with the noisy com-
plete dataset. Now, test the missing values for imputa-
tion. The reconstruction data present imputation data.
We construct the model as simply as possible because
of the small dataset and reduce the overfitting.
4.3 LSTM Autoencoder
The denoising LSTM autoencoder is an extension of
DAE (Coto-Jimenez et al., 2018). Figure 1 depicts an
alternative DAE model, specifically built as a Denois-
ing Long Short-Term Memory Autoencoder (LSTM
DAE). It’s important to note that the primary dis-
tinction between the first and second models lies in
the design of the encoder and decoder. The initial
DAE model employed conventional neurons for its
autoencoder, while the LSTM DAE model was con-
structed using LSTM components. Notwithstanding
this disparity, all other configurations and parameters
of the models remain consistent. Similarly, the second
block, corresponding to missing value imputations,
follows the same procedure. Another consideration
pertains to data preparation for LSTM; details on this
aspect are provided in the data preparation section.
5 DATA DESCRIPTION AND
PREPARATION
5.1 Dataset
In this research study, we harnessed emergency med-
ical data, which is not publicly accessible, and aimed
to address the challenge of missing value imputa-
tion. The data originated from emergency medical
cases involving numerous patients, thus constituting a
multifaceted dataset encompassing various attributes
and parameters, including categorical, numerical, and
text-based data. This dataset exhibited substantial
heterogeneity. Our focus primarily centred on numer-
ical data, such as blood pressure, oxygen levels, and
various measurements, which are inherently specific
to individual diseases. The dataset encompasses ap-
proximately 200 distinct diseases with varying preva-
lence. While some diseases are commonly encoun-
tered, others are quite rare, resulting in a pronounced
class imbalance within the dataset.
The dataset, comprising about 16,000 samples,
has an incidence of approximately 50% missing val-
ues. Of these samples, 9,834 are complete, and the
rest exhibit missing data. The missing values ap-
pear randomly, with little discernible interrelationship
among variables, suggesting a lack of inherent asso-
ciations. These omissions result from the data gen-
eration process, where telenotary doctors input in-
formation during emergency cases via telephone and
video calls without mandatory field checks or re-
minder functions. A synthetic dataset mirroring the
actual distribution is available in the appendix.
5.2 Data Preparation
At first, we separated the dataset into two parts: miss-
ing and non-missing datasets. To train a denoising
autoencoder, we need one noisy train dataset, which
will be reconstructed into original data. Traditionally,
extra noise in the training dataset and train the model.
We add the noise in a slightly different way, not using
random noise on the training dataset.
Figure 2 demonstrates the process of adding noise.
In this figure, there are two blocks presents. The first
block shows a simple block diagram of how the pro-
cess is realized. The second block presents the same
as the first block but in the form of a table representa-
tion of missing imputation. At first, we have to iden-
tify the location of the missing values on missing sam-
ples. Then, we place the exact location on the com-
plete dataset and replace the mean of the non-missing
dataset. Now, this non-missing dataset gets corrupted
or noisy(Ma et al., 2020).
We employed various missing data percentages
for imputation within the dataset. These missing data
percentages encompassed values of 20%, 30%, 40%,
and 50%.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1232
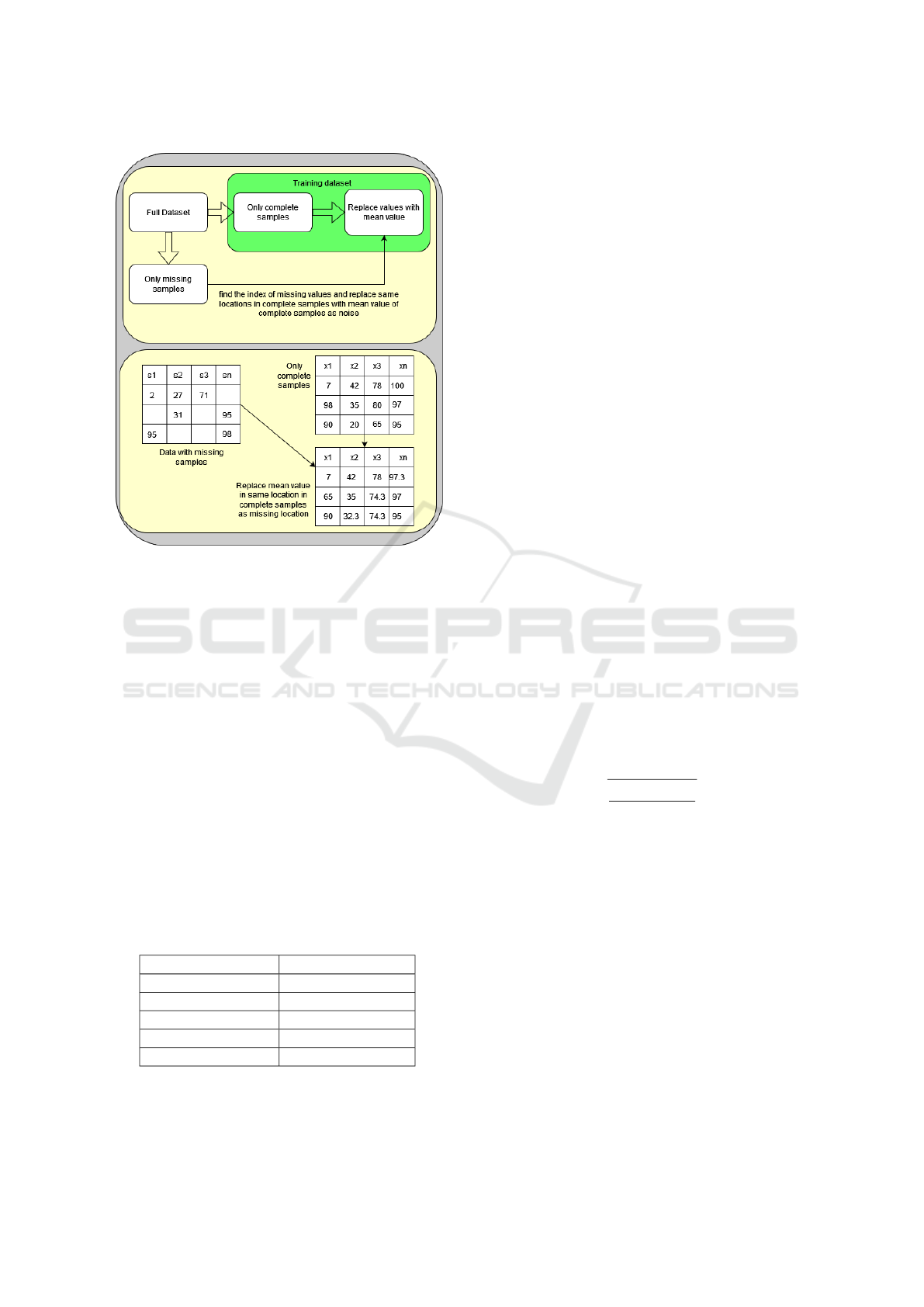
Figure 2: Data preparation for denoising autoencoder.
6 EXPERIMENTS
This experiment section describes two model configu-
rations: the activation function, the evaluation matrix
and the results.
6.1 Model Configuration
From the complete dataset with no missing values,
12% was assigned for creating the ground truth, and
the rest constituted the training dataset. Within the
training dataset, 70% was used for training, and the
remaining 30% served for validation. To optimize re-
sults, we adopted a trial-and-error approach, leading
to a variable training epoch. Given the highly class
imbalance and presence of outliers, we implemented
early stopping to address potential overfitting.
Table 1: Model hyperparameter and hyperparameter range.
Parameter value range
Learning rate 0.0001 ≤ n ≤ 0.01
Batch size 16 ≤ n ≤ 64
Hidden layers 0 ≤ n ≤ 2
Hidden layers units 16 ≤ n ≤ 64
Activation function Relu and Softmax
Table 1 outlines the hyperparameters and their
corresponding ranges utilized in both models, care-
fully selected based on superior performance com-
pared to alternative configurations. Key hyperparam-
eters influencing model construction include hidden
layers, units within hidden layers, activation func-
tions, learning rate, and batch size. Learning rates
spanned from 0.0001 to 0.01, with batch sizes rang-
ing from 16 to 64. An extended exploration con-
sidered learning rates from 0.00001 to 0.1 and batch
sizes from 4 to 512, with larger batch sizes incorpo-
rating batch size normalization layers. Hidden layer
parameters were optimized through exploration, vary-
ing from 0 to 2 layers and an additional experiment
ranging from 0 to 5 layers. Hidden layer units ranged
from 16 to 64, with an extensive analysis consider-
ing units from 4 to 512. The Relu activation function
was predominantly used in hidden layers, while soft-
max was exclusively employed in the dense output
layer. Various activation functions were evaluated for
their efficacy in hidden layer presentations. The mean
squared error (MSE) loss function served as the train-
ing metric, assessing loss across both training and val-
idation datasets. This paper introduces two models
with 24 distinct hyperparameter combinations for in-
dividual missing percentages, each trained separately.
The identified hyperparameter ranges are crucial for
model training and missing imputation.
6.2 Evaluation Matrix
In the test experiment, several evaluation matrices
were performed to test the ground truth and recon-
struction of ground truth. Other measures have also
been analysed, but the RSME provides the most re-
liable scores. This paper includes only root mean
square error (RMSE). The RMSE is defined as
r
∑
n
i=1
( ˆy
i
− y
i
)
2
n
In the equation above, n refers to the number of
observations. ˆy
i
and y
i
are presents as predicted and
original data respectively (Tyagi et al., 2022).
6.3 Results
In this section, we analyze the experiment outcomes,
emphasizing the optimal performance achieved with
diverse hyperparameters. Table 2 exclusively show-
cases the top result among 24 unique model configu-
rations. For each missing percentage category (e.g.,
20%, 30%, etc.), the best-performing model is high-
lighted from the 24 configurations.
Overall, eight results were displayed from 96
models. So, we present only two figures for ground
truth and predicted ground truth. We choose for 30%
missing imputation results and comparison.
A Deep Analysis for Medical Emergency Missing Value Imputation
1233
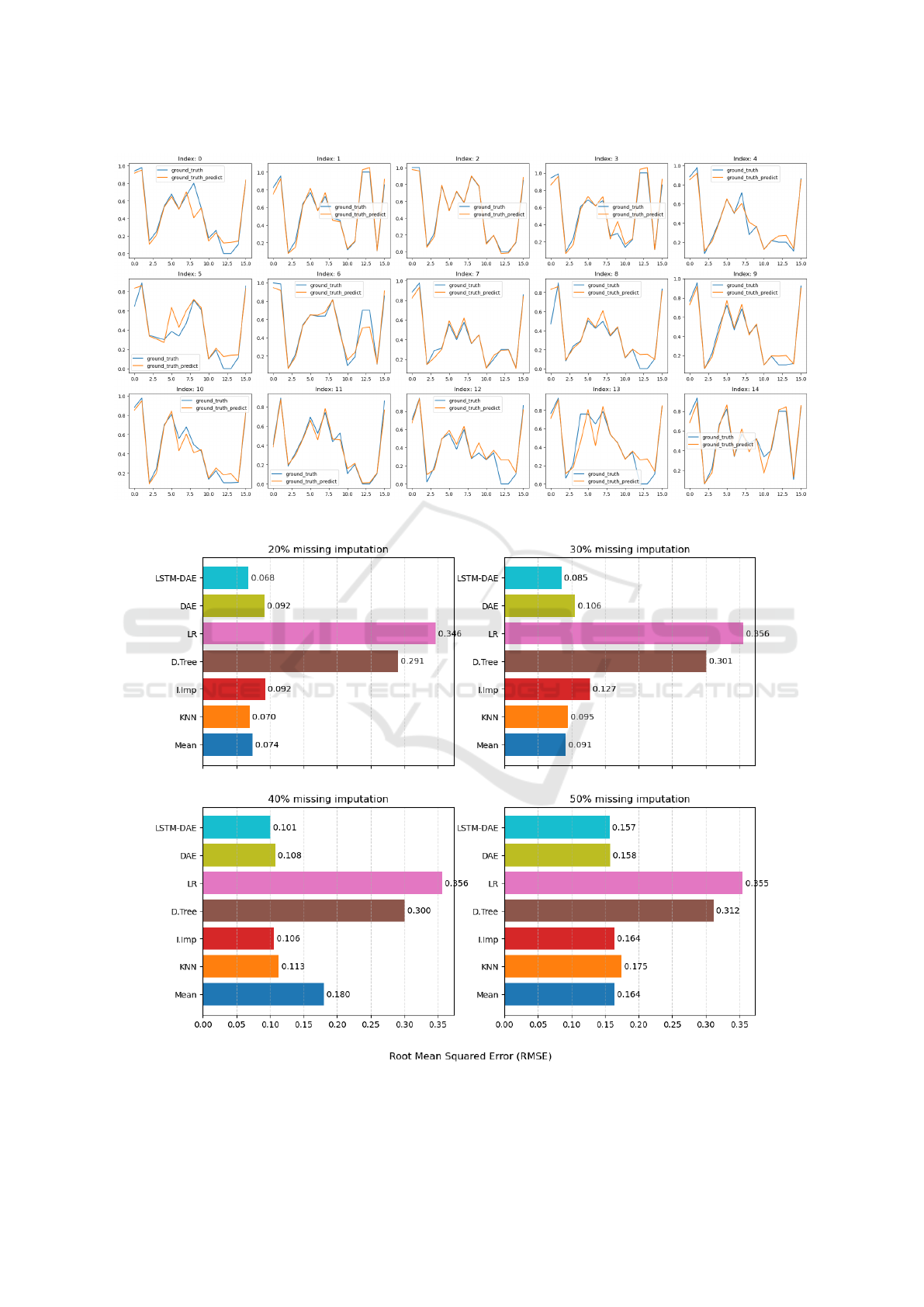
Figure 3: Comparision of ground truth and predicted ground truth for LSTM DAE.
Figure 4: Comparision of seven different missing imputation methods.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1234
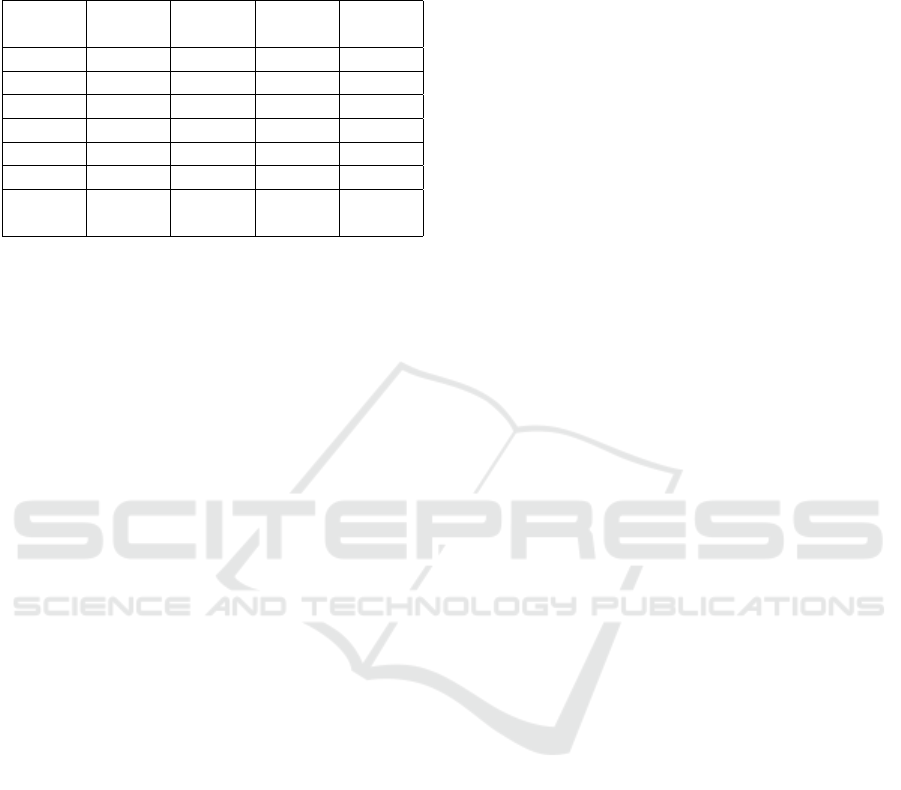
Table 2: LSTM DAE model compared with several ma-
chine learning and statistical methods for missing imputa-
tion. Here is the RMSE value, which is compared to the
ground truth generated.
model
\missing
20% 30% 40% 50%
Mean 0.07369 0.09149 0.18007 0.16381
KNN 0.06960 0.09472 0.11314 0.17491
I.Imp 0.09231 0.12739 0.10607 0.14498
D.Tree 0.29109 0.30084 0.30034 0.31181
LR 0.34629 0.35624 0.35604 0.35461
DAE 0.09180 0.10596 0.10763 0.15755
LSTM
DAE
0.06792 0.08523 0.10083 0.15702
Figure 3 illustrates the comparison between the
actual ground truth and the predicted values based on
the ground truth. The figure comprises 15 subplots,
with each subplot representing distinct features. In
Figure 3, the original ground truth is depicted by blue
lines, while the orange lines indicate the predicted
values.
7 DISCUSSION
The results section presents the outcomes of the miss-
ing values imputation across various missing per-
centage scenarios, ranging from 20% to 50% miss-
ing. Notably, the LSTM DAE consistently outper-
forms the standard DAE in every missing data case.
The proposed models perfectly capture the class-
imbalanced data.
The optimal DAE model for 20% missing imputa-
tion features a single hidden layer with 64 units for
both the encoder and decoder, outperforming other
configurations with a learning rate of 0.0001 and a
batch size of 64. Similarly, for 30%, 40%, and 50%
missing imputation, the DAE model performs best
with the same configuration, adjusting the learning
rate slightly to 0.001. A lower learning rate enhances
model accuracy as the missing imputation increases.
For the LSTM DAE, the rise in missing values cor-
responds to a reduction in the number of hidden layer
units. Conversely, an increase in learning rate and
batch size improves performance as missing values
decrease. Specifically, configurations with 64 units
for the hidden layer, a learning rate of 0.0001, and
a batch size of 16 exhibit superior performance for
20%, 30%, and 40% missing values compared to all
other setups. Conversely, the model for 50% missing
values imputation outperforms when configured with
32 hidden layer units, a learning rate of 0.001, and a
batch size of 32.
Both models demonstrate their peak performance
when the proportion of missing values remains below
30%; beyond this threshold, their efficacy declines.
Importantly, LSTMs, typically applied to time series
data, prove effective for numerical data even without
a temporal dimension, surpassing the performance of
traditional DAE models in such scenarios.
Moreover, we conducted a comparative analysis
of several machine-learning models, which are shown
in Figure 4. Both Mean imputation and KNN demon-
strate a performance proximity to LSTM-DAE when
tasked with imputing 20% and 50% missing values.
Conversely, DAE yields results comparable to the It-
erative Imputer (I.Imp) technique. For the imputa-
tion of 30% missing values, Mean and KNN tech-
niques exhibit similar performance to LSTM-DAE,
with KNN demonstrating a decrease in accuracy as
missing values escalate. Throughout all scenarios,
Decision Tree (D.Tree) and Linear Regression (LR)
consistently exhibit lower performance relative to the
other techniques. Furthermore, LSTM-DAE consis-
tently outperforms all other models in all instances of
missing data.
In this experiment, we identified limitations in
both data preprocessing and the employed models.
In the data preprocessing stage, mean values were
introduced as noise at the locations of missing val-
ues within the complete dataset. However, this ap-
proach may have limitations when the missing dataset
is larger than the non-missing dataset. It proves effec-
tive only when the missing value dataset is consis-
tently smaller than the non-missing dataset.
8 CONCLUSIONS
Imputing missing data in emergency medical datasets
presents a significant challenge, especially when deal-
ing with class imbalance and outliers, where missing
values occur randomly. To tackle this, we utilized
two models: the denoising autoencoder and the de-
noising LSTM autoencoder. Optimal hyperparameter
tuning is essential to effectively address class imbal-
ance during imputation. We compare our proposed
models with various machine learning models, and
our results demonstrate that the denoising LSTM au-
toencoder surpasses all other models in this context.
Future investigations will explore additional deep-
learning models, such as variational autoencoder,
generative adversarial network, and transformer, for
imputing missing values in both categorical and
continuous numerical data. The resulting imputed
datasets will be utilized in multi-label classification
tasks to assess the effectiveness of various imputation
A Deep Analysis for Medical Emergency Missing Value Imputation
1235

techniques, with classification performance serving as
the evaluation metric.
ACKNOWLEDGEMENTS
This work was funded by the German Ministry of
Education and Research (Bundesministerium f
¨
ur Bil-
dung und Forschung) as part of the project “KI-
unterst
¨
utzter Telenotarzt (KIT2)” under grant number
13N16402. We would like to thank them for their sup-
port.
REFERENCES
Aachen, U. (2023). https://www.telenotarzt.de/. Teleno-
tarzt.
Abiri, N., Linse, B., Ed
´
en, P., and Ohlsson, M. (2019). Es-
tablishing strong imputation performance of a denois-
ing autoencoder in a wide range of missing data prob-
lems. Neurocomputing, 365:137–146.
Adhikari, D., Jiang, W., Zhan, J., He, Z., Rawat, D. B.,
Aickelin, U., and Khorshidi, H. A. (2022). A Com-
prehensive Survey on Imputation of Missing Data
in Internet of Things. ACM Computing Surveys,
55(7):133:1–133:38.
Chang, Y.-W., Natali, L., Jamialahmadi, O., Romeo, S.,
Pereira, J. B., Volpe, G., and Initiative, f. t. A. D. N.
(2022). Neural network training with highly incom-
plete medical datasets. Machine Learning: Science
and Technology, 3(3):035001. Publisher: IOP Pub-
lishing.
Chen, S. and Guo, W. (2023). Auto-encoders in deep learn-
ing—a review with new perspectives. Mathe-
matics, 11(8).
Coto-Jimenez, M., Goddard-Close, J., Di Persia, L.,
and Leonardo Rufiner, H. (2018). Hybrid Speech
Enhancement with Wiener filters and Deep LSTM
Denoising Autoencoders. In 2018 IEEE Interna-
tional Work Conference on Bioinspired Intelligence
(IWOBI), pages 1–8.
Emmanuel, T., Maupong, T., Mpoeleng, D., Semong, T.,
Mphago, B., and Tabona, O. (2021). A survey on
missing data in machine learning. Journal of Big
Data, 8(1):140.
Hameed, W. M. and Ali, N. A. (2023). Missing value im-
putation techniques: A survey. UHD J. Sci. Technol.,
7(1):72–81.
Ho, N.-H., Yang, H.-J., Kim, J., Dao, D.-P., Park, H.-R., and
Pant, S. (2022). Predicting progression of Alzheimer’s
disease using forward-to-backward bi-directional net-
work with integrative imputation. Neural Networks,
150:422–439.
Kabir, S. and Farrokhvar, L. (2022). Non-linear missing
data imputation for healthcare data via index-aware
autoencoders. Health Care Management Science,
25(3):484–497.
Liu, M., Li, S., Yuan, H., Ong, M. E. H., Ning, Y., Xie,
F., Saffari, S. E., Shang, Y., Volovici, V., Chakraborty,
B., and Liu, N. (2023a). Handling missing values in
healthcare data: A systematic review of deep learning-
based imputation techniques. Artificial Intelligence in
Medicine, 142:102587.
Liu, M., Li, S., Yuan, H., Ong, M. E. H., Ning, Y., Xie,
F., Saffari, S. E., Shang, Y., Volovici, V., Chakraborty,
B., and Liu, N. (2023b). Handling missing values in
healthcare data: A systematic review of deep learning-
based imputation techniques. Artificial Intelligence in
Medicine, 142:102587.
Lu, X., Yuan, L., Li, R., Xing, Z., Yao, N., and Yu,
Y. (2023). An Improved Bi-LSTM-Based Missing
Value Imputation Approach for Pregnancy Examina-
tion Data. Algorithms, 16(1):12. Number: 1 Pub-
lisher: Multidisciplinary Digital Publishing Institute.
Ma, Q., Lee, W.-C., Fu, T.-Y., Gu, Y., and Yu, G. (2020).
MIDIA: exploring denoising autoencoders for missing
data imputation. Data Mining and Knowledge Discov-
ery, 34(6):1859–1897.
Macias, E., Serrano, J., Vicario, J. L., and Morell, A.
(2021). Novel Imputation Method Using Average
Code from Autoencoders in Clinical Data. In 2020
28th European Signal Processing Conference (EU-
SIPCO), pages 1576–1579. ISSN: 2076-1465.
Mongia, A., Sengupta, D., and Majumdar, A. (2020).
deepMc: Deep Matrix Completion for Imputation of
Single-Cell RNA-seq Data. Journal of Computational
Biology, 27(7):1011–1019. Publisher: Mary Ann
Liebert, Inc., publishers.
Pan, H., Ye, Z., He, Q., Yan, C., Yuan, J., Lai, X., Su, J.,
and Li, R. (2022). Discrete Missing Data Imputation
Using Multilayer Perceptron and Momentum Gradi-
ent Descent. Sensors, 22(15):5645. Number: 15 Pub-
lisher: Multidisciplinary Digital Publishing Institute.
Psychogyios, K., Ilias, L., and Askounis, D. (2022). Com-
parison of Missing Data Imputation Methods us-
ing the Framingham Heart study dataset. In 2022
IEEE-EMBS International Conference on Biomed-
ical and Health Informatics (BHI), pages 1–5.
arXiv:2210.03154 [cs].
Sun, Y., Li, J., Xu, Y., Zhang, T., and Wang, X. (2023).
Deep learning versus conventional methods for miss-
ing data imputation: A review and comparative study.
Expert Systems with Applications, 227:120201.
Tyagi, K., Rane, C., Harshvardhan, and Manry, M. (2022).
Regression analysis. In Artificial Intelligence and Ma-
chine Learning for EDGE Computing, pages 53–63.
Elsevier.
Vincent, P., Larochelle, H., Bengio, Y., and Manzagol, P.-
A. (2008). Extracting and composing robust features
with denoising autoencoders. In Proceedings of the
25th international conference on Machine learning -
ICML ’08, pages 1096–1103, Helsinki, Finland. ACM
Press.
Vincent, P., Larochelle, H., Lajoie, I., Bengio, Y., and Man-
zagol, P.-A. Stacked Denoising Autoencoders: Learn-
ing Useful Representations in a Deep Network with a
Local Denoising Criterion.
Wang, Y., Yao, H., and Zhao, S. (2016). Auto-encoder
based dimensionality reduction. Neurocomputing,
184:232–242.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
1236
