
Counting Red Blood Cells in Thin Blood Films: A Comparative Study
Alan Klinger Sousa Alves
1 a
and Bruno Motta de Carvalho
2 b
1
Instituto Federal de Educac¸
˜
ao, Ci
ˆ
encia e Tecnologia do Rio Grande do Norte - Campus Nova Cruz,
Av. Jos
´
e Rodrigues de Aquino Filho, Nova Cruz, Brazil
2
Departamento de Inform
´
atica e Matem
´
atica Aplicada, Universidade Federal do Rio Grande do Norte,
Rua Cel. Jo
˜
ao Medeiros, Natal, Brazil
Keywords:
Malaria, Computer Vision, RBC Counting.
Abstract:
Malaria is a disease caused by a parasite that is transmitted to humans through the bites of infected mosquitoes.
There were an estimated 247 million cases of malaria in 2021, with an estimated number of 619,000 deaths.
One of the tasks in diagnosing malaria and prescribing the correct course of treatment is the computation of
parasitemia, that indicates the level of infection. The parasitemia can be computed by counting the number of
infected Red Blood Cells (RBCs) per µL or the percentage of infected RBCs in 500 to 2,000 RBCs, depending
on the used protocol. This work aims to test several techniques for segmenting and counting red blood cells
on thin blood films. Popular methods such as Otsu, Watershed, Hough Transform, combinations of Otsu with
Hough Transform and convolutional neural networks such as U-Net, Mask R-CNN and YOLO v8 were used.
The results obtained were compared with two other published works, the Malaria App and Cell Pose. As a
result, one of our implemented methods obtained a higher F1 score than previous works, especially in the
scenario where there are clumps or overlapping cells. The methods with the best results were YOLO v8 and
Mask R-CNN.
1 INTRODUCTION
Malaria is a disease caused by a parasite of the genus
Plasmodium and it is transmitted by some mosquitoes
of the genus Anopheles (Giacometti et al., 2021). The
main method for diagnosing malaria infections is op-
tical microscopy (Organization et al., 2021). In order
to have the right course of treatment, it is necessary
to determine the specie of the plasmodium, the stage
of the disease and parasitemia of the sample. Thus,
it is important to make a proper estimate, as both an
overestimation or an underestimation can be equally
harmful (Eziefula et al., 2012). The parasitemia is
computed as a percentage of infected Red Blood Cells
(RBCs) relative to the total number of red blood cells
(Organization et al., 2021). Thus, a high accuracy in
the red blood cell counting process is very important
for achieving accurate parasitemia results.
Estimating parasitemia from thin blood films has
been addressed in several studies (Rehman et al.,
2018). However, there is still room for improvement,
specially in the case of clumped or overlapped cells
a
https://orcid.org/0000-0003-2501-4751
b
https://orcid.org/0000-0002-9122-0257
a) Cell Pose b) Malaria App
Figure 1: Both tools present difficulty in individually iden-
tifying each RBC in the scenario with overlapped cells.
Figure 2: The Malaria App makes a rectangle around the
cells and then estimates the number of cells in the area. This
estimate may not be very efficient.
(Rehman et al., 2018). Figure 1 shows the result of
the RBC segmentation of two published methods for
the same sample, on the left side, Figure 1a, we have
the segmentation generated by Cell Pose (Stringer
Alves, A. and Motta de Carvalho, B.
Counting Red Blood Cells in Thin Blood Films: A Comparative Study.
DOI: 10.5220/0012455000003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 687-694
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
687

et al., 2021) and on the right side, Figure 1b, we have
the segmentation from Malaria App (Oliveira, 2019),
Both present poor results in the scenario where the
cells are very close to each other, with the red dots
indicating which cells were identified. In this case
many cells went unnoticed. This problem is reported
in several other works (Rehman et al., 2018).
In the case of the Malaria App (Oliveira, 2019),
there is still a worst case scenario that can be seen in
the Figure 2, that happens when the algorithm cannot
break down clumps of red blood cells and produces an
estimative of the number of cells in an area marked by
a rectangle. In this case, it is possible that the method
produces a lower quality estimate, with a larger error,
that can affect the computed parasitemia.
The objective of this work is to propose alternate
methods for counting RBCs in thin blood films and
analyze their results, in order to obtain more accu-
rate parasitemia estimates. Section 2 presents some
related works while Section 3 briefly describes the im-
plemented methods. Section 4 presents the results and
a discussion about them. Finally, Section 5 presents
our conclusion and suggests future work for this task.
2 RELATED WORK
One of the main related works is the Malaria App
(Oliveira et al., 2018) (Oliveira, 2019), which em-
ployed an adaptive version of the Otsu threshold
(Otsu, 1979) to produce binary images, segmenting
the RBCs from the background. Morphology oper-
ations such as hole filling, dilation and erosion were
performed to fill up cells and break down connected
cells, these operations are already known and recom-
mended (Wu et al., 2010). Finally, clumps of cells are
identified and the number of RBCs in each clump are
estimated based on the area and border length of the
clump. The infected cells and white cells are detected
using a Hue, Saturation and value (HSV) filter, since
the malaria samples are stained with the Giemsa dye.
Another important related work is Cell Pose
(Stringer et al., 2021) which used a generalist ap-
proach through a convolutional neural network based
on an U-Net. Approximately 510 images of mi-
croscopic objects were used for training, but not all
of them are of blood cells. Cell Pose was tested
and compared to other state-of-the-art tools, obtain-
ing results superior to all. The comparisons were
performed against the results produced by Stardist
(Schmidt et al., 2018), Mask R-CNN (He et al., 2017)
(Abdulla, 2017) and the U-Net (Ronneberger et al.,
2015) with two and three classes. A test was carried
out with 111 images similar to our study, and Cell
Pose achieved an accuracy of 93% in the Intersec-
tion Over Union (IoU) metric, while Mask R-CNN
achieved 82% and Stardist achieved 75%.
In another study, (Devi et al., 2018) proposed a
method for detecting infected erythrocytes that uses
Otsu’s thresholding to obtain binary images of the
thin blood samples, followed by a Watershed segmen-
tation. Then, they extracted 54 features that are fed to
several feature selection techniques to remove irrel-
evant data. The selected features are used to train
several different classifiers (SVM, k-NN and Naive
Bayes) that are also combined in hybrid classifiers.
It is important to note that that method only detects
the infected cells and does not count all the RBCs.
The RBC and white blood cells (WBC) counts are
also important for the diagnosis of other diseases such
as leukemia, allergies, tissue injuries, bronchogenic
infection, anemia and even kidney tumors and lung
diseases (Society, 2023). For this reason, there are
several studies that seek to count RBC or WBC. The
techniques for both are similar, where some of the
techniques being used are based on separation by col-
ors on the Hue, Saturation and Intensity (HSI) color
space or the L*a*b* color space, or on machine learn-
ing techniques such as K-Means, Fuzzy C-Means or
convolutional neural networks (CNNs) (Tran et al.,
2018).
A combination of Ycbcr space Color conversion
was used in conjunction with a circle Hough Trans-
form and watershed algorithm, with this method ob-
taining an accuracy of 90.98% in Acute lymphoblastic
leukemia image database (ALL-IDB) (Yeldhos et al.,
2018). Using the same data set, another work applied
a SegNet obtaining an accuracy rate of 85.50% for
WBC and 82.9% for RBC (Tran et al., 2018).
The network You Only Look Once (YOLO) ver-
sion 2 was also used for WBC detection, achieving
a detection accuracy of 96% (Abas et al., 2021), to
100% (Wang et al., 2018). It is important to note that
the WBC detection problem is simpler than RBC seg-
mentation, as some samples have many objects over-
lapping each other in the latter case.
In this work, we also use convolutional networks
such as the U-Net (Ronneberger et al., 2015) which
has the advantage of producing better results when
working with fewer samples when compared to other
convolutional neural networks (CNNs), the Mask R-
CNN (Abdulla, 2017), which is an extension of a R-
CNN that adds the object mask prediction and the
newest version of the YOLO, the YOLO v8 (Jocher
et al., 2023).
HEALTHINF 2024 - 17th International Conference on Health Informatics
688

3 METHODOLOGY
This work began by obtaining the same data set
used in the Malaria App project (Oliveira, 2019),
then we started with the implementation of tradi-
tional methods that did not use neural networks us-
ing only Python and OpenCV. The data set consists
of 30 samples that were stained with the Giemsa dye
and imaged using a standard optical microscope using
a 1000× zoom, the standard protocol for diagnosing
malaria. The images were originally captured with a
resolution of 2592 × 1944, and reduced to 640 × 480
in our case, since the detection and counting of RBCs
do not need a high resolution image.
Now, we proceed by briefly describing the used
algorithms.
3.1 Otsu
Otsu’s method (Otsu, 1979) is an algorithm for de-
termining an optimal threshold for separating pixels
from an image into two classes. It has been exten-
sively used in its original form and in adaptive ways,
usually by subdividing the image into blocks, thus,
avoiding problems caused by non-uniform illumina-
tion and contrast.
It was noticed that in this method the blurring fil-
ter was bringing together the edges of some cells. It is
possible to see an example of this effect in Figure 3,
we have a group indicated by the yellow arrow with
the wrong estimate, they should have 4 cells but 3
were estimated. In Figure 4, fewer cells were included
in the group, thus, the estimate obtained the correct
answer, in this Figure we have the same method, but
without the blur. In general, even without blurring the
image, this method generates many groups of cells,
which cannot be segmented individually, having to
resort to making an estimate, similar to what was im-
plemented in Malaria App. This estimate can be quite
wrong when the group contains many cells.
In both Figures it is also possible to notice another
problem: this method captures practically any object,
for example, the spot in the center of both Figures 3
and 4 that was identified as a group of cells, therefore,
this will generate many false positives. In this work
we generate results for Otsu Adaptive, which in the
tables appears labeled as Otsu and for Otsu Adapta-
tive with Gaussian blur, which in the tables appears
labeled as Otsu + Gaussian blur.
3.2 Watershed
The Watershed algorithm views an image as a topo-
graphic surface in which high intensities denote peaks
Figure 3: Segmentation with Otsu + Gaussian blur, the spots
are identified as cells and the estimation of the group iden-
tified by the yellow arrow is flawed.
Figure 4: Segmentation with Otsu, the groups of cells iden-
tified by the yellow arrow became smaller and the estima-
tion improved.
and hills while low intensities denote valleys, and
gradually fills the valleys until the water from differ-
ent regions merge. Then, an “artificial dam” is built
between them and determine boundaries between ob-
jects.
Figure 5 shows the result of the Watershed algo-
rithm, that generates something similar to a relief map
of the image (left) and the RBC labeling on the right.
Note that although some cells appear to be together,
it manages to separate them. One of the problems no-
ticed is that it has difficulty finding cells that are very
clear and in some cases it generates groups of cells.
Figure 5: Watershed segmentation can individually identify
some cells that are very close to each other, but has difficulty
identifying very light cells.
Watershed also doesn’t make a good distinction
between sample objects. In Figure 6 with sample C,
we can see 8 false positives.
Counting Red Blood Cells in Thin Blood Films: A Comparative Study
689

Figure 6: Yellow arrows show incorrect classifications with
Watershed, any object is incorrectly classified as a cell.
3.3 Circle Hough Transform
This method uses the circle Hough Transform from
Python’s OpenCV library to identify circles in the
sample. The difficulty of the method is the config-
uration of the parameters. Figure 7 shows the result
of the method with each circle that was found in sam-
ple B, the perceived problem is that this method finds
circles in places where there are no cells, generating
false positives. We indicate this problem with yel-
low arrows. Furthermore, this method has difficulty
capturing cells that are not in an approximate circular
shape.
Figure 7: Empty spaces can be classified as cells if objects
in the surrounding environment form something like a cir-
cle, the yellow arrows point out these problems.
In Figure 8 we have a sample with 7 false posi-
tives, in which we can observe that the circle Hough
Transform will capture any object that is in a circular
format, making no distinction between contaminated
cells, WBCs and RBCs.
Figure 8: Incorrect classification with circle Hough Trans-
form, any circular shaped object is identified as a cell, as
highlighted by the yellow arrows.
3.4 Otsu + Circle Hough Transform
We noted that the circle Hough Transform and Otsu
could overcome some of each other’s deficiencies,
since the circle Hough Transform can identify cells
individually, even if they are very close, while Otsu
can identify cells that are in different formats, so we
decided to test two methods that would unite Otsu and
the circle Hough Transform.
The first method would be, after using Otsu, in-
stead of trying to make an estimate to identify the
number of cells in the groups that could not be sep-
arated, use circle Hough Transform to break down
the groups of cells generated. In this way we imple-
mented Otsu + Hough 1, whose result for a sample
can be seen in Figure 9. The Figure 9a shows that
Otsu created several groups, while in Figure 9b there
is the same sample with the result generated by Otsu
+ Hough 1. there, it is possible to see that all groups
were individually segmented with only 8 false posi-
tives indicated by the yellow arrows.
a) Otsu b) Otsu + Hough 1
Figure 9: Segmentation with Otsu + Hough 1, figure (a)
shows the result of Otsu, figure (b) shows the result of Otsu
after breaking the cell groups with the circle Hough Trans-
form. Yellow arrows highlight false positives.
The second method would simply be to combine
the Otsu result with circle Hough Transform, we call
this method Otsu + Hough 2, this second method gen-
erates fewer false positives, but finds fewer cells than
the previous method. A result of its use can be seen
in Figure 10.
Figure 10: Segmentation with Otsu + Hough 2, this combi-
nation is the simple sum of the results of the two methods,
consequently there will be errors present in both methods.
Yellow arrows highlight false positives.
HEALTHINF 2024 - 17th International Conference on Health Informatics
690

3.5 Mask R-CNN
Mask R-CNN (Abdulla, 2017) was the slowest
method, with its execution taking about 1 minute and
30 seconds to segment just one sample. This time
does not preclude its use, but it is important to high-
light it. This network obtained good results, with two
examples being shown on Figure 11. It is possible to
see that it generates few false positives and does not
generate groups of cells.
a) Sample A b) Sample B
Figure 11: Mask R-CNN was not able to accurately differ-
entiate cells from spots and others objects. Yellow arrows
highlight problems.
Despite the good results with images with clumps
of cells, Mask R-CNN also presented a problem: it
considers most objects as RBCs. In sample C pre-
sented in Figure 12, it is possible to observe at least
11 objects that were erroneously classified as RBCs.
The problem occurs in most samples and can also be
seen in Figure 11.
Figure 12: Wrong classification with Mask R-CNN, this
neural network is also not capable of differentiating cells
from other objects in the samples. Yellow arrows highlight
problems.
3.6 U-Net
U-Net is also a convolutional network, with a big ad-
vantage that it is fast to train and requires fewer im-
ages to obtain a good result when compared to other
CNNs. Figure 13 exemplifies its use with two sam-
ples. It was one of the only methods that did not
generate false positives, managing to precisely differ-
entiate the cells from spots. In Figure 13b, does not
generate false positives, however, it finds fewer cells
than actually exist in the image.
a) Sample A b) Sample B
Figure 13: For these samples, U-Net can differentiate spots
and others objects from cells.
Like Mask R-CNN, U-Net also captures many ob-
jects such as RBCs, but in smaller quantities, Figure
14 is the same sample as Figure 12 but with only 6
false positives. Just by looking at Figure 13 it would
not be possible to observe this problem.
Figure 14: U-Net cannot differentiate cells from other ob-
jects when they are very similar to them. False positives are
indicated with yellow arrows.
3.7 YOLO v8
The version 8 of YOLO at the moment is the latest
one of this convolutional network. This network had
a reasonable training time, but the segmentation exe-
cution takes less than 1 second. Figure 15 shows that
it generated some false positives in sample A, but it
was the only method that found all RBCs in the sam-
ple B.
a) Sample A b) Sample B
Figure 15: The segmentation with YOLO v8 made only a
few false positives in the image (a) highlighted by the yel-
low arrows and managed to find all the red blood cells in
the image (b).
Unlike the two previous convolutional networks,
YOLO v8 correctly classifies most objects, an exam-
ple can be seen in Figure 16, where there are no false
positives.
Counting Red Blood Cells in Thin Blood Films: A Comparative Study
691

Figure 16: Good classification with YOLO v8. It was pos-
sible to differentiate even cell-like objects.
4 EXPERIMENTS
In this session we describe the experiments per-
formed, the metrics used for comparing the results
and present the results.
After testing traditional algorithms for segmenta-
tion, we started testing with convolutional networks
such as U-Net, Mask R-CNN and YOLO v8. In order
to train a more general model, an additional 400 mi-
croscopic images were obtained, of which 100 would
be used only for validation and comparison of the
methods. With these 330 images, data augmentation
was carried out with rotations and mirroring, generat-
ing 2,860 images. The experiments were performed
on a computer equipped with 40 Intel Xeon E5-2660
CPUs and with 64GB of RAM.
Resolutions of 256, 384, 416, 512 and 1024 were
tested for the convolutional networks, however data
will only be presented for the resolutions that pro-
vided the best results.
The U-Net took 28 hours to train for 1,500 epochs
using Tensorflow (Abadi et al., 2015), Adam opti-
mizer (Kingma and Ba, 2014) and binary cross en-
tropy as the loss function with the resolution of 256 ×
256. A total of 2,145 images were used for training
and 715 for validation.
For the Mask R-CNN training, we used the same
parameters used in U-Net, 2,145 images for train-
ing and 715 for validation, the best resolution was
1024× 1024. The initial weights were the same as the
ones used on ImageNet, and we used 20 epochs for
the net head and 40 epochs for the other layers. Each
epoch had 100 steps of training, using the ResNet50’s
backbone. The whole training took approximately 6
days.
For the YOLO v8, we used 1,425 images for train-
ing, 804 for validation and 631 for testing. The train-
ing used 2,000 epochs with a batch size of 25 and the
best resolution was 512 × 512, in about 59 hours.
4.1 Comparing Results
Our objective was not to perform an exact segmen-
tation, but rather a precise count of the RBCs in the
sample, for this reason, we did not use the Intersec-
tion over Union (IoU) metric.
We created a tool to compare the evaluated image
and the ground truth, based on the position of each
RBC. All segmentation algorithms mark each RBC
with a red circle, the comparison tool searches for
these circles, comparing the position of each one with
the closest RBC to the same position in the ground
truth. When an RBC is found in the same position
in both the ground truth and the evaluated image, a
green circle is made above and a number is added, so
that it is possible to check exactly whether the RBC
in that position corresponds to the RBC in the same
position in the other image. This comparison can be
seen in Figure 17, where on the left side (a) we have
the image being evaluated and on the right side (b) we
have the ground truth.
Some algorithms, such as the Malaria App or Wa-
tershed, generate groups of cells and make an esti-
mate for each group. In these cases, a yellow square is
added with the count of the group’s RBCs in the eval-
uated image, the example can be seen in Figure 17(a),
where we have 1 purple rectangle with the count high-
lighted in a yellow square and in the ground truth im-
age the RBCs within the group area are painted light
green, which can also be seen in Figure 17(b).
a) Malaria App b) Ground truth
Figure 17: (a) Results from (Oliveira, 2019) . (b) Ground
truth image, dark green circles mark cells that were cor-
rectly identified, the light green circles in identify the cells
that were “extracted” from RBC groups, while the smaller
red circles identify the cells that were not detected.
Therefore, the red circles that remain in the im-
age being evaluated are considered false positives and
those that remain in the ground truth image are false
negatives. Additionally, a 50px border was created
around the image to avoid considering RBCs that are
very close to the edge, because the count made by an
expert would not evaluate these RBCs as they are only
partially seen.
HEALTHINF 2024 - 17th International Conference on Health Informatics
692
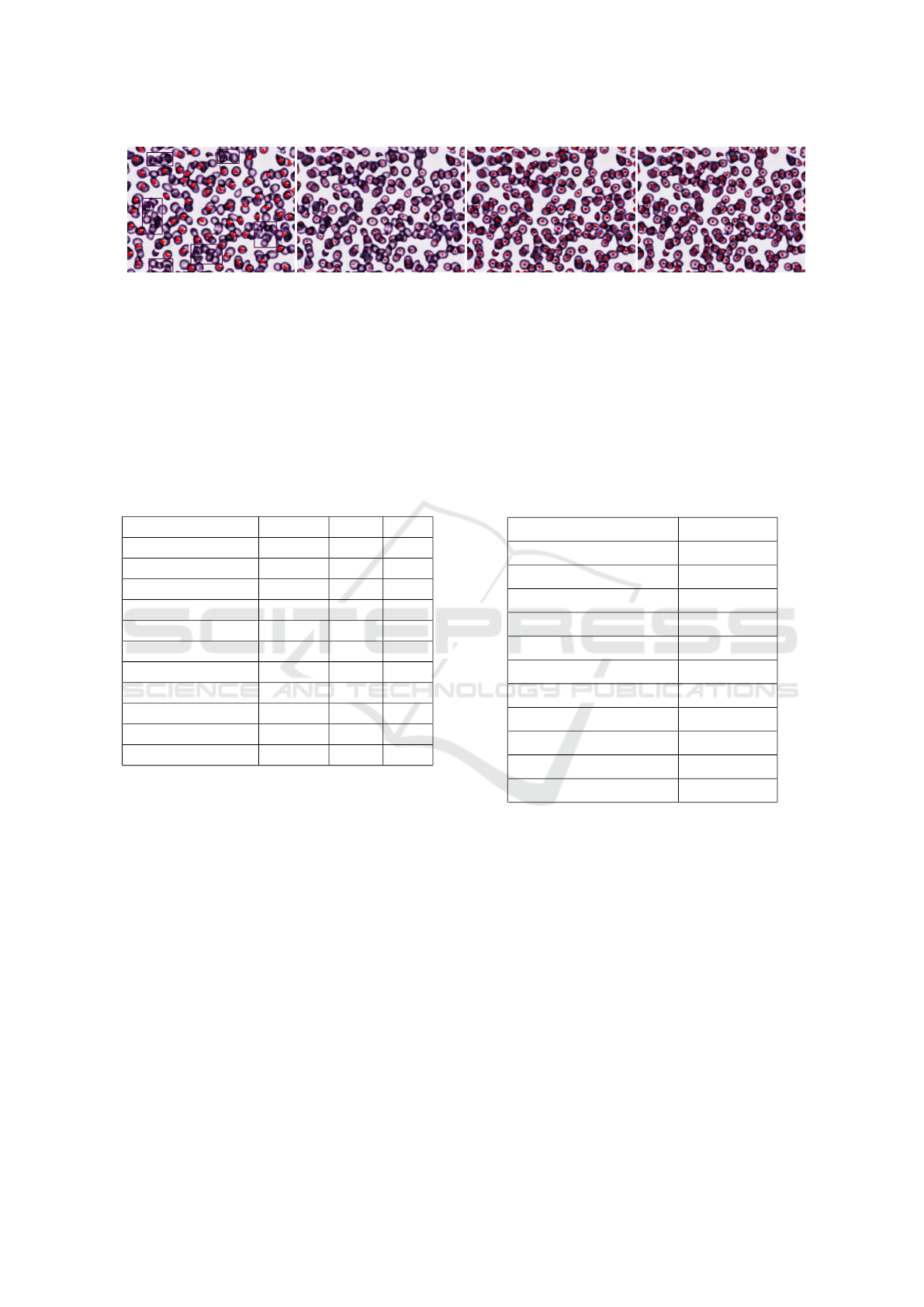
.
(a) Malaria App (b) Cell Pose (c) R-CNN (d) YOLO v8
Figure 18: Segmentation of a complex sample with 199 RBCs: Malaria App found 84 RBCs, Cell Pose found 113 RBCs,
Mask R-CNN found 172 RBCs and YOLO v8 found 183 RBCs.
4.2 Results
As most methods only took average execution times
between 0.5 and 1.5 seconds per sample, we consider
the time differences as negligible and do not report on
them. Table 1 presents the precision, recall and F1
scores accuracies for all methods tested.
Table 1: Precision, recall and F1 scores.
Method name Precision Recall F1
Hough Transform 0.928 0.817 0.861
Otsu + Gaussian blur 0.898 0.763 0.817
OTSU + Hough 1 0.869 0.887 0.872
OTSU + Hough 2 0.907 0.874 0.886
OTSU 0.902 0.771 0.824
Mask R-CNN 0.919 0.980 0.948
U-Net 0.916 0.842 0.873
Watershed 0.895 0.904 0.894
YOLO v8 0.942 0.976 0.958
Malaria App 0.897 0.842 0.862
Cell Pose 0.936 0.943 0.936
Among the methods tested by us, YOLO v8 ob-
tained the best result in precision and F1 score (0.942)
and (0.958) respectively, followed by circle Hough
Transform with the second highest precision (0.928)
and Mask R-CNN with the second best F1 score
(0.948). Regarding Recall, Mask R-CNN obtained
the best result (0.980), followed by YOLO v8 (0.976).
However, we have to emphasize that the para-
sitemia computation is an easier task than the clas-
sification task which we are reporting. In the count-
ing task, a false positive and a false negative in the
same sample cancel each other, while in the classifi-
cation task, they count as two errors. Therefore, we
present Table 2 that shows the average percentage of
errors for each implemented method. The errors are
counted in a simpler way: if in the sample there are
10 cells and the algorithm identified 12 or 8 the er-
ror will be 2. To calculate this percentage, the dif-
ference between the expected count for each sample
and the count carried out by each method was com-
puted, and only then the percentage of this difference
was calculated. According to Table 2, of the methods
tested by us, YOLO v8 managed to have the best error
average with just 5.02% followed by Mask R-CNN
(8.28%), while Malaria App had an average error rate
of 11.22% and Cell Pose had an error rate of 6.81%.
Table 2: Methods error percentage average.
Method Percent error
Circle Hough Transform 15.539
Otsu + Gaussian blur 18.066
Otsu + Hough 1 13.659
Otsu + Hough 2 10.754
Otsu 18.375
Mask R-CNN 8.281
U-Net 11.461
Watershed 10.596
YOLO v8 5.025
Malaria App 11.220
Cell Pose 6.812
In simple samples, most methods present a good
result, and the big difference occurs in samples with
clumped or overlapped cells. Figure 18 presents the
result of segmentation by the two published methods
and the two best methods implemented by us, in one
of the more challenging samples. This sample has
199 RBCs, while Malaria App found only 84 and
Cell Pose found 113, our Mask R-CNN and YOLO
v8 found 172 and 183 respectively.
Cell Pose presents an smaller percent error value
than Mask R-CNN because most of the samples used
in this study are simple. If the majority were more
complex, the Mask R-CNN’s percent error would
probably be smaller than Cell Poses’s percent error.
Counting Red Blood Cells in Thin Blood Films: A Comparative Study
693

5 CONCLUSIONS
The results show the YOLO v8 and Mask R-CNN
as the most accurate ones. The big difference be-
tween these two methods compared to Malaria App
and Cell Pose is in dealing with clumped and over-
lapped RBCs, in which cases both tools present lower
accuracies than YOLO v8 and Mask R-CNN.
As future work, we intend to train convolution
networks to deal specifically with the problem of
clumped groups of RBCs, the main problem in this
task. Moreover, we believe that the problem may
be simplified with a better designed pre-processing
phase, with more robust hole filling and cell separa-
tion procedures. This approach will also be further
investigated.
To have an even more generic CNN, we could add
more images to the training dataset. The difficulty lies
in obtaining and labeling each image, as it is a time-
consuming process. Furthermore, other datasets will
need to be created to identify other diseases, so the
labeling process needs to be improved. So, as we al-
ready have a CNN capable of identifying RBCs, we
can use the prediction result as a starting point to la-
bel more samples in a semi-automatic approach, dras-
tically reducing the labeling time.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., et al. (2015). Tensor-
Flow: Large-scale machine learning on heterogeneous
systems. Software available from tensorflow.org.
Abas, S. M., Abdulazeez, A. M., and Zeebaree, D. (2021).
A yolo and convolutional neural network for the de-
tection and classification of leukocytes in leukemia.
Indones. J. Electr. Eng. Comput. Sci, 25(1).
Abdulla, W. (2017). Mask R-CNN for object detection and
instance segmentation on keras and tensorflow. https:
//github.com/matterport/Mask RCNN.
Devi, S. S., Roy, A., Singha, J., Sheikh, S. A., and Laskar,
R. H. (2018). Malaria infected erythrocyte classifi-
cation based on a hybrid classifier using microscopic
images of thin blood smear. Multimed. Tools Appl.,
77(1):631–660.
Eziefula, A. C., Gosling, R., Hwang, J., Hsiang, M. S.,
Bousema, T., Von Seidlein, L., and Drakeley, C.
(2012). Rationale for short course primaquine in
africa to interrupt malaria transmission. Malaria J.,
11:1–15.
Giacometti, M., Milesi, F., Coppadoro, P. L., Rizzo, A., Fa-
giani, F., Rinaldi, C., Cantoni, M., Petti, D., Albisetti,
E., Sampietro, M., et al. (2021). A lab-on-chip tool
for rapid, quantitative, and stage-selective diagnosis
of malaria. Adv. Sci., page 2004101.
He, K., Gkioxari, G., Doll
´
ar, P., and Girshick, R. (2017).
Mask R-CNN. In Proceedings of the ICCV, pages
2961–2969.
Jocher, G., Chaurasia, A., and Qiu, J. (2023). YOLO by
Ultralytics.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. ArXiv preprint
arXiv:1412.6980.
Oliveira, A. D., Carvalho, B. M., Prats, C., Espasa, M.,
i Prat, J. G., Codina, D. L., and Albuquerque, J.
(2018). An automatic system for computing malaria
parasite density in thin blood films. In Progress in Pat-
tern Recognition, Image Analysis, Computer Vision,
and Applications, pages 186–193. Springer Interna-
tional Publishing.
Oliveira, A. D. d. (2019). MalariaApp: Um sistema de
baixo custo para diagn
´
ostico de mal
´
aria em l
ˆ
aminas
de esfregac¸o sangu
´
ıneo usando dispositivos m
´
oveis.
PhD thesis, Universidade Federal do Rio Grande do
Norte.
Organization, W. H. et al. (2021). Who guideline for
malaria, 13 july 2021. Technical report, World Health
Organization.
Otsu, N. (1979). A threshold selection method from gray-
level histograms. IEEE Trans. on Syst. Man Cybern.,
9(1):62–66.
Rehman, A., Abbas, N., Saba, T., Mehmood, Z., Mah-
mood, T., and Ahmed, K. T. (2018). Microscopic
malaria parasitemia diagnosis and grading on bench-
mark datasets. Mic. Res. Tech., 81(9):1042–1058.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
Net: Convolutional networks for biomedical image
segmentation. In Pro. of MICCAI, pages 234–241.
Springer.
Schmidt, U., Weigert, M., Broaddus, C., and Myers, G.
(2018). Cell detection with star-convex polygons. In
Proc. of MICCAI, pages 265–273. Springer.
Society, C. C. (2023). Complete blood count (CBC). https:
//cancer.ca/en/treatments/tests-and-procedures/co
mplete-blood-count-cbc#ixzz5JmJ8jOk9. Accessed:
2023-08-28.
Stringer, C., Wang, T., Michaelos, M., and Pachitariu, M.
(2021). Cellpose: A generalist algorithm for cellular
segmentation. Nat. Methods, 18(1):100–106.
Tran, T., Kwon, O.-H., Kwon, K.-R., Lee, S.-H., and Kang,
K.-W. (2018). Blood cell images segmentation using
deep learning semantic segmentation. In Proc. of the
ICECE, pages 13–16. IEEE.
Wang, X., Xu, T., Zhang, J., Chen, S., and Zhang, Y.
(2018). So-yolo based wbc detection with fourier
ptychographic microscopy. IEEE Access, 6:51566–
51576.
Wu, Q., Merchant, F., and Castleman, K. (2010). Micro-
scope Image Processing. Elsevier.
Yeldhos, M. et al. (2018). Red blood cell counter using
embedded image processing techniques. Res. Rep., 2.
HEALTHINF 2024 - 17th International Conference on Health Informatics
694
