
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear
Group Accumulators in Batch Mode
William J Buchanan
1
, Sam Grierson
1
and Daniel Uribe
2
1
Blockpass ID Lab, Edinburgh Napier University, Edinburgh, U.K.
2
GenoVerse.io
Keywords:
Accumulators, Genomic Privacy, Bilinear Group.
Abstract:
Biometric data is often highly sensitive, and a leak of this data can lead to serious privacy breaches. Some of
the most sensitive of this type of data relates to the usage of DNA data on individuals. A leak of this type of
data without consent could lead to privacy breaches of data protection laws. Along with this, there have been
several recent data breaches related to the leak of DNA information, including from 23andMe and Ancestry. It
is thus fundamental that a citizen should have the right to know if their DNA data is contained within a DNA
database and ask for it to be removed if they are concerned about its usage. This paper outlines a method of
hashing the core information contained within the data stores - known as Single-Nucleotide Polymorphisms
(SNPs) - into a bilinear group accumulator in batch mode, which can then be searched by a trusted entity
for matches. The time to create the witness proof and to verify were measured at 0.86 ms and 10.90 ms,
respectively.
1 INTRODUCTION
Biometric information often is sensitive as it can re-
veal personally identifiable information about a per-
son. This could include the recognition of a face,
a finger-recognition or a retina. Overall, there is a
spectrum of sensitivity without these biometrics, with
retina scan and fingerprint recognition often requiring
high levels of privacy, whereas it is often difficult to
protect one’s face from being kept private. However,
one of the most sensitive areas of biometric matching
relates to the storage and matching of DNA informa-
tion. With this, there have been several major data
breaches related to DCT-GT (Direct Consumer Test-
ing - Genetic Testing), such as with 23andMe and An-
cestry (Garner and Kim, 2018). Along with privacy
issues, there is a strong business model for companies
selling DNA-related data, such as in 2018 when Glax-
oSmith Kline purchased the personal data of thou-
sands of customers from 23AndMe for $300 million
(DeFrancesco and Klevecz, 2019).
Within DNA analysis, Single-Nucleotide Poly-
morphism (SNP) information can fully identify a per-
son and which must be thus kept securely. If a per-
son has their SNPs stored in a data store, they often
have the right to search for the data in a data store.
These types of requests, though, must be kept in a
privacy-aware way, and where it is possible to search
for SNP information without revealing other informa-
tion on the contents of a data store of SNP informa-
tion.
Along with this, this matching could be useful
within the Health and Life Insurance industry, as it
is possible for a citizen to prove that they do not have
a genetic carrier that increases their chances of devel-
oping a disease, such as Alzheimer’s disease, without
disclosing their entire DNA dataset.
1.1 Background
DNA (Deoxyribonucleic acid) is contained in the nu-
cleus of cells and defines the genetic information of a
person (BBC, 2023). It is a large and complex poly-
mer which has two strands of a double helix, and
where, apart from identical twins, each person has a
unique DNA structure. With the cell’s nucleus, we
have chromosomes which are long threads of DNA.
These are then made from genes that contain a code
related to a sequence of amino acids - these may then
be copied and passed onto the next generation. Each
gene can have different forms - which are called al-
leles. For example, we can have an allele for blue
eye colour and another allele for brown eye colour. A
genotype is then a collection of these alleles that de-
226
Buchanan, W., Grierson, S. and Uribe, D.
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear Group Accumulators in Batch Mode.
DOI: 10.5220/0012454300003648
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 10th International Conference on Information Systems Security and Privacy (ICISSP 2024), pages 226-233
ISBN: 978-989-758-683-5; ISSN: 2184-4356
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

fine a phenotype. Overall, a person inherits one chro-
mosome from their mother and another from their fa-
ther - and where the pair carry the same gene in the
same location.
2 RELATED WORK
Alsaffar et al. (Alsaffar et al., 2022) outline the risks
related to the querying, sharing and genomic testing
stages and related countermeasures:
• Querying genome data. These risks include the
aggregation of data, aggregation of statistics, and
correlation attacks. Mitigations include differen-
tial privacy, range query limits, homomorphic en-
cryption, and privacy-preserving computing (Al-
saffar et al., 2022).
• Sharing genome data. These risks include belief
propagation attacks (
¨
Oks
¨
uz et al., 2021), multi-
party data sharing, inference attacks, likelihood
ratio (LHR) tests (Von Thenen et al., 2019), and
linkage attacks. The mitigations include statis-
tically aggregated data, multiparty data sharing,
encryption, statistical results, multiparty secret
sharing, distributed computations, and multiparty
queries (Alsaffar et al., 2022).
• Direct to consumer testing. These risks include
terms of usage and website vulnerabilities. The
mitigations include anonymising genome data and
best practice guides (Alsaffar et al., 2022).
Naveed et al. (Naveed et al., 2015) defined that
genome sequencing technology has advanced at a fast
pace and is often focused on: associating with traits
and certain diseases, identifying individuals (such as
in forensics applications); and in revealing family re-
lationships. Shringarpure et al. (Shringarpure and
Bustamante, 2015) found that in a dataset with 65 in-
dividuals, it was possible to pinpoint a certain person
from just 250 SNPs.
Bonomi et al. (Bonomi et al., 2020) outline that
sharing genomic data will allow for the enhancement
of precision medicine and support providing person-
alized treatments but privacy concerns and data mis-
use provide barriers to this sharing. The protection
of DNA data provides a challenge for research work
as it contains some of the most sensitive attributes of
a citizen’s identity. In the US, this will typically fo-
cus on federal status and regulations such as HIPAA
(Health Insurance Portability and Accountability Act)
and GINA (Genetic Information Nondiscrimination
Act) (Clayton et al., 2019).
3 ACCUMULATORS
A cryptographic accumulator, as originally proposed
by (Benaloh and De Mare, 1993), is a construction
that can accumulate a finite set of values into a single
succinct accumulator. Accumulators have the advan-
tageous property of being able to efficiently compute
a witness, which verifies the membership of any ac-
cumulated value in that accumulator. More formally,
given a finite set X = {x
1
,...,x
n
}, acc
X
is an accumu-
lator of X if every x ∈ X has an efficiently computable
witness wit
x
which certifies membership of x ∈ X and
it is computationally infeasible to find a wit
y
for any
non-accumulated value y /∈ X (Derler et al., 2015).
Since the original proposal, the basic notion of
a cryptographic accumulator has been extended and
iterated upon, resulting in some additional proper-
ties. Dynamic accumulators are one such extension,
where values can be dynamically added and deleted
to and from an accumulator (Camenisch and Lysyan-
skaya, 2002). Furthermore, additional security prop-
erties such as undeniability and indistinguishability
were proposed (Derler et al., 2015). The undeniabil-
ity security property ensures that it should be infeasi-
ble for two contradicting witnesses to be computed,
certifying that wit
x
∈ acc
X
and wit
x
/∈ acc
X
. Infor-
mally, the indistinguishability security property spec-
ifies that the accumulator acc
X
and witnesses wit
x
for
x ∈ X leak no information about the accumulated set
X. An accumulator that satisfies the indistinguisha-
bility property allows computation of a witness that
certification of a value in zero-knowledge.
The original application of a cryptographic accu-
mulator was timestamping of records to ensure their
existence as specific times (Benaloh and De Mare,
1993). Over time the use of accumulators has ex-
panded to numerous applications, including ring sig-
natures (Xu et al., 2010), group signatures (Tsudik
and Xu, 2003), encrypted searches (Ge et al., 2020),
revoking anonymous credentials (Xu et al., 2019) and
vector commitments (Catalano and Fiore, 2013). Fur-
thermore, the indistinguishability property of some
accumulator constructions that allows them to prove
in zero-knowledge membership or non-membership
of values in the accumulator has been used for re-
vocation of group signatures and anonymous creden-
tials (Camenisch and Lysyanskaya, 2002) such as in
the Zerocash protocol, now the Zcash cryptocurrency
(Ben Sasson et al., 2014).
3.1 Hidden Order Group Accumulators
The original scheme of (Benaloh and De Mare, 1993)
and its refined variant by (Bari
´
c and Pfitzmann,
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear Group Accumulators in Batch Mode
227

1997) was heavily based on the RSA cryptosystem
(Rivest et al., 1978). The security of Benahloah and
de Mare’s scheme is derived from the strong RSA
assumption (s-RSA) (Bari
´
c and Pfitzmann, 1997),
which states that given two primes p and q of bit-
length ℓ such that N = pq and a uniformly chosen
c ∈ Z
∗
N
then it holds that for all probabilistic poly-
nomial time adversaries A that
Pr[(m,e) ← A (c,n) s.t. m
e
= c mod n] ≤
1
2
ℓ
.
Using the s-RSA assumption, the RSA accumula-
tor of (Benaloh and De Mare, 1993) and (Bari
´
c and
Pfitzmann, 1997) is defined as follows. Let the func-
tion H : {0,1}
∗
→ Z
∗
N
be a collision-resistant hash
function mapping bit strings of arbitrary length to
primes in the hidden order set Z
∗
N
. Given a gener-
ator g of Z
∗
N
, the RSA accumulator for the set X =
{x
1
,...,x
n
} is
acc
A
= g
∏
x∈X
H(x)
mod N.
A proof of membership that x
i
∈ X can be created by
computing the witness that wit
x
i
∈ acc
X
using the fol-
lowing
wit
x
i
= g
∏
x∈X\{x
i
}
H(x)
mod N.
Note that the witness is just the RSA accumulator of
the set X \{x
i
} or the H(x
i
)
th
root of acc
X
. Therefore,
verification of the witness is done by checking that the
equality acc
X
= wit
H(x
i
)
x
i
mod N is satisfied.
The base RSA accumulator was later extended by
(Camenisch and Lysyanskaya, 2002) to support dy-
namically deleting and adding values to and from
the accumulator, which became the first known dy-
namic accumulator construction. Their scheme sup-
ported updates of existing witnesses without the re-
quired knowledge of the RSA trapdoor. In the work
of (Li et al., 2007), further improvements were made
to the construction by enabling efficient computation
of witnesses that function as non-membership proofs.
In an exciting improvement, (Wang et al., 2007) con-
structed an RSA accumulator that enabled batching
of various operations; however, this was shown to be
insecure by (Camacho and Hevia, 2010).
3.2 Hash-Based Accumulators
Another common way to construct accumulators is
by using symmetric primitives that satisfy the defi-
nitions of a collision-resistant hash function, i.e. a
hash function H : {0,1}
∗
→ {0, 1}
n
defined for n ∈ N
is collision-resistant if for all efficient probabilistic
polynomial time adversaries A
Pr[(x,y) ← A s.t. x ̸= y and H(x) = H(y)] ≤
1
2
n
.
The first constructions that used collision-resistant
hash functions were based on hash trees or Merkle
trees (Merkle, 1989) such as the work of Buldas
et. al. (Buldas et al., 2000) who aimed to solve
the problems of accountable certificate management.
In essence, the hash-based accumulator constructions
use a Merkle root to prove the membership of a value
within the accumulator (Baldimtsi et al., 2017).
In more recent work by (Camacho et al., 2008),
the authors developed an accumulator scheme relying
only on the collision-resistant hash function assump-
tion (a far weaker assumption than s-RSA). The con-
struction itself uses a hash tree similar to those pro-
posed before it and allows for additions and deletions
of values to and from the accumulator. However, de-
spite the greater security guarantees of the collision-
resistant hand function assumption, the hash tree ac-
cumulator proposed by (Camacho et al., 2008) was
significantly less efficient than the RSA accumula-
tors discussed previously. More recent work has been
done to improve the performance of addition and re-
moval operations (Reyzin and Yakoubov, 2016); how-
ever, as shown in (Camacho and Hevia, 2010), batch-
ing is not possible on a number of accumulator oper-
ations.
4 KNOWN ORDER GROUP
ACCUMULATORS WITH
OPERATION BATCHING
A dynamic accumulator construction was proposed
by (Nguyen, 2005) based on bilinear maps between
groups of prime order. Using the notation of (Boneh
et al., 2001) A bilinear map or pairing operation is the
map e : G
1
× G
2
→ G
T
in which G
1
, G
2
, and G
T
are
cyclic groups of prime order p if given the group gen-
erators g ∈ G
1
and h ∈ G
2
for all a, b ∈ F
∗
p
where F
∗
p
is a finite field over prime p the map e satisfies:
• e(g
a
,h
b
) = e(g,h)
ab
= e(g
b
,g
a
) (bilinarity)
• e(g,h) generates G
T
(non-degeneracy)
Note that the above is written as for multiplicative
groups but can be trivially modified to work with ad-
ditive cyclic groups of prime order.
The security of Nguyen’s accumulator is based
on the q-Strong Diffie Hellman (q-SDH) assumption.
The q-SDH assumption, as defined by (Boneh and
Boyen, 2004), states that given a prime p of bit-length
ℓ, a finite cyclic group G of order p, a generator ele-
ment g ∈ G, a uniformly chosen x ∈ F
∗
p
and a q ∈ N
then for all probabilistic polynomial time adversaries
ICISSP 2024 - 10th International Conference on Information Systems Security and Privacy
228
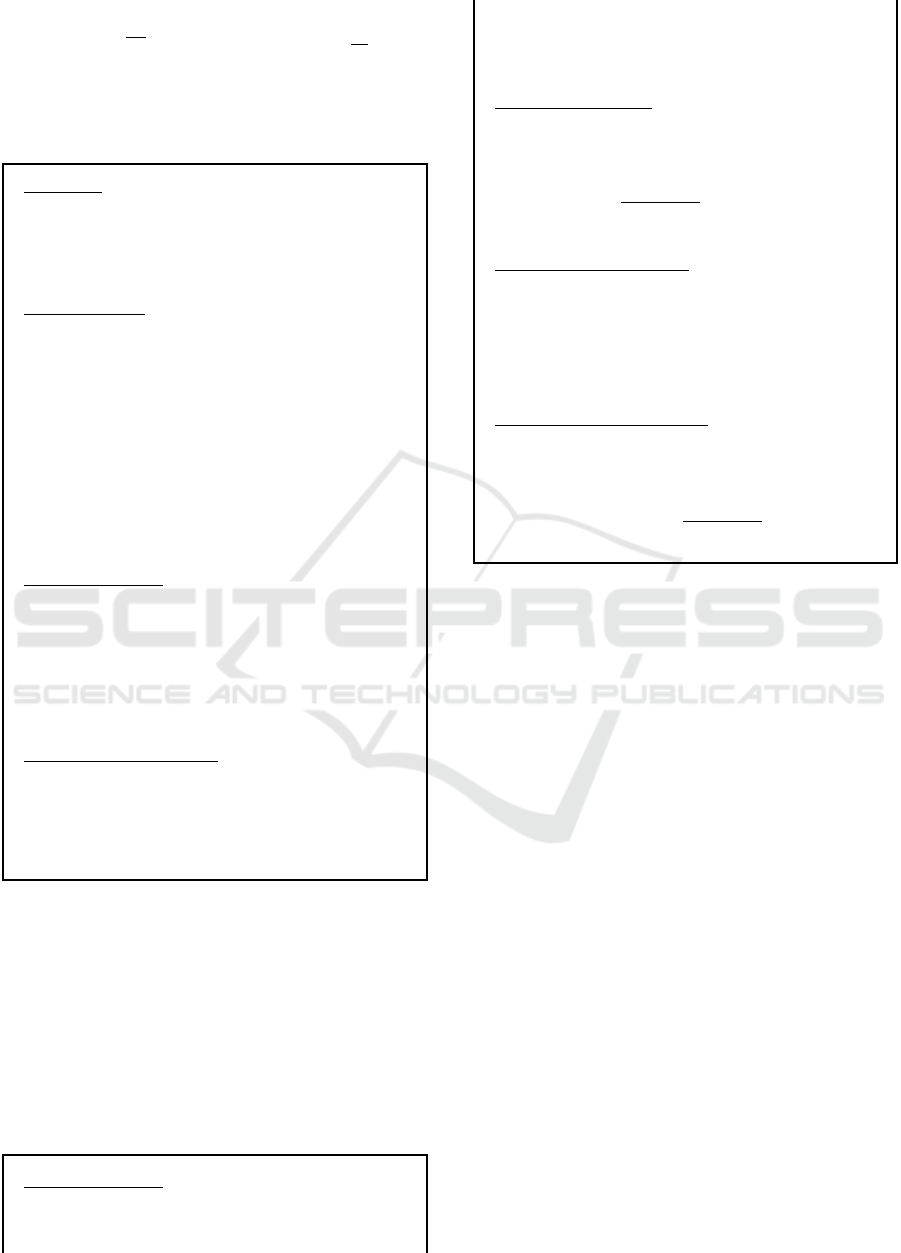
A
Pr
h
(c,g
1
x+c
) ← A (g,g
x
,...,g
x
q
)
i
≤
1
2
ℓ
where c ∈ F
∗
p
is a scalar of A’s choosing.
The bilinear accumulator, as defined by (Nguyen,
2005), acc
X
for the finite set X = {x
1
,...,x
n
} is com-
puted as follows:
acc.setup
Set the public parameters of the bilinear map op-
eration p, G
1
, G
2
, G
T
, e, g, h and uniformly
choose a back door s ∈ Z
∗
p
and compute h
s
for
use in the verification process.
acc.commit(X)
Define H : {0,1}
∗
→ F
∗
p
to be a collision-
resistant hash functions that maps arbitrary
length binary strings to elements in the finite
field F
∗
p
. The accumulator for the set X is com-
puted by taking the generator for G
1
and com-
puting
acc
X
= g
∏
x∈X
(s+H(x))
which is essentially computing an accumulator
polynomial in the exponent of the q-SDH pa-
rameters.
acc.prove(X,x
i
a)
The proof of membership witness for the ele-
ment x
i
∈ X in the accumulator is computed as
wit
x
i
= g
∏
x∈X\{x
i
}
(s+H(x))
which is a commitment to the same accumulator
polynomial but without the root at x
i
.
acc.verify(acc
X
,x
i
,wit
x
i
)
Verification that the element x
i
∈ X is done by
checking that the following pairing operation
equailty holds
e(wit
x
i
,h
x
i
h
s
) = e(acc
X
,h).
4.1 Addition and Deletion Operations
In subsequent work by (Damg
˚
ard and Triandopoulos,
2008) and (Au et al., 2009), the authors expanded
the functionality of the (Nguyen, 2005), enabling dy-
namic operations such as the addition and deletion of
elements to and from the accumulator as well as non-
membership proofs. For use in the rest of the rest of
this paper, the addition and deletion operations are de-
fined below.
acc.add(acc
X
,Y )
The set of elements X ∩Y =
/
0 can be added to
the accumulator by simply updating the accu-
mulator through the computation
acc
′
X
= acc
∏
y∈Y
(s+H(y))
X
= g
∏
z∈X∪Y
(s+H(z))
acc.delete(acc
X
,Y )
The set of elements Y ⊆ X can be deleted from
the accumulator by simply updating the accu-
mulator through the computation
acc
′
X
= acc
1
∏
y∈Y
(s+H(y))
X
= g
∏
z∈X\Y
(s+H(z))
acc.witnessadd(wit
x
i
,x
i
)
The membership witness wit
x
i
can be updated on
the addition of set Y to correspond to acc
X∪Y
by
computing
wit
′
x
i
= wit
∏
y∈Y
(s+H(y))
x
i
.
acc.witnessdelete(wit
x
i
,x
i
)
The membership witness wit
x
i
can be updated on
the deletion of set Y to correspond to acc
x\Y
by
computing
wit
′
x
i
= wit
1
∏
y∈Y
(s+H(y))
x
i
.
4.2 Operation Batching
The result of (Camacho and Hevia, 2010) shows that
batch witness updates with update data of size in-
dependent from the number of elements involved is
impossible. In many cases, witness updates are im-
portant since witnesses that are already issued would
become invalid when the accumulator is updated. In
work by (Vitto and Biryukov, 2022) the authors define
support for batched operations circumnavigating the
impossibility result of (Camacho and Hevia, 2010)
and providing an optimal batch witness update pro-
tocol. The batched operations improve the efficient
of the dynamic operations of an accumulator for large
updates.
5 SNPS
Table 1 outlines the format of the SNPs (Cheng,
2022), and which includes:
• RSID field. This field is a unique identifier related
to the record. It is not possible to have the same
RSID in a raw genome file. Overall, identifiers
that start with an ’rs’ relate to the dbSNP database
sourced from the National Center for Biotechnol-
ogy Information (Wheeler et al., 2001).
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear Group Accumulators in Batch Mode
229

• Chromosome. This field can range from 1 to 22
and can also contain coding for X, Y, XY, and MT.
• Position. This field typically ranges from 3 to
249,218,992, and where there can be multiple
matches for the same chromosome and position
fields.
• Genotype. This field relates to the genotype of the
SNP.
Table 1: SNP data format (Cheng, 2022).
RSID Chromosome Position Genotype
rs12564807 1 734462 AA
rs3131972 1 752721 AG
rs148828841 1 760998 CC
rs12124819 1 776546 AA
rs115093905 1 787173 GG
For instance, with the SNP record of
”rs367789441 1 68082 TT”, the private element
of this data is the genotype ”TT”, and where the two
bases (”TT”) come from a person’s ”X” and ”Y”
allele.
Overall, the main matches that relate to these
searches often relate to genetic relatedness, kinship,
parentship and medical ailments. For example, ”rsID
rs429358” is related to the APOE-ε4 allele and which
has a strong influence on the risk of Alzheimer’s dis-
ease (Lundberg et al., 2023).
6 PRIVACY-AWARE SEARCHING
The number of SNPs stored within a data store can
vary based on the applications. A personalized hair
treatment use case can range between 30 to 40 SNPs,
and ancestry applications require around 350,000
SNPs. Overall, a person will have around 650,000
SNPs.
Overall, Company X could hash all of its SNPs,
such as for ”rs12564807-1-734462-AA”, and store
them in an accumulator and pass this to Trent. A ba-
sic use case could be where a person (Alice) wants
to know if Company X has their SNPs on their data
store. Company X then fills a privacy-aware store
with the SNPs they have and gives it to a trusted entity
(Trent) - each SNP is then hashed into the accumula-
tor. Alice sends hashed versions of the SNPs and asks
Trent to search for them. Trent then returns proof that
all the SNPs are contained in the store or not. This can
be 25 matches for a paternity test and 44 for a person.
The use of an accumulator, too, could provide a way
of showing Alice that her SNPs have been removed
from Company X’s datastore.
Figure 1 outlines the privacy-aware framework.
The system splits into two main systems: Matcher;
and Resolver, and has three main stages:
• Setup. Initially, the gathered SNPs are encrypted
and stored with a unique identifier. This identifier
is then passed to a resolution service and which
stores the details related to the gathered data for
the SNPs. Each gathered set of SNPs is split either
into hash values or split into secret shares and then
added into an accumulator.
• Matching. When Alice wants to see if her SNPs
have been registered on the system, she submits
these and is split into hash values or shares. The
Bloom filter will then say for definite if SNPs are
not stored. With a high probability of success, Al-
ice will be informed if there is a successful dis-
covery. Alice’s encrypted set of SNPs is then sub-
mitted to be checked against a full set of the pre-
viously encrypted SNPs. A hash search will find
the matches.
• Resolving. On one or more matches, Alice will
be delivered a privacy ticket which will be used to
resolve the actual details of SNPs gathering within
the resolver service.
7 RESULTS
The RSA accumulator method would struggle with
performance in adding and removing so many hashed
values of SNPs, but the pairing-based method using
the BLS 12381 curve (Barreto et al., 2003) provides
a more efficient way. For this, we hash our data
onto the curve and produce a scalar value which can
then be added into the accumulator (Buchanan, 2023).
Table 2 shows the results of using the method de-
fined in (Vitto and Biryukov, 2022) and running on
a t2.medium instance in AWS (two vCPUs and 4GB
of memory) for the operations of hashing to the curve
and adding to the accumulator, and also where we
have pre-computed hashes and then just add these to
the accumulator. In this case, one hash is added to
the accumulator at a time. We see, though, that the
addition of the hashing of the SNP onto the curve has
a minimum effect compared with the adding of the
hashed value into the accumulator. For this, the exper-
iment gives 68.23 seconds for the addition of 100,000
SNPs.
In Table 3, we use the batch mode of the Vitto et
al. method (Vitto and Biryukov, 2022), and where we
can add in batches of 10 and 100. We can see that
batch processing considerably enhances the speed of
ICISSP 2024 - 10th International Conference on Information Systems Security and Privacy
230
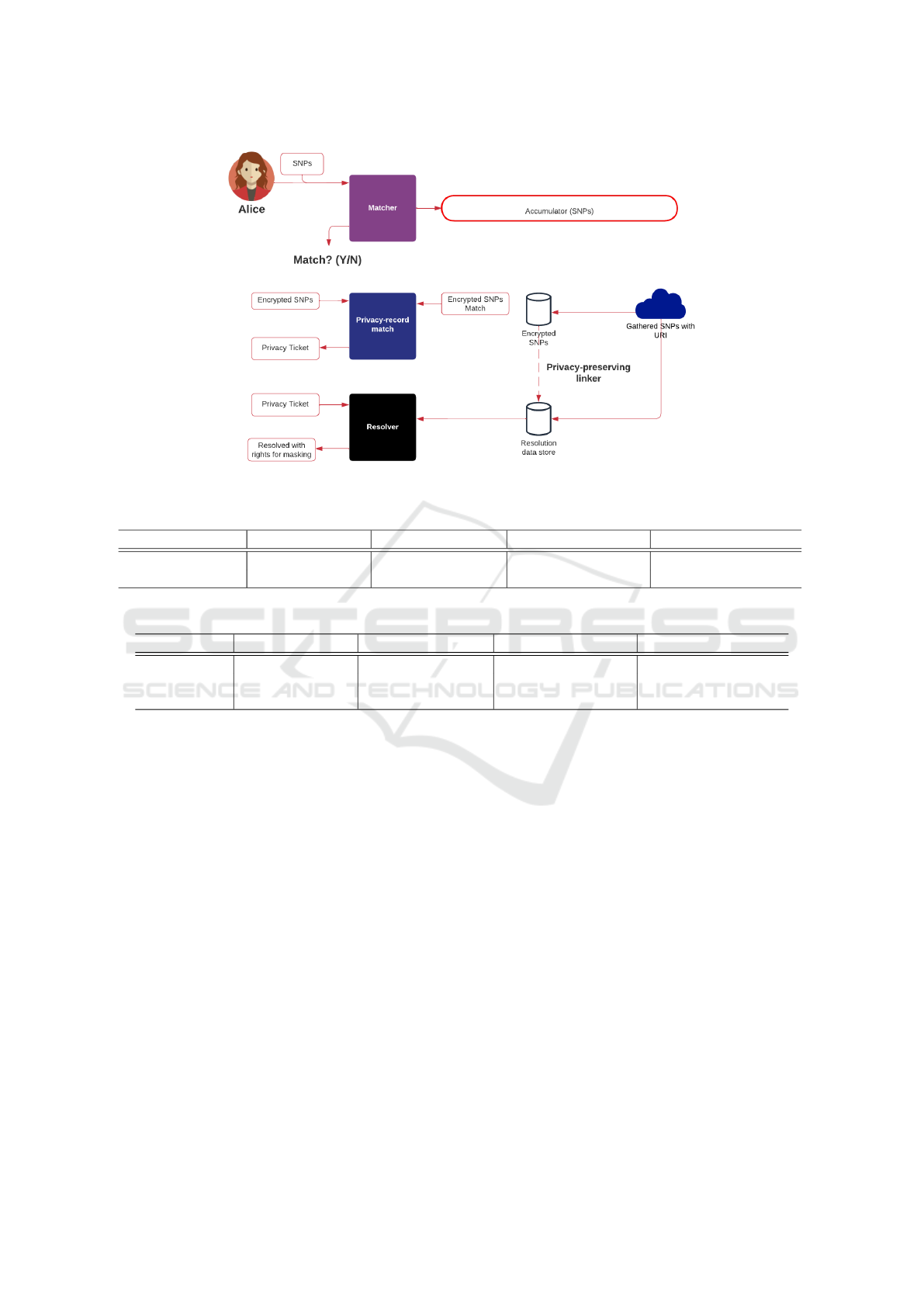
Figure 1: Privacy-aware framework for SNP matching.
Table 2: Results for adding to accumulator.
Method Adding 100 (ms) Adding 1,000 (ms) Adding 10,000 (ms) Adding 100,000 (ms)
Adding with hash 0.1 0.9 7.28 69.44
Adding no hash 0.1 0.87 7.01 68.23
Table 3: Results for adding to accumulator using batch mode.
Method Adding 100 (ms) Adding 1,000 (ms) Adding 10,000 (ms) Adding 100,000 (ms)
Batch of 1 0.1 0.66 6.95 67.3
Batch of 10 0.01 0.1 0.86 7.61
Batch of 100 0.006 0.01 0.12 0.87
operation of adding the SNP hashes to the accumula-
tor. ‘Now, rather than 68.23 seconds for the addition
of 100,000 SNPs, the time to compute drops to 0.87
seconds.
Overall, it can be seen that for the range of exam-
ples, there is an almost linear relationship in adding
the SNP into the accumulator, and has an approximate
overhead of around 0.774 ms for adding a single SNP
hash to the accumulator, and 8.7 µS when processed
in batches of 100. For batches of 100, it would mean
that a single user with 640,000 SNPs onto the accu-
mulator - in this case - would take around 5.65 sec-
onds. The time to create the witness proof and to ver-
ify does not vary as much as the number of values in
the accumulator, and these were measured at 0.86 ms
and 10.90 ms, respectively.
8 CONCLUSIONS
While Bloom filters and homomorphic encryption
provide strong levels of privacy, they can be affected
by scalability issues. The usage of accumulators can
provide one method of preserving the data values in a
data store, and for this, to have a fixed data size width.
The hashing of the SNP onto an elliptic curve point on
the BLS 12381 curve has a relatively small overhead
when compared with the addition of the point into the
accumulator. The time taken to build the accumula-
tor is thus mostly dependent on the time to add the
data point into the accumulator. The batch mode used
in (Ayday et al., 2017) considerably reduces the pro-
cessing overhead when adding the SNPs when used in
batches. Overall, one of the core advantages of using
an accumulator is that we can provide witness proof
that a data entity is contained within a data set, along
with the entity being removed.
REFERENCES
Alsaffar, M. M., Hasan, M., McStay, G. P., and Sedky, M.
(2022). Digital dna lifecycle security and privacy: an
overview. Briefings in Bioinformatics, 23(2):bbab607.
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear Group Accumulators in Batch Mode
231

Au, M. H., Tsang, P. P., Susilo, W., and Mu, Y. (2009). Dy-
namic universal accumulators for ddh groups and their
application to attribute-based anonymous credential
systems. In Fischlin, M., editor, Topics in Cryptology
– CT-RSA 2009, pages 295–308, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Ayday, E., Tang, Q., and Yilmaz, A. (2017). Cryptographic
solutions for credibility and liability issues of genomic
data. IEEE Transactions on Dependable and Secure
Computing, 16(1):33–43.
Baldimtsi, F., Camenisch, J., Dubovitskaya, M., Lysyan-
skaya, A., Reyzin, L., Samelin, K., and Yak-
oubov, S. (2017). Accumulators with applications
to anonymity-preserving revocation. In 2017 IEEE
European Symposium on Security and Privacy (Eu-
roS&P), pages 301–315. IEEE.
Bari
´
c, N. and Pfitzmann, B. (1997). Collision-free accumu-
lators and fail-stop signature schemes without trees.
In Fumy, W., editor, Advances in Cryptology — EU-
ROCRYPT ’97, pages 480–494, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Barreto, P. S., Lynn, B., and Scott, M. (2003). Constructing
elliptic curves with prescribed embedding degrees. In
Security in Communication Networks: Third Interna-
tional Conference, SCN 2002 Amalfi, Italy, Septem-
ber 11–13, 2002 Revised Papers 3, pages 257–267.
Springer.
BBC (2023). Genetic inheritance. https://tinyurl.com/3sts
zk39. Accessed: November 20, 2023.
Ben Sasson, E., Chiesa, A., Garman, C., Green, M., Miers,
I., Tromer, E., and Virza, M. (2014). Zerocash: De-
centralized anonymous payments from bitcoin. In
2014 IEEE Symposium on Security and Privacy, pages
459–474.
Benaloh, J. and De Mare, M. (1993). One-way accumula-
tors: A decentralized alternative to digital signatures.
In Workshop on the Theory and Application of of
Cryptographic Techniques, pages 274–285. Springer.
Boneh, D. and Boyen, X. (2004). Short signatures without
random oracles. In Cachin, C. and Camenisch, J. L.,
editors, Advances in Cryptology - EUROCRYPT 2004,
pages 56–73, Berlin, Heidelberg. Springer Berlin Hei-
delberg.
Boneh, D., Lynn, B., and Shacham, H. (2001). Short sig-
natures from the weil pairing. In Boyd, C., editor,
Advances in Cryptology — ASIACRYPT 2001, pages
514–532, Berlin, Heidelberg. Springer Berlin Heidel-
berg.
Bonomi, L., Huang, Y., and Ohno-Machado, L. (2020).
Privacy challenges and research opportunities for ge-
nomic data sharing. Nature genetics, 52(7):646–654.
Buchanan, W. J. (2023). Witnesses and accumulators in
kryptology for zkp. https://asecuritysite.com/zero/wit
ness. Accessed: November 20, 2023.
Buldas, A., Laud, P., and Lipmaa, H. (2000). Accountable
certificate management using undeniable attestations.
In Proceedings of the 7th ACM Conference on Com-
puter and Communications Security, CCS ’00, page
9–17, New York, NY, USA. Association for Comput-
ing Machinery.
Camacho, P. and Hevia, A. (2010). On the impossibility
of batch update for cryptographic accumulators. In
Abdalla, M. and Barreto, P. S. L. M., editors, Progress
in Cryptology – LATINCRYPT 2010, pages 178–188,
Berlin, Heidelberg. Springer Berlin Heidelberg.
Camacho, P., Hevia, A., Kiwi, M., and Opazo, R. (2008).
Strong accumulators from collision-resistant hashing.
In Wu, T.-C., Lei, C.-L., Rijmen, V., and Lee, D.-T.,
editors, Information Security, pages 471–486, Berlin,
Heidelberg. Springer Berlin Heidelberg.
Camenisch, J. and Lysyanskaya, A. (2002). Dynamic ac-
cumulators and application to efficient revocation of
anonymous credentials. In Yung, M., editor, Advances
in Cryptology — CRYPTO 2002, pages 61–76, Berlin,
Heidelberg. Springer Berlin Heidelberg.
Catalano, D. and Fiore, D. (2013). Vector commitments
and their applications. In Public-Key Cryptography–
PKC 2013: 16th International Conference on Prac-
tice and Theory in Public-Key Cryptography, Nara,
Japan, February 26–March 1, 2013. Proceedings 16,
pages 55–72. Springer.
Cheng, J. (2022). 23andme to plink. https://www.jade-c
heng.com/au/23andme-to-plink/. Accessed: Nov 15,
2023.
Clayton, E. W., Evans, B. J., Hazel, J. W., and Rothstein,
M. A. (2019). The law of genetic privacy: applica-
tions, implications, and limitations. Journal of Law
and the Biosciences, 6(1):1–36.
Damg
˚
ard, I. and Triandopoulos, N. (2008). Supporting non-
membership proofs with bilinear-map accumulators.
Cryptology ePrint Archive, Paper 2008/538. https:
//eprint.iacr.org/2008/538.
DeFrancesco, L. and Klevecz, A. (2019). Your dna broker.
Nat Biotechnol, 37(10):842–7.
Derler, D., Hanser, C., and Slamanig, D. (2015). Revisiting
cryptographic accumulators, additional properties and
relations to other primitives. In Topics in Cryptology—
CT-RSA 2015: The Cryptographer’s Track at the RSA
Conference 2015, San Francisco, CA, USA, April 20-
24, 2015. Proceedings, pages 127–144. Springer.
Garner, S. A. and Kim, J. (2018). The privacy risks of
direct-to-consumer genetic testing: A case study of
23andme and ancestry. Wash. UL Rev., 96:1219.
Ge, C., Susilo, W., Liu, Z., Xia, J., Szalachowski, P.,
and Fang, L. (2020). Secure keyword search and
data sharing mechanism for cloud computing. IEEE
Transactions on Dependable and Secure Computing,
18(6):2787–2800.
Li, J., Li, N., and Xue, R. (2007). Universal accumulators
with efficient nonmembership proofs. In Katz, J. and
Yung, M., editors, Applied Cryptography and Network
Security, pages 253–269, Berlin, Heidelberg. Springer
Berlin Heidelberg.
Lundberg, M., Sng, L. M., Szul, P., Dunne, R., Bayat,
A., Burnham, S. C., Bauer, D. C., and Twine, N. A.
(2023). Novel alzheimer’s disease genes and epista-
sis identified using machine learning gwas platform.
Scientific Reports, 13(1):17662.
Merkle, R. C. (1989). A certified digital signature. In Con-
ICISSP 2024 - 10th International Conference on Information Systems Security and Privacy
232

ference on the Theory and Application of Cryptology,
pages 218–238. Springer.
Naveed, M., Ayday, E., Clayton, E. W., Fellay, J., Gunter,
C. A., Hubaux, J.-P., Malin, B. A., and Wang, X.
(2015). Privacy in the genomic era. ACM Comput-
ing Surveys (CSUR), 48(1):1–44.
Nguyen, L. (2005). Accumulators from bilinear pairings
and applications. In Cryptographers’ track at the RSA
conference, pages 275–292. Springer.
¨
Oks
¨
uz, A. C¸ ., Ayday, E., and G
¨
ud
¨
ukbay, U. (2021). Privacy-
preserving and robust watermarking on sequential
genome data using belief propagation and local dif-
ferential privacy. Bioinformatics, 37(17):2668–2674.
Reyzin, L. and Yakoubov, S. (2016). Efficient asynchronous
accumulators for distributed pki. In Zikas, V. and
De Prisco, R., editors, Security and Cryptography for
Networks, pages 292–309, Cham. Springer Interna-
tional Publishing.
Rivest, R. L., Shamir, A., and Adleman, L. (1978). A
method for obtaining digital signatures and public-key
cryptosystems. Commun. ACM, 21(2):120–126.
Shringarpure, S. S. and Bustamante, C. D. (2015). Privacy
risks from genomic data-sharing beacons. The Amer-
ican Journal of Human Genetics, 97(5):631–646.
Tsudik, G. and Xu, S. (2003). Accumulating com-
posites and improved group signing. In Advances
in Cryptology-ASIACRYPT 2003: 9th International
Conference on the Theory and Application of Cryptol-
ogy and Information Security, Taipei, Taiwan, Novem-
ber 30–December 4, 2003. Proceedings 9, pages 269–
286. Springer.
Vitto, G. and Biryukov, A. (2022). Dynamic universal ac-
cumulator with batch update over bilinear groups. In
Cryptographers’ Track at the RSA Conference, pages
395–426. Springer.
Von Thenen, N., Ayday, E., and Cicek, A. E. (2019).
Re-identification of individuals in genomic data-
sharing beacons via allele inference. Bioinformatics,
35(3):365–371.
Wang, P., Wang, H., and Pieprzyk, J. (2007). A new dy-
namic accumulator for batch updates. In Qing, S.,
Imai, H., and Wang, G., editors, Information and
Communications Security, pages 98–112, Berlin, Hei-
delberg. Springer Berlin Heidelberg.
Wheeler, D. L., Church, D. M., Lash, A. E., Leipe,
D. D., Madden, T. L., Pontius, J. U., Schuler, G. D.,
Schriml, L. M., Tatusova, T. A., Wagner, L., et al.
(2001). Database resources of the national center for
biotechnology information. Nucleic acids research,
29(1):11–16.
Xu, L., Xu, C., Liu, Z., Wang, Y., Wang, J., et al. (2019).
Enabling comparable search over encrypted data for
iot with privacy-preserving. Computers, Materials &
Continua, 60(2):675–690.
Xu, Z., Tian, H., Liu, D., and Lin, J. (2010). A ring-
signature anonymous authentication method based on
one-way accumulator. In 2010 Second International
Conference on Communication Systems, Networks
and Applications, volume 2, pages 56–59. IEEE.
Privacy-Aware Single-Nucleotide Polymorphisms (SNPs) Using Bilinear Group Accumulators in Batch Mode
233
