
Minimalist CNN for Medical Imaging Classification with Small Dataset:
Does Size Really Matter and How?
Marie
´
Economid
`
es
1,2 a
and Pascal Desbarats
2 b
1
GE Healthcare, F-78530 Buc, France
2
Univ. Bordeaux, CNRS, Bordeaux INP, LaBRI, UMR 5800, F-33400 Talence, France
Keywords:
Deep Learning, Medical Imaging, MRI, Classification, Convolutional Neural Network.
Abstract:
Deep learning has become a key method in computer vision, and has seen an increase in the size of both the
networks used and the databases. However, its application in medical imaging faces limitations due to the size
of datasets, especially for larger networks. This article aims to answer two questions: How can we design a
simple model without compromising classification performance, making training more efficient? And, how
much data is needed for our network to learn effectively? The results show that we can find a minimalist
CNN adapted to a dataset that gives results comparable to larger architectures. The minimalist CNN does not
have a fixed architecture. Its architecture varies according to the dataset and various criteria such as overall
performance, training stability, and visual interpretation of network predictions. We hope this work can serve
as inspiration for others concerned with these challenges.
1 INTRODUCTION
Last past years, Deep Learning (DL) methods demon-
strated high performances in computer vision tasks
like classification, detection, or segmentation. Classi-
fication challenges on large datasets like MNIST (Le-
Cun et al., 2010), ImageNet (Fei-Fei et al., 2009) or
CIFAR10- CIFAR100 (Krizhevsky et al., 2009) have
driven the development of powerful neural networks
such as ResNet (He et al., 2016), VGG (Simonyan and
Zisserman, 2014), and AlexNet (Krizhevsky et al.,
2012). However, as the network size and complexity
increased, the need for larger datasets increased too.
In medical imaging, DL methods demonstrated
high performance in various applications like knee
abnormalities classification and detection (Rizk et al.,
2021). However, medical datasets frequently lack
sufficient volume compared to the architectures em-
ployed. Medical imaging dataset size is limited be-
cause of the complex process of collecting data en-
suring patient’s rights and the fastidious and time-
consuming annotation process. Confronted with such
small, often imbalanced datasets (Gao et al., 2020),
common strategies rely on data augmentation meth-
ods, like MRNet work (Bien et al., 2018) using ro-
a
https://orcid.org/0009-0001-3576-1526
b
https://orcid.org/0000-0003-4267-492X
tations or horizontal flips. Additionally, a common
practice often involves pre-training models on non-
medical images (Kim et al., 2022). This introduces
irrelevant features and unnecessary parameters, lead-
ing to high computational costs.
An alternative approach involves designing more
suitable networks (Zavalsız et al., 2023), (Albelwi and
Mahmood, 2017). For instance, the framework pro-
posed by (Cao, 2015) relies on deconvolution for fea-
ture visualization and correlation coefficient calcula-
tion for architecture optimization. Another work by
(Wasay and Idreos, 2020) introduces a design frame-
work centered on controlling the number of param-
eters in the network, along with a thorough analysis
of numerous parameters. These works offer valuable
insights into CNN design. However, they may not
necessarily address adaptation to a small dataset and
limited computational resources.
In this study, we aim to provide a methodology
to design the smallest possible Convolutional Neural
Network (CNN) adapted to a specific dataset while
maintaining accuracy comparable to more complex
CNNs, guided by the comprehension of learned fea-
tures.
This paper presents a preliminary study on the
minimalist CNN design based on preliminary results
on MRI classification tasks and a comparison between
different architectures. In the first step, a way of de-
Économidès, M. and Desbarats, P.
Minimalist CNN for Medical Imaging Classification with Small Dataset: Does Size Really Matter and How?.
DOI: 10.5220/0012452700003660
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024) - Volume 2: VISAPP, pages
755-762
ISBN: 978-989-758-679-8; ISSN: 2184-4321
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
755
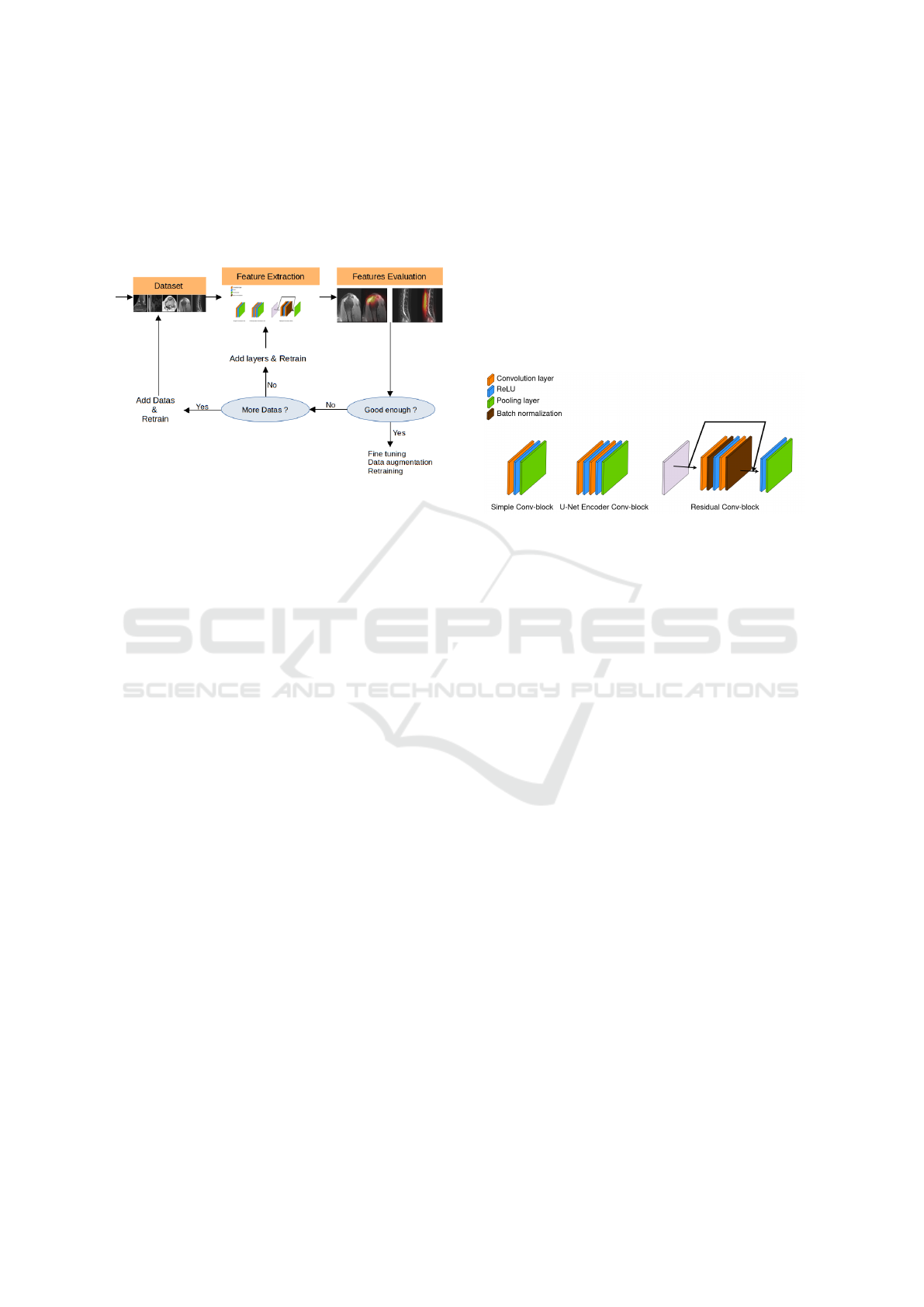
signing a minimalist CNN adapted to the dataset size
is introduced. Secondly, we present the dataset and
training parameters. Then, we focus on the compar-
ison between different architectures using different
criteria, especially comprehension of learned features
along with a discussion.
Figure 1: Illustration of the process of designing a minimal-
ist CNN.
2 METHOD
A Convolutional Neural Network (CNN) is composed
of a feature extractor and classifier. Classical met-
rics will be employed for performance evaluation dur-
ing and after training. Visual explanations, specif-
ically GradCam, will provide a visual interpretation
of activations. Depending on the outcomes of previ-
ous training iterations, the model’s architecture can
be adjusted and enhanced by incorporating additional
conv-blocks.
2.1 Design Architecture
We design our minimalist CNN using convolutional
blocks (conv-blocks) for feature extraction and a clas-
sifier for final predictions. This minimalist CNN aims
to be as small and lightweight as possible while per-
forming at least as well as larger models found in the
literature.
Design of Features Extractor. The features extrac-
tor is composed of conv-blocks. A convolutional
block in deep learning is a fundamental unit com-
prising convolutional layers, activation functions, op-
tional pooling, and normalization operations. It al-
lows feature extraction and processing at different
levels of abstraction, enabling convolutional neural
networks to learn hierarchical representations of vi-
sual information.
The first layers allow for the extraction of high-
level features, corresponding to the global character-
istics of the image, while the deeper layers, known as
low-level layers, extract more specific features of the
image. This implies that a minimum number of con-
volutional layers is necessary to construct our model.
Figure 2 illustrates the different conv-block that
will serve to design a minimalist CNN and compare
it to bigger architectures. We take inspiration from
well-known models like LeNet5 (LeCun et al., 1998),
U-Net (Ronneberger et al., 2015) and ResNet (He
et al., 2016).
Figure 2: Illustration of different conv-blocks.
The first conv-block is inspired by the LeNet5 ar-
chitecture. This CNN architecture was one of the first
to demonstrate its effectiveness in classifying small
images and simple tasks such as digit recognition. In
a LeNet5 network, there are three convolutional lay-
ers followed by a tanh activation function, with two
of them followed by a pooling layer. We use this
architecture as a reference for the minimum number
of convolutional layers needed for feature extraction.
We define a simple conv-block as a convolutional
layer followed by an activation function and a pooling
layer. Based on this simple conv-block, we design a
feature extractor composed of 4 simple conv-blocks.
For clarity in the paper, we decide to refer to this net-
work as miniCNN.
The second illustrated conv-block is based on the
U-Net architecture, renowned for its superior perfor-
mance and precise predictions, even when working
with small datasets. Shaped like a ’U’, U-Net com-
prises an encoder that captures the contextual infor-
mation of the image, a decoder for accurate localiza-
tion and information expansion to restore the initial
image size, and a bottleneck connecting the encoder
and decoder. While its primary application is in seg-
mentation tasks, a closer examination of the encoder
component is interesting. A conv-block consists of
two consecutive convolutional layers, each followed
by a ReLU activation function, and concludes with a
pooling layer. Based on this conv-block, we design
a feature extractor composed of 5 conv-blocks. As it
refers to the encoder part of U-Net, we decide to refer
to this network as UEnc.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
756

In related work by RadImageNet (Mei et al.,
2022), various well-known CNN architectures were
explored including the ResNet architecture. The ef-
fectiveness of this model to train very deep networks
has made it a popular choice across many applica-
tions. As our minimalist CNN aims to approach or
match the performance of larger networks with lim-
ited data, we decide to include this architecture in our
tests.
ResNet’s strength lies in its use of residual conv-
blocks and skip connections. It allows more direct
flow of gradients and mitigates the Vanishing Gradi-
ent Problem. Moreover, it improves training speed
and convergence compared to different architectures.
ResNet also enables the training of larger models that
improve feature representation. However, it’s impor-
tant to note that this approach may increase com-
plexity and require more computational resources. It
may also be more sensitive to overfitting with smaller
datasets and interpretation of learned features can be
more difficult. That is the reason why two versions
of ResNet will be trained: ResNet18 (RN18) and
ResNet50 (RN50).
Classifier. After the feature extraction process, the
classifier takes the extracted features as input and uti-
lizes them to assign labels and make predictions. The
classifier is composed of one or more fully connected
layers, also known as dense layers. In addition to
the fully connected layers, an activation function like
softmax for multi-class classification can be used to
convert outputs into probability scores for each class
and the highest probability corresponds to the pre-
dicted class. The classifier architecture complexity
depends on the task and data. To improve perfor-
mance and generalization additional layers such as
dropout or normalization can be used with one or sev-
eral dense layers.
Considering the ResNet classifier is composed of
1 Fully Connected layer followed by softmax, our
minimalist CNN classifier will adopt the same struc-
ture.
Commonly used Cross Entropy loss function for
classification tasks is given to measure model perfor-
mance.
2.2 Model Explanation
Obtaining a deeper understanding of our model’s
learning process is crucial for validating our mini-
malist CNN to ensure high precision and extraction
of meaningful features. To achieve this, we focus on
the Gradient Class Activation Mapping (Grad-CAM)
technique (Selvaraju et al., 2017). Additionally, we
enhance this visual interpretation of prediction by us-
ing a quantitative quality score for GradCam evalua-
tion.
Visual Explaination. Grad-CAM provides a visu-
alization that allows for the interpretation of CNN
predictions by indicating which parts of an image
have contributed the most to the classification of an
image and the prediction of a specific class. Gradi-
ents at the output of a convolutional layer are com-
puted for a specific class. They are then weighted to
generate a heat map to highlight the most activated ar-
eas of the image. Multiple GradCAM methods have
been deployed. We choose to use ablationCAM (Ra-
maswamy et al., 2020), an improved version of clas-
sical GradCAM.
3 EXPERIMENTAL SETUP
We apply our method to a medical imaging problem,
specifically focusing on the classification of osteoar-
ticular MRI. This section presents the dataset used,
training parameters, and details about the equipment
employed.
3.1 Dataset
In medical imaging, datasets are typically small and
imbalanced. Here, we choose to design a minimalist
model for osteoarticular MRI classification, specifi-
cally focusing on structure classification. A typical
dataset consists of around 55 examinations. An ex-
amination is a series of images acquired in a specific
orientation, averaging about 32 images per series, to-
taling approximately 1760 images. For a given model,
training is conducted by varying the amount of data
without data augmentation. Our goal is to determine
if we can find an architecture that is large enough
to achieve performance comparable to datasets with
thousands of images with this amount of data, yet as
small as possible to reduce training time and compu-
tational costs.
RadImageNet. Dataset (Mei et al., 2022) consists
of over a million images from various acquisitions,
modalities, and anatomical structures. From this
dataset, we choose to extract MRI osteoarticular data
which corresponds to 5 classes: Ankle, Hip, Knee,
Shoulder, and Spine.
Sub Datasets. To create the training, validation,
and test datasets, we initially split the dataset with a
Minimalist CNN for Medical Imaging Classification with Small Dataset: Does Size Really Matter and How?
757

Figure 3: Images samples from dataset A- Ankle, B- Hip,
C- Knee, D- Shoulder, E- Spine.
Figure 4: Dataset classes repartition.
distribution of 65%, 25%, and 10%. Subsequently,
to form the sub-datasets, we perform a random shuf-
fle within the base training dataset to extract the de-
sired quantity of data. Following this, we conduct an-
other random shuffle within the validation dataset to
obtain 30% of the sub-training set for the final valida-
tion dataset.
Table 1: Number of datas in each training.
Percentage Training set Validation set
100 348 824 134 161
70 244 176 73 252
50 174 412 52 323
30 104 647 31394
10 34 882 10 464
5 17 441 5232
3 10 464 3139
1 3 488 1046
0.5 1 744 523
3.2 Training Parameters
For training, data augmentation is not employed. The
input image size for the network is set to 224x224,
with a fixed batch size of 32. The learning rate is
established at 1e-4, and models are trained for 30
epochs. The optimizer used is Adam, and Cross
Entropy serves as the loss function. The metrics
monitored during training include loss, accuracy, and
AUC.
Training involves shuffling the dataset, while vali-
dation does not shuffle. The best model is saved when
the validation loss decreases. During inference on the
test dataset, the best model is used.
3.3 Setup
We aim to design a minimalist CNN for high-
performance training with mainstream computational
resources. All training is conducted on a laptop
equipped with an Intel Core i9 12th generation pro-
cessor, 32 GB of RAM, and an NVidia GeForce
RTX3070ti GPU with 8 GB of video memory.
4 RESULTS
Table 2: Training time per epoch for each model and each
subdataset in hour and minutes.
%Data miniCNN UEnc RN18 RN50
0.5 00:04 00:12 00:05 00:08
1 00:06 00:23 00:07 00:14
3 00:12 01:05 00:16 00:41
5 00:20 01:52 00:25 01:10
10 00:39 03:32 00:49 02:35
30 01:57 10:10 02:25 06:32
50 03:27 19:13 04:01 11:34
70 05:22 26:23 05:23 14:41
100 08:23 38:05 10:45 27:47
Training Time & Performances. There is a signif-
icant increase in training time depending on the net-
work and the amount of data used, as illustrated in
Figure 2. With the same amount of data, UEnc takes
more time to train than other architectures. Overall,
all models exhibit nearly similar performances for a
specific amount of data, as shown in Table 3.
However, when comparing training times to the
number of parameters in each model, RN50 outper-
forms UEnc with more parameters. Surprisingly,
miniCNN, despite its simpler architecture, demon-
strates comparable performance to RN50.
Performances & Training Stability. If the per-
formances of miniCNN are comparable to those of
RN50, it is essential to verify that models are not over-
fitting or underfitting. For the same amount of data,
both miniCNN and RN18 exhibit less stable learning
than the U-Net encoder or RN50. Regardless of the
data quantity, RN18 displays the least stable learning
and struggles to converge easily, Figure 7.
Examining the training stability of miniCNN con-
cerning the data quantity reveals that the learning pro-
cess becomes more stable with at least 10% of the
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
758

Table 3: Results of 4 models trained with different amount of datas evaluated with classical metrics (precision, AUC).
Models
miniCNN UEnc RN18 RN50
# param 948 293 18 848 325 11 179 077 23 518 277
Dataset AUC ACC AUC ACC AUC ACC AUC ACC
0.5 95.48 79.23 98.20 90.23 99.46 94.42 99.22 92.63
1 97.15 84.25 99.55 95.46 99.82 97.03 99.76 96.61
3 99.28 93.09 99.92 98.40 99.92 97.69 99.89 97.48
5 99.62 95.63 99.95 98.82 99.97 99.03 99.93 99.28
10 99.89 97.49 99.98 99.48 99.98 99.43 99.98 99.38
30 99.98 99.13 99.99 99.64 99.99 99.74 99.99 99.73
50 99.98 99.31 99.99 99.58 99.99 99.85 99.99 99.84
70 99.99 99.40 99.99 99.84 99.99 99.87 99.99 99.88
100 99.99 99.68 99.98 99.47 99.99 99.92 99.99 99.89
dataset. The same observation can be done for the
UEnc and RN50.
Performances & Grad-CAMs. The Grad-CAMs 6
associated with the last conv-block of each network
for different data quantities are presented. We aim to
determine if the learned features remain relevant with
a smaller network and fewer data. We use an image
from the test dataset to present a visual analysis of
activation maps, Figure 5. A general observation in-
dicates that the Grad-CAM activations of Uenc with a
data quantity greater than 30% of the dataset are more
precise. In contrast, the Grad-CAMs of miniCNN are
much less accurate, regardless of the data quantity.
Figure 5: Hip MRI from test dataset from RadImageNet.
5 DISCUSSION
Conv-Block Choice. For simplicity in our model
selection, we focused on U-Net and ResNet architec-
tures. However, we could explore other well-known
models like DenseNet, AlexNet, VGG in future inves-
tigations in order to provide additional comparisons.
Dataset & Classification Tasks. We chose a spe-
cific classification task, but it would be interesting to
test on a more complex task, such as lesion classifica-
tion. This would allow us to compare our results with
RadImageNet or with other related works in medical
imaging, providing a more comprehensive and com-
plete evaluation.
Training Parameters. In this study, all parameters
were fixed to evaluate the performance of different
models consistently. However, with a limited dataset,
training may be unstable. The batch size initially set
at 32 could be reduced to aid the network in better
convergence. In cases where the dataset cannot be ex-
panded due to a lack of data, data augmentation meth-
ods could enhance learning stability. Furthermore,
training was conducted for 30 epochs, but increasing
the number of epochs and incorporating early stop-
ping methods would be beneficial.
Minimalist CNN as the Best Compromise. Our
minimalist CNN is not a specific network. In fact, it
represents a model from a sufficient compromise be-
tween the dataset, training time, model performance,
learning quality, and the features learned by the net-
work. For instance, in our sub-dataset containing
0.5% of the dataset, the performance of miniCNN and
UEnc is lower than RN18 and RN50. The training
curves show smoother learning curves for miniCNN
than RN18 and RN50. The Grad-CAMs of miniCNN,
UEnc, and RN50 are not as relevant as those of RN18
in this context, making RN18 the minimalist CNN.
Moving on to our sub-dataset with 30% of the data, all
four architectures exhibit excellent and similar perfor-
mances, and training appears stable. In comparison to
our reference image, we observe that the Grad-CAMs
are more relevant with UEnc than with the other mod-
els. However, concerning training time, UEnc is the
Minimalist CNN for Medical Imaging Classification with Small Dataset: Does Size Really Matter and How?
759
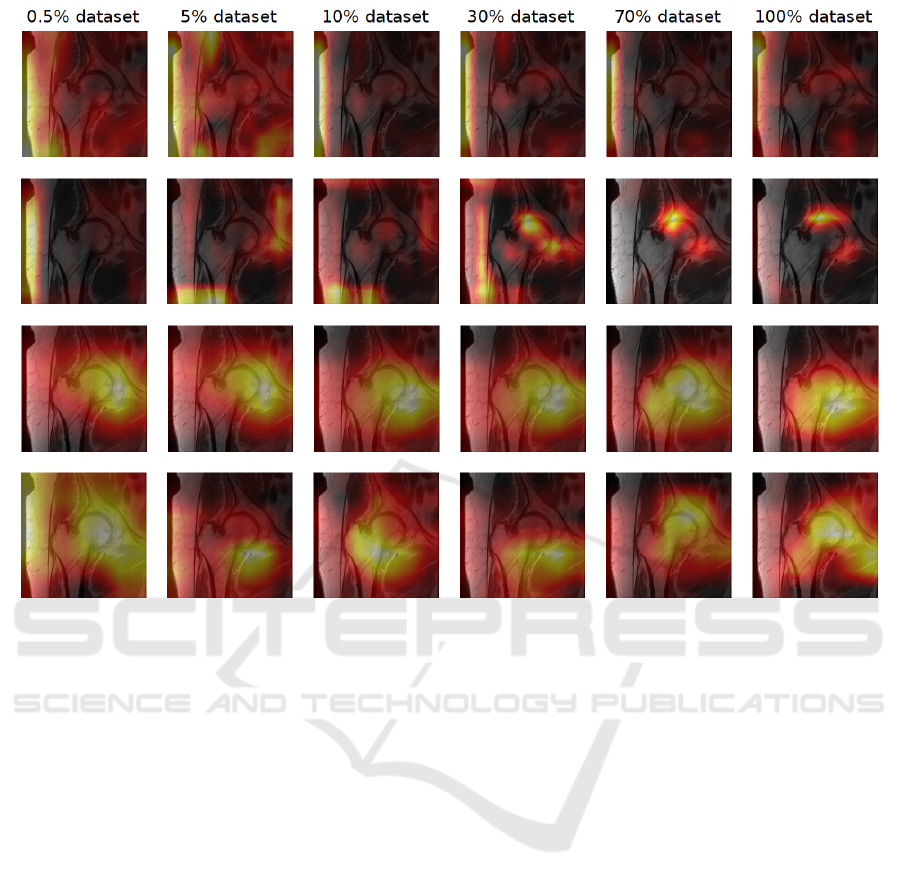
Figure 6: Grad-CAM on the last epoch for each model. Row1 - miniCNN, Row2 - UEnc, Row3- RN18, Row4 - RN50.
slowest model to train. Therefore, to determine our
minimalist CNN, a compromise must be done be-
tween training time and the quality of explanations
provided by Grad-CAMs.
Grad-CAMs. Provide a first comprehension ele-
ment about how the network predicts a class. In ad-
dition to visual analysis, a quantitative evaluation of
Grad-CAMs could be conducted to ensure the rele-
vance of the produced Grad-CAMs. There are various
evaluation methods, some involving model retraining
and others not. For example, an image perturbation
method known as ROAD (Remove and Debias) (Rong
et al., 2022), combines Most Relevant First (MORF)
and Least Relevant First (LERF) methods. MORF re-
moves the highest attention pixels first, while LERF
removes the least attention pixels first.
6 CONCLUSIONS
In this preliminary study, we have demonstrated our
ability to design a minimalist CNN adapted to a spe-
cific medical image classification task and dataset.
The results indicate that our minimalist CNN achieves
comparable performance to larger CNN architectures
with mainstream computational resources. This high-
lights the potential of our approach in establishing a
pipeline to design a minimalist CNN. Moving for-
ward, we aim to further refine our minimalist CNN
by incorporating more comparison criteria in the pro-
cess such as explainability method. This could help
ensure not only performance but also a deeper un-
derstanding of the learned features. More precisely,
we could use the quality evaluation of the features
learned by the network and integrate this evaluation
as an additional metric to improve the architecture of
our network, keeping it as small as possible.
In future works, we will apply our Minimalist
CNN design to other medical image datasets and dif-
ferent classification tasks such as specific or general
abnormalities classification. To address the challenge
of limited data availability we will evaluate the in-
fluence of data augmentation and transfer learning
on our model. Furthermore, we need to test more
CNN architecture to design our minimalist CNN.
Even though a lot of current studies are based on
big complex architectures trained to answer multiple
tasks, it’s interesting to design smaller models. We
will compare our minimalist CNN with bigger models
on the same tasks and dig deeper into our evaluation
process to ensure interpretability and robustness.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
760
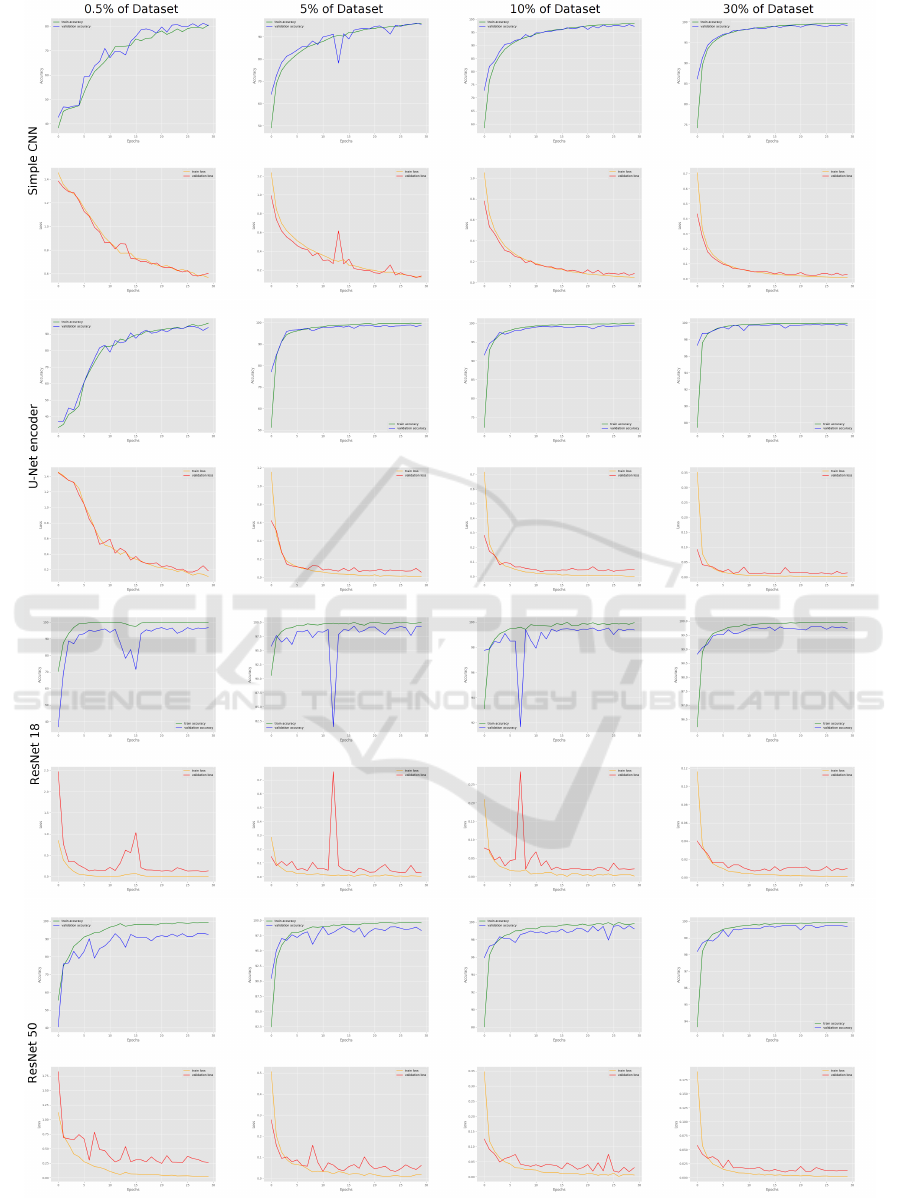
Figure 7: Training curves.
Minimalist CNN for Medical Imaging Classification with Small Dataset: Does Size Really Matter and How?
761

ACKNOWLEDGEMENTS
This research was supported by GE Healthcare. We
thank Nicolas Gogin for comments that greatly im-
proved this paper.
We would also like to show our gratitude to the
reviewers for their comments and advices.
REFERENCES
Albelwi, S. and Mahmood, A. (2017). A framework for de-
signing the architectures of deep convolutional neural
networks. Entropy.
Bien, N., Rajpurkar, P., Ball, R. L., Irvin, J., Park, A., Jones,
E., Bereket, M., Patel, B. N., Yeom, K. W., Shpan-
skaya, K., et al. (2018). Deep-learning-assisted di-
agnosis for knee magnetic resonance imaging: devel-
opment and retrospective validation of mrnet. PLoS
medicine.
Cao, X. (2015). A practical theory for designing very deep
convolutional neural networks. Unpublished Techni-
cal Report.
Fei-Fei, L., Deng, J., and Li, K. (2009). Imagenet: Con-
structing a large-scale image database. Journal of vi-
sion.
Gao, L., Zhang, L., Liu, C., and Wu, S. (2020). Handling
imbalanced medical image data: A deep-learning-
based one-class classification approach. Artificial in-
telligence in medicine.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Kim, H. E., Cosa-Linan, A., Santhanam, N., Jannesari, M.,
Maros, M. E., and Ganslandt, T. (2022). Transfer
learning for medical image classification: a literature
review. BMC medical imaging.
Krizhevsky, A., Hinton, G., et al. (2009). Learning multiple
layers of features from tiny images.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
agenet classification with deep convolutional neural
networks. Advances in neural information processing
systems, 25.
LeCun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
LeCun, Y., Cortes, C., and Burges, C. (2010). Mnist hand-
written digit database. ATT Labs [Online]. Available:
http://yann.lecun.com/exdb/mnist.
Mei, X., Liu, Z., Robson, P. M., Marinelli, B., Huang, M.,
Doshi, A., Jacobi, A., Cao, C., Link, K. E., Yang, T.,
et al. (2022). Radimagenet: an open radiologic deep
learning research dataset for effective transfer learn-
ing. Radiology: Artificial Intelligence.
Ramaswamy, H. G. et al. (2020). Ablation-cam: Vi-
sual explanations for deep convolutional network via
gradient-free localization. In proceedings of the
IEEE/CVF winter conference on applications of com-
puter vision.
Rizk, B., Brat, H., Zille, P., Guillin, R., Pouchy, C., Adam,
C., Ardon, R., and d’Assignies, G. (2021). Menis-
cal lesion detection and characterization in adult knee
mri: a deep learning model approach with external
validation. Physica Medica.
Rong, Y., Leemann, T., Borisov, V., Kasneci, G., and Kas-
neci, E. (2022). A consistent and efficient evalua-
tion strategy for attribution methods. arXiv preprint
arXiv:2202.00449.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18. Springer.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. In Proceedings of the IEEE international
conference on computer vision.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Wasay, A. and Idreos, S. (2020). More or less: When and
how to build convolutional neural network ensembles.
In International Conference on Learning Representa-
tions.
Zavalsız, M. T., Alhajj, S., Sailunaz, K.,
¨
Ozyer, T., and Al-
hajj, R. (2023). A comparative study of different pre-
trained deeplearning models and custom cnn for pan-
creatic tumor detection. International Arab Journal of
Information Technology.
VISAPP 2024 - 19th International Conference on Computer Vision Theory and Applications
762
