
Open Platform for the De-Identification of Burned-in Texts in Medical
Images Using Deep Learning
Quentin Langlois
1 a
, Nicolas Szelagowski
2 b
, Jean Vanderdonckt
1,2 c
and S
´
ebastien Jodogne
1 d
1
Institute for Information and Communication Technologies, Electronics and
Applied Mathematics (ICTEAM), UCLouvain, Belgium
2
Louvain Research Institute in Management and Organizations (LRIM), UCLouvain, Belgium
Keywords:
Medical Imaging, Deep Learning, Text Detection, Image de-identification, Open-Source Software.
Abstract:
While the de-identification of DICOM tags is a standardized, well-established practice, the removal of pro-
tected health information burned into the pixels of medical images is a more complex challenge for which
Deep Learning is especially well adapted. Unfortunately, there is currently a lack of accurate, effective, and
freely available tools to this end. This motivates the release of a new benchmark dataset, together with free
and open-source software leveraging dedicated Deep Learning algorithms, with the goal of improving patient
confidentiality. The proposed methods consist of adapting scene-text detection models (SSD and TextBoxes)
to the task of image de-identification. Results have shown that fine-tuning such generic text detection models
on medical images significantly improves performance. The developed algorithms can be applied either from
the command line or using a Web interface that is tightly integrated with a free and open-source PACS server.
1 INTRODUCTION
Deep Learning (DL) methods have been applied to
a variety of different medical tasks, such as tumor
segmentation (Bakas et al., 2018), organ segmen-
tation (Wasserthal et al., 2023), or free-text analy-
sis (Johnson et al., 2020). Since hospitals generally
have neither the technical expertise nor the computa-
tional resources in-house, they need to be able to ex-
port their patient data to researchers for the training of
DL models. More generally, collecting medical data
as open-access databases has become a major chal-
lenge for the development of artificial intelligence in
the healthcare sector. Collecting such data requires
not only patient consent but also the use of automated
tools that can prevent the revelation of patient identity.
The de-identification of electronic health records
(EHR) is a well-established practice (Vithya et al.,
2020). In the context of medical imaging, the of-
ficial DICOM standard defines multiple so-called
“Application-Level Confidentiality Profiles” that list
which DICOM tags must be removed or cleared
a
https://orcid.org/0009-0006-7135-3809
b
https://orcid.org/0009-0001-5335-7682
c
https://orcid.org/0000-0003-3275-3333
d
https://orcid.org/0000-0001-6685-7398
to preserve patient confidentiality. However, imag-
ing modalities can store protected health information
(PHI) not only in the DICOM tags but also as texts
that are directly burned into the raw pixels of medi-
cal images. This for instance frequently happens with
radiography, mammography, and ultrasound imaging
modalities. Therefore, the detection and removal of
PHI that is burned into the raw pixels data of medical
images is an important concern to prevent the leak-
age of personal information through the raw pixels of
medical images. Yet, this process is rarely applied in
the clinical routine, because of a lack of dedicated,
easy-to-use, freely available tools.
The de-identification of burned-in PHI is intri-
cately linked to the task of detecting texts in 2D
scenes. Indeed, once a PHI text has been detected, its
bounding box can easily be wiped out from the pixel
data to generate a de-identified DICOM instance. In
recent years, numerous open-access scene-text detec-
tion benchmarks have been released, such as the IC-
DAR competition databases (Lucas et al., 2003) and
the SynthText generator (Gupta et al., 2016), along
with dedicated evaluation protocols, which has en-
abled the development of new state-of-the-art DL ar-
chitectures. Nonetheless, the performance of such
DL models have not been widely studied in the con-
text of PHI detection and removal in medical images
Langlois, Q., Szelagowski, N., Vanderdonckt, J. and Jodogne, S.
Open Platform for the De-identification of Burned-in Texts in Medical Images using Deep Learning.
DOI: 10.5220/0012430300003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 297-304
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
297

yet: The main challenge that currently prevents the
direct application of DL-based models to burned-in
text detection in medical images is a lack of ded-
icated, annotated databases. Indeed, most existing
open-access medical image databases, notably The
Cancer Imaging Archive (TCIA) (Clark et al., 2013),
contain only a few instances of burned-in texts, which
are not specifically annotated.
According to this discussion, this paper presents
a new benchmark dataset for text detection in med-
ical images, along with effective fine-tuned versions
of the “single-shot multi-box detector” (SSD) (Liu
et al., 2016) and TextBoxes (Liao et al., 2017) algo-
rithms. The developed models have been evaluated
against pre-trained versions of “Efficient and Accu-
rate Scene-Text detector” (EAST) (Zhou et al., 2017)
and DPText-DETR (Ye et al., 2022) architectures,
demonstrating significant improvements for burned-
in text detection in medical images. This highlights
the importance of fine-tuning such DL models on
task-specific databases, which also justifies our devel-
opment and release of open-access databases to fos-
ter further research in this domain. As a final con-
tribution, two free and open-source tools to automat-
ically remove burned-in texts in medical images are
released, the former working from the command line,
while the latter is a Web interface that is integrated
with the Orthanc PACS server (Jodogne, 2018).
2 RELATED WORK
This section introduces different techniques used for
burned-in text detection in medical images, both be-
fore and after the rise of DL for scene-text detection.
2.1 Image Processing for Burned-in
Text Detection
One of the earliest methods applicable to PHI detec-
tion and extraction in images consisted of detecting
texts using Daubechies wavelets (Wang et al., 1997).
This method is based on the characteristic diagonal
variations in the frequency domain observed in most
Roman characters and Arabic numbers.
More recent work performs text detection using
heuristic observations on the properties of medical
images. A first technique proposes to separate texts
from the background by analyzing the variance of
pixel values in some regions of interest (Yingxuan
et al., 2010). Based on similar observations, another
multi-step pipeline has been proposed to isolate texts
by progressively applying low- and high-threshold fil-
ters, along with morphological transformations such
as dilation (Newhauser et al., 2014). Both the lat-
ter techniques use an Optical Character Recognition
(OCR) post-processing to filter out false detections
(i.e., detected regions without texts). A major limi-
tation of these approaches is that they are not easily
reproducible, as neither their code nor a benchmark
dataset is publicly available.
2.2 Deep Learning for PHI Detection
Since the rise of DL, most new PHI algorithms pri-
oritize text recognition (OCR) over text detection.
The objective is to reduce false positives while retain-
ing valuable non-PHI data, such as positional annota-
tions, as emphasized by the Canadian Association of
Radiologists (William et al., 2021). Recent advance-
ments differ in OCR model training, using either real-
world images (Vcelak et al., 2019) or a manually an-
notated private database of medical images (Monteiro
et al., 2017). In the latter methods, contour analysis
implemented by OpenCV (Bradski, 2000) is used for
the initial text detection.
This paper concentrates solely on text detection in
medical images, not on recognition. A plausible rea-
son for the prevalent focus on OCR might be the lack
of a comprehensive, openly accessible benchmark
dataset for burned-in text detection. Existing medi-
cal de-identification datasets, like those mentioned by
Rutherford et al. (Rutherford et al., 2021), are focused
on DICOM tags anonymization, while pre-existing
burned-in text is ignored, which highlights a lack of
resources for training PHI detection DL models.
2.3 Deep Learning for Generic Text
Detection
Contrary to PHI detection in medical images, the gen-
eral task of scene-text detection has received much
attention in recent years, leading to the development
of numerous model architectures, evaluation method-
ologies, and benchmark datasets. In turn, this large
amount of annotated data has enabled the devel-
opment of powerful DL-based methods for scene-
text detection. These models are often separated in
two main categories: region-proposal models such as
SSD (Liu et al., 2016), TextBoxes (Liao et al., 2017),
or DPText-DETR (Ye et al., 2022), and segmentation-
based models that perform prediction at a pixel-level,
such as EAST (Zhou et al., 2017). This work will fo-
cus on the four aforementioned models, as they are
all available as free and open-source software, which
enables an independent comparison of their applica-
bility to text detection in medical images, with and
without fine-tuning.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
298

The two first models, SSD (initially designed
for object detection) and TextBoxes (the extension
of SSD to text detection), operate through a single
pass of a deep Convolutional Neural Network (CNN),
which balances speed and accuracy by using multiple
convolutional layers to detect objects or texts at vari-
ous scales. A notable aspect of these models is their
use of “default boxes” of varying shapes and scales.
During inference, they assess the confidence score
(i.e., the likelihood of an anchor containing an object
or text), and the relative offset to adjust the anchor
to the specific object or text. As shown in Figure 1,
a non-maximal suppression step is performed to re-
move overlapping boxes. Such a technique is largely
used in object detection models, including EAST.
In contrast, segmentation-based models such as
EAST predict the text position at a pixel level. Sim-
ilarly to SSD, EAST uses CNN to detect text regions
directly, although without the need for anchors. Fi-
nally, recent architectures, like DPText-DETR, tend
to use Transformers-based architectures to predict
text regions with dynamic points, through an encoder-
decoder architecture. This work studies the impact
of fine-tuning the SSD and TextBoxes models on our
new benchmark dataset for text detection in medi-
cal images, and compares their performances against
more advanced generic scene-text detection models
(EAST and DPText-DETR) without fine-tuning.
2.4 Text Detection Evaluation Protocols
In addition to benchmark datasets for generic scene-
text detection, dedicated methodologies to evaluate
the performance of the text detection algorithms have
been proposed, with variations in the computation of
the precision (P), recall (R), and F-beta (F) scores.
The ICDAR and DetEval evaluation protocols are
nowadays widely accepted
1
.
2.4.1 ICDAR Detection Protocol
The ICDAR protocol defines the best match m(r,R)
for a rectangle r in a set of rectangles R as:
m(r,R) = max m
p
r,r
′
| r
′
∈ R (1)
In this formula, m
p
(r,r
′
) represents the match be-
tween two rectangles r and r
′
, calculated as their area
of intersection divided by the area of the minimum
bounding box containing both rectangles. Based on
this definition, P, R, and F scores can be computed as:
P =
∑
r
e
∈E
m(r
e
,T )
|E|
, R =
∑
r
t
∈T
m(r
t
,E)
|T |
, (2)
1
This paper directly uses the software scripts for IC-
DAR and DetEval that are published on the Robust Reading
Competition Website, with their default settings.
F =
1
α/P + (1 − α)/R
, (3)
where T (resp. E) represents the set of ground truth
(resp. estimated) rectangles, while r
t
(resp. r
e
) corre-
sponds to a ground truth (resp. estimated) rectangle.
In these definitions, α is a weighting parameter, which
is typically set to 0.5 in the official evaluation script.
2.4.2 DetEval Detection Protocol
The ICDAR protocol might not effectively handle
the one-to-many (i.e., splitting a single prediction
to match multiple targets) and many-to-one matches
(i.e., merging multiple predictions to match a single
target), which may lead to an underestimation of the
algorithm performance. The DetEval protocol was in-
troduced to incorporate area overlap and object-level
evaluation (Wolf et al., 2006).
In the context of the DetEval protocol, the metrics
of interest are P
′
and R
′
and are based on an analysis
of the “overlapping matrices”, where a non-zero value
at index (i, j) indicates an overlap between the detec-
tion D
i
and the ground truth G
j
(Liang et al., 1998):
P
′
=
∑
i
Match
D
(D
i
,G,t
r
,t
p
)
|D|
,
R
′
=
∑
j
Match
G
(G
j
,D,t
r
,t
p
)
|D|
,
(4)
In this definition, the Match
D
and Match
G
are func-
tions that consider the distinct types of matches. The
parameters t
r
and t
p
are thresholds that define the min-
imal area proportion of G
i
(resp. D
i
) that should over-
lap with ground truths (resp. predictions).
3 METHODS
As motivated by the discussions above, this sec-
tion first introduces a new, semi-synthetic benchmark
dataset for burned-in text detection in medical im-
ages. Secondly, adaptations to the generic SSD and
TextBoxes architectures are proposed to improve their
performances on medical images.
3.1 Dataset Generation
The dataset creation methodology was directly in-
spired by the SynthText (Gupta et al., 2016) and the
DICOM dataset (Rutherford et al., 2021) generators.
After selecting real medical images, random synthetic
text was generated and burned into the pixel data. Fig-
ure 2 depicts some examples from our dataset.
Open Platform for the De-identification of Burned-in Texts in Medical Images using Deep Learning
299
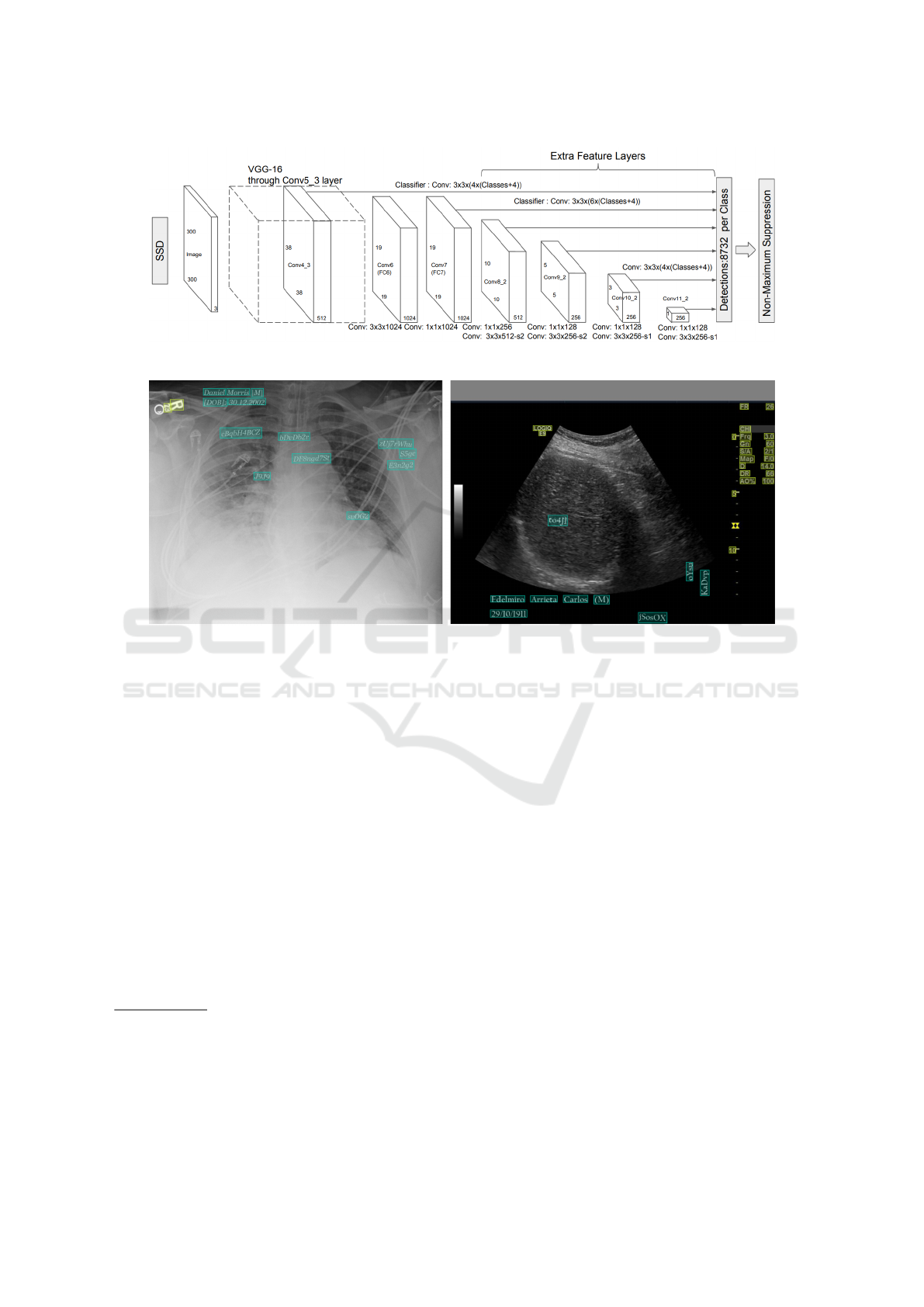
Figure 1: The SSD architecture (Liu et al., 2016).
Figure 2: Examples from our dataset. Cyan annotations represent synthetic text, while yellow ones denote pre-existing text.
3.1.1 Original Image Collection
The first step has been to collect a large variety of
real-world DICOM instances. To this end, the TCIA
project has been taken as a starting point (Clark
et al., 2013). A total of 1,944 images were selected
from various studies corresponding to medical imag-
ing modalities prone to having burned-in text, no-
tably digital radiography (DX), computed radiogra-
phy (CR), and medical ultrasound (US)
2
A conversion
process was undertaken to transcode the original DI-
COM format into the JPEG format, making medical
images easier to integrate to popular DL frameworks.
3.1.2 Training, Validation, and Test Subsets
Upon visual inspection of the 1,944 accumulated
images, 121 images have been identified as free
of burned-in texts, while 1,823 images contained
2
The following TCIA datasets were used: ACRIN
6667 (Lehman et al., 2007), ACRIN 6668 (Machtay et al.,
2013), B-mode-and-CEUS-Liver, COVID-19-AR (Desai
et al., 2020)(Jenjaroenpun et al., 2021), COVID-19-NY-
SBU, LIDC-IDRI (Armato et al., 2011), Pseudo-PHI-
DICOM-Data (Rutherford et al., 2021), RIDER Pilot,
TCGA-BLCA, TCGA-KIRC, and TCGA-UCEC.
burned-in text. From the latter, 276 images have been
manually selected to ensure a broad diversity of back-
grounds and modalities. These 397 images have been
randomly divided into training, validation, and test
sets with specific criteria: The test set includes 100
images with burned-in texts, the training set com-
prises 60% of both the remaining images with text
and those without text, and the validation set consists
of the remaining images.
3.1.3 Text Annotation
The bounding boxes of the already existing burned-in
texts in the 276 images have been manually annotated
using the online annotation tool makesense.ai. The
annotated regions were then exported using the popu-
lar COCO file format (Lin et al., 2014). In this format,
the boxes are stored as (x, y,w, h) tuples, where (x,y)
denotes the coordinates of the upper-left pixel, and w
(resp. h) denotes the width (resp. height) of the box.
3.1.4 Synthetic Text Generation
To augment the amount of data and the variety of
text fonts, sizes, colors, and locations, random syn-
thetic text has been added to the images in the train-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
300

Table 1: Statistics of the semi-synthetic dataset.
Training Validation Test
# background 177 120 100
# images 17 280 11 730 100
# texts 297 281 198 300 448
# synthetic (%) 85.20 85.14 0
ing and validation sets. Synthetic PHI text has also
been burned in the image, such as patient name, gen-
der, and date of birth, using the Python Faker library.
Both PHI and random texts have been placed at ran-
dom positions with various fonts, sizes, colors, and
orientations for diversity while ensuring that texts do
not overlap, do not exceed the image, and have a min-
imal contrast against the background. The test set has
not been modified to keep it as realistic and represen-
tative as possible. Table 1 provides statistics about the
resulting semi-synthetic dataset. The resulting dataset
is publicly available for download.
3.2 Refinements to Model Architectures
This section describes the modifications that have
been applied to the SSD and TextBoxes model archi-
tectures to better fit the specific aspects of burned-in
text detection in medical images compared to regular
scene-text detection.
3.2.1 Model Truncation
In the traditional object detection context, deeper lay-
ers of the neural network are responsible for detecting
larger-scale objects or text (cf. Figure 1). However, in
the context of medical images, large-scale texts never
occur. Consequently, the proposed architecture only
keeps the three first detection blocks and removes the
last ones. This modification led to slight improve-
ments in the performance metrics.
3.2.2 Modification of TextBoxes Anchors
In both models, each prediction layer is associated
with a set of default boxes or anchors, of various
shapes and sizes, that are carefully designed to deal
with specific tasks. TextBoxes is designed to focus
on horizontal texts: It has wider default boxes along
with a second row of boxes with a vertical offset that
targets paragraphs. However, experiments revealed
that this second row hindered text detection in med-
ical images, in which PHI rarely takes the form of
a paragraph. Removing these vertically offset boxes
from TextBoxes significantly improved performance
on medical images. This modification is specific to
TextBoxes, as offset boxes do not exist in SSD.
Table 2: Performance metrics of the DL models. The “T ”
superscript denotes the truncated version of the respective
models, as explained in Section 3.2.1.
Model Validation set Test set
ICDAR DetEval ICDAR DetEval
F1 (%) F1 (%) F1 (%) F1 (%)
SSD
T
512 92.54 92.75 84.70 85.83
SSD 512 92.91 93.09 82.33 83.95
TB
T
512 91.15 91.38 84.18 85.79
TB 512 91.11 91.37 82.34 83.60
EAST 9.25 8.26 64.77 65.95
DPText-DETR 47.62 47.79 78.19 79.25
4 RESULTS
This section illustrates the benefits of the developed
DL models, by analyzing their performance on the
validation and test sets of the newly developed bench-
mark database. A comparison against the regu-
lar scene-text detection models (EAST and DPText-
DETR) is then proposed to highlight the benefits of
fine-tuning the models on medical images. As a last
contribution, free and open-source tools implement-
ing the models are discussed and compared to existing
open and proprietary tools.
4.1 General Outcomes
The results shown in Table 2 illustrate the benefits of
adapting and fine-tuning the models on medical im-
ages. The original TextBoxes algorithm is not rep-
resented as its performance did not improve during
the fine-tuning, leading to an F-score lower than 1%.
Models have been fine-tuned for 45 epochs, with the
Adam optimizer (Diederik et al., 2017) with a learn-
ing rate of 0.001. Figure 3 shows predictions obtained
with the truncated version of SSD on real-world ex-
amples (i.e., without synthetic text) from our test set.
It is important to notice that EAST and DPText-
DETR have been taken as is from free and open-
source projects, and were only trained on manually
annotated detection datasets
3
. This is the reason
why these models perform worse than the SSD and
TextBoxes models after their fine-tuning on medical
images. Furthermore, the fact that they were trained
on manually annotated datasets, like ICDAR2019,
can explain the inferior performance on the semi-
synthetic validation set, as compared to our manually
annotated test set. Indeed, based on a complementary
3
The EAST model is available at https://github.com/
SakuraRiven/EAST, and DPText-DETR at https://github.
com/ymy-k/DPText-DETR
Open Platform for the De-identification of Burned-in Texts in Medical Images using Deep Learning
301
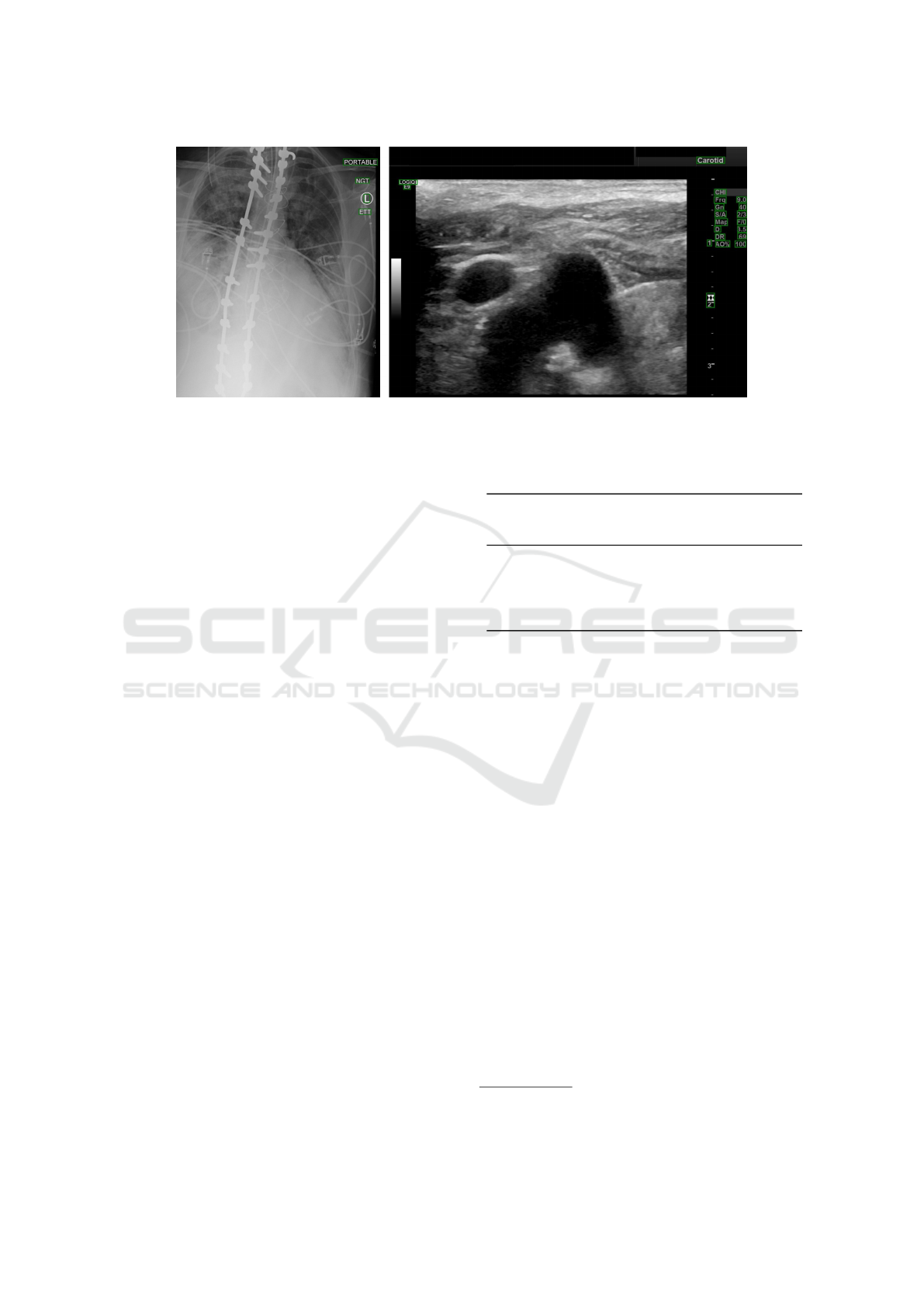
Figure 3: SSD
T
predictions on 2 samples from our test set.
evaluation, the Intersection-over-Union between syn-
thetic and predicted boxes rarely exceeds 60%, and is
thus evaluated as non-matching, lowering actual de-
tection performances. Nonetheless, the results of the
test set clearly illustrate the specificity of medical im-
ages as compared to world scene images, which calls
for the development of additional benchmark datasets
dedicated to the detection of texts burned in medical
images that could be used to fine-tune DL models.
4.2 Evaluation
The results shown in Table 2 have been obtained with
a confidence threshold of 0.2 for all models except
for SSD, for which the confidence threshold was set
to 0.35. The selection of an appropriate confidence
threshold is critical depending on the application, as
it defines the balance between precision and recall: A
higher threshold will filter out predictions with low
scores, which increases the model precision, at the
cost of a lower recall. Considering the sensitive nature
of patient data, the focus should be on maximizing the
recall, ensuring that most of the PHI text is detected.
This therefore reduces the risk of leaking patient in-
formation during the de-identification process, at the
cost of increasing false positives.
Table 3 depicts the impact of this threshold on
the DetEval performance metrics for our best model,
which justifies the choice of the 0.2 threshold as a
good balance between precision and recall. Figure 4
compares the precision and the recall for the tested
models and for the various thresholds of the best
model, respectively. Since all conditions do not fol-
low a normal distribution (all conditions did not pass a
Kolmogorov–Smirnov test with p ≤ 0.0001), a Fried-
man non-parametric one-way analysis of variance for
matched images with Dunn’s correction for multiple
comparisons for both precision and recall revealed a
Table 3: DetEval metrics at varying confidence thresholds
for the SSD truncate model. A comparable trend is observed
under the ICDAR protocol metrics.
Confidence
threshold
(%)
Precision (%) Recall (%) F1 (%)
10 71.98 85.27 78.07
20 88.98 82.90 85.83
30 90.44 81.56 85.77
40 91.18 78.93 84.61
50 91.90 77.86 84.30
significant difference between our fine-tuned models
and generic pre-trained models, but not among thresh-
olds of the best model. For both precision (F = 31.73)
and recall (F = 217.8), we obtained p < 0.0001
∗∗∗∗
for n = 100 images, n
T
= 6. The level of significance
between conditions is represented as follows: p <
0.05
∗
, p < 0.01
∗∗
, p < 0.001
∗∗∗
, and p < 0.0001
∗∗∗∗
.
4.3 Software
One of the main concerns of this work is to make
patient confidentiality more accurate and accessible,
with an open-science perspective. To this end, in
addition to the benchmark dataset described in Sec-
tion 3.1.4, the weights of the DL models are available
as open data alongside the new benchmark dataset.
Furthermore, to make the models easily applicable on
real-world medical images, two new free and open-
source tools are released
4
.
Existing Tools. Some de-identification tools are al-
ready available, such as Microsoft Presidio Image
Redactor and Google Healthcare API. Presidio, still
in beta as of writing, is an open-source, configurable
4
Their code is available at: https://forge.uclouvain.be/
QuentinLanglois/medical-image-de-identification-tools
BIOIMAGING 2024 - 11th International Conference on Bioimaging
302
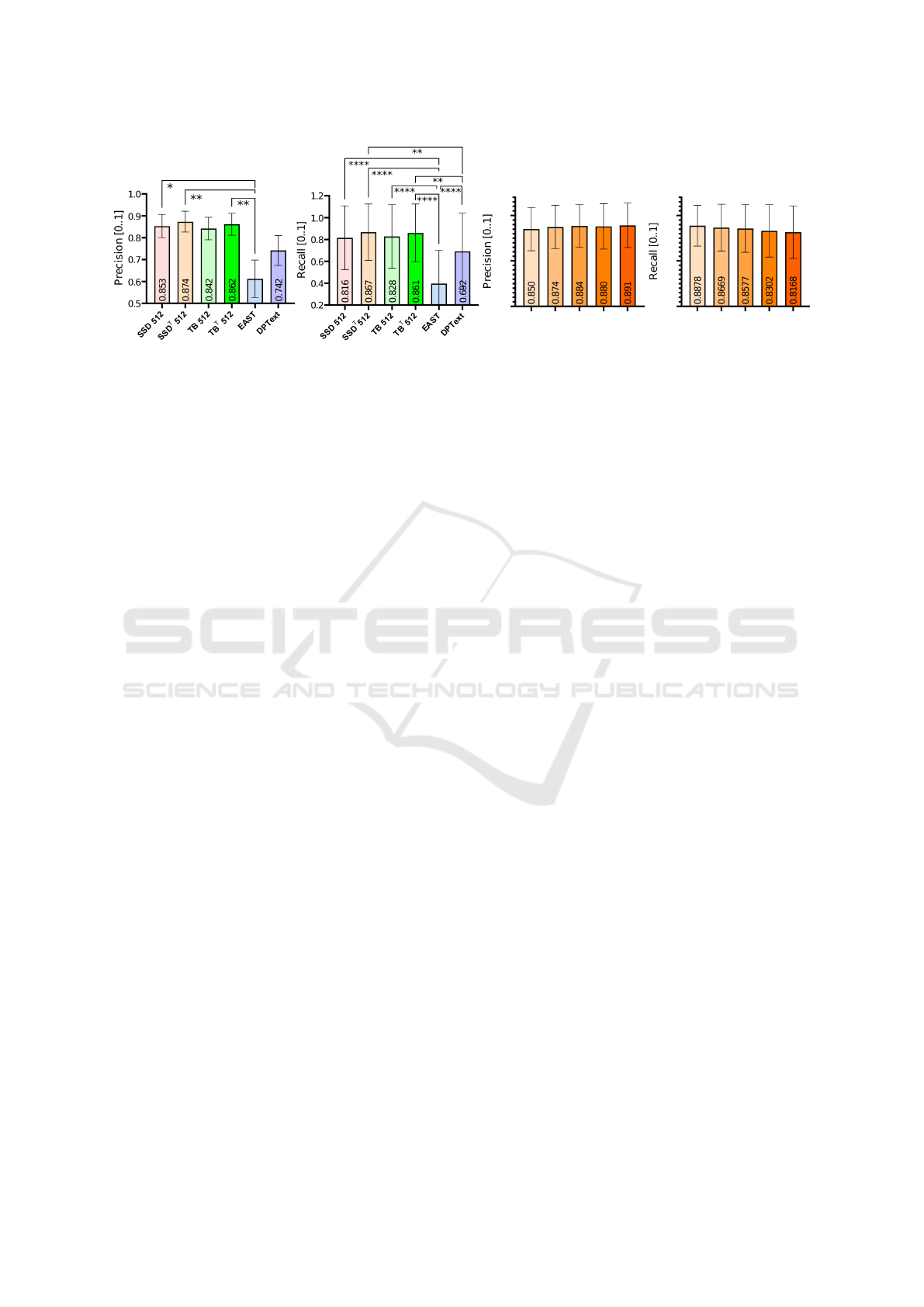
Figure 4: P and R rates of the models (1-2) and of the best model thresholds (3-4). Error bars show a confidence interval of
95%.
tool that detects and redacts PHI in medical images
using the Tesseract OCR framework. Its main limi-
tation is the efficiency on medical images, hindered
by a lack of dedicated fine-tuning. Google Health-
care API, in contrast, is a proprietary solution that
offers extensive documentation and allows for exten-
sive customization in the de-identification process.
However, it incurs significant operational costs and
its closed-source nature restricts its reproducibility.
Proposed Tools. To address these limitations, we
introduce a new free and open-source software for
burned-in text removal. Our tools do not rely on
OCR for non-PHI filtering and remove all detected
texts, which is generally preferred to avoid sensi-
tive information leaks. Future enhancements may
integrate OCR to overcome this limitation. The
command-line tool offers an efficient solution for
the batch de-identification of folders containing im-
ages in various file formats (JPEG, PNG, DICOM).
The browser-based tool leverages Orthanc, a free,
lightweight, standalone DICOM server with a REST-
ful API (Jodogne et al., 2013). Our MedTextCleaner
(MTC) is an interactive Web application, developed
as an Orthanc plugin. It uses Python and Vue.js
framework for automated text detection, allowing
users to validate and manually edit as needed. A de-
tailed demonstration video is available.
5 CONCLUSION
The contributions of this paper are threefold. Firstly,
a new semi-synthetic dataset dedicated to the detec-
tion and removal of texts burned in medical images is
published in open access. This new dataset will hope-
fully provide a benchmark to foster further research
on DL applied to the task of image de-identification.
Secondly, it has been shown that generic DL models
for scene text detection can be fine-tuned on medical
images, leading to vastly improved performance. This
approach contrasts with recent work, which tends to
focus on OCR techniques instead of text detection
methods. Finally, pre-trained models are released
as open data, together with free and open-source
software implementing a platform for text detection
and removal in medical images. The latter contribu-
tion subscribes to the open-science paradigm by pro-
moting transparency, reproducibility, and knowledge
sharing for the de-identification of medical images.
This work is specifically focused on the improve-
ment of the text detection techniques, without any fil-
tering of the detected texts using OCR. Nevertheless,
such filtering may be useful to keep relevant informa-
tion, such as location tags or medical results, which
may be relevant for some applications. Consequently,
future work will explore the combination of the de-
veloped DL models with an OCR engine. The new
benchmark database will also be used to assess the
performance of more medical image de-identification
algorithms, whether based on Deep Learning or on
more traditional computer vision approaches.
ACKNOWLEDGMENTS
The results published are in part based on data gen-
erated by the TCGA Research Network. The au-
thors acknowledge the National Cancer Institute and
the Foundation for the National Institutes of Health,
and their critical role in the creation of the free
publicly available LIDC/IDRI Database used in this
study. Computational resources have been provided
by the Consortium des
´
Equipements de Calcul In-
tensif (C
´
ECI), funded by the Fonds de la Recherche
Scientifique de Belgique (F.R.S.-FNRS) under Grant
No. 2.5020.11 and by the Walloon Region. Quentin
Langlois is funded by the Walloon Region through a
F.R.S.-FNRS FRIA (Fund for Research training in In-
dustry and Agriculture) grant.
Open Platform for the De-identification of Burned-in Texts in Medical Images using Deep Learning
303

REFERENCES
Armato, S. G. et al. (2011). The lung image database con-
sortium (LIDC) and image database resource initiative
(IDRI): A completed reference database of lung nod-
ules on CT scans. Medical Physics, 38(2):915–931.
Bakas, S. et al. (2018). Identifying the best machine learn-
ing algorithms for brain tumor segmentation, progres-
sion assessment, and overall survival prediction in the
BRATS challenge. ArXiv, abs/1811.02629.
Bradski, G. (2000). The OpenCV Library. Dr. Dobb’s Jour-
nal of Software Tools.
Clark, K. et al. (2013). The Cancer Imaging Archive
(TCIA): Maintaining and operating a public reposi-
tory. Journal of Digital Imaging, 26(6):1045–1057.
Desai, S. et al. (2020). Chest imaging representing a
COVID-19 positive rural u.s. population. Scientific
Data, 7(1).
Diederik, P. et al. (2017). Adam: A method for stochastic
optimization.
Gupta, A. et al. (2016). Synthetic data for text localisation
in natural images. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2315–2324.
Jenjaroenpun, P. et al. (2021). Two SARS-CoV-2 genome
sequences of isolates from rural U.S. patients harbor-
ing the D614G mutation, obtained using nanopore se-
quencing. Microbiology Resource Announcements,
10(1):10.1128/mra.01109–20.
Jodogne, S. (2018). The Orthanc ecosystem for medical
imaging. Journal of Digital Imaging, 31(3):341–352.
Jodogne, S. et al. (2013). Orthanc - a lightweight, restful DI-
COM server for healthcare and medical research. In
2013 IEEE 10th International Symposium on Biomed-
ical Imaging, pages 190–193. IEEE.
Johnson, A. et al. (2020). Deidentification of free-text med-
ical records using pre-trained bidirectional transform-
ers. In Proceedings of the ACM Conference on Health,
Inference, and Learning, CHIL ’20, page 214–221,
New York, NY, USA. Association for Computing Ma-
chinery.
Lehman, C. D. et al. (2007). MRI evaluation of the con-
tralateral breast in women with recently diagnosed
breast cancer. New England Journal of Medicine,
356(13):1295–1303.
Liang, J. et al. (1998). Performance evaluation of docu-
ment layout analysis algorithms on the UW data set.
Proceedings of SPIE - The International Society for
Optical Engineering.
Liao, M. et al. (2017). TextBoxes: A fast text detector with
a single deep neural network. Proceedings of the AAAI
Conference on Artificial Intelligence, 31(1).
Lin, T.-Y. et al. (2014). Microsoft COCO: Common objects
in context. In Computer Vision–ECCV 2014: 13th Eu-
ropean Conference, Zurich, Switzerland, September
6-12, 2014, Proceedings, Part V 13, pages 740–755.
Springer.
Liu, W. et al. (2016). SSD: Single shot multibox detec-
tor. In Computer Vision–ECCV 2016: 14th Euro-
pean Conference, Amsterdam, The Netherlands, Octo-
ber 11–14, 2016, Proceedings, Part I 14, pages 21–37.
Springer.
Lucas, S. et al. (2003). ICDAR 2003 robust reading compe-
titions. In Seventh International Conference on Doc-
ument Analysis and Recognition, 2003. Proceedings.,
pages 682–687.
Machtay, M. et al. (2013). Prediction of survival by
[18F]Fluorodeoxyglucose positron emission tomog-
raphy in patients with locally advanced non–small-
cell lung cancer undergoing definitive chemoradia-
tion therapy: Results of the ACRIN 6668/RTOG 0235
trial. Journal of Clinical Oncology, 31(30):3823–
3830.
Monteiro, E. et al. (2017). A de-identification pipeline for
ultrasound medical images in DICOM format. J. Med.
Syst., 41(5):1–16.
Newhauser, W. et al. (2014). Anonymization of DICOM
electronic medical records for radiation therapy. Com-
puters in biology and medicine, 53:134—140.
Rutherford, M. et al. (2021). A DICOM dataset for eval-
uation of medical image de-identification. Scientific
Data, 8(1).
Vcelak, P. et al. (2019). Identification and classification
of DICOM files with burned-in text content. Interna-
tional Journal of Medical Informatics, 126:128–137.
Vithya, Y. et al. (2020). A review of automatic end-to-end
de-identification: Is high accuracy the only metric?
Applied Artificial Intelligence, 34(3):251–269.
Wang, J. Z. et al. (1997). A textual information detection
and elimination system for secure medical image dis-
tribution. Proceedings : a conference of the American
Medical Informatics Association. AMIA Fall Sympo-
sium, page 896—896.
Wasserthal, J. et al. (2023). TotalSegmentator: Robust seg-
mentation of 104 anatomic structures in CT images.
Radiology: Artificial Intelligence, 5(5).
William, P. et al. (2021). Canadian association of radi-
ologists white paper on de-identification of medical
imaging: Part 1, general principles. Canadian Asso-
ciation of Radiologists Journal, 72(1):13–24. PMID:
33138621.
Wolf, C. et al. (2006). Object count/area graphs for the
evaluation of object detection and segmentation algo-
rithms. International Journal of Document Analysis
and Recognition (IJDAR), 8:280–296.
Ye, M. et al. (2022). DPText-DETR: Towards better scene
text detection with dynamic points in transformer.
Yingxuan, Z. et al. (2010). An automatic system to
detect and extract texts in medical images for de-
identification. In Liu, B. J. and Boonn, W. W., editors,
Medical Imaging 2010: Advanced PACS-based Imag-
ing Informatics and Therapeutic Applications, volume
7628, page 762803. International Society for Optics
and Photonics, SPIE.
Zhou, X. et al. (2017). EAST: an efficient and accurate
scene text detector. In Proceedings of the IEEE con-
ference on Computer Vision and Pattern Recognition,
pages 5551–5560.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
304
