
Semantic Textual Similarity Assessment in Chest X-ray Reports Using a
Domain-Specific Cosine-Based Metric
Sayeh Gholipour Picha
a
, Dawood Al Chanti
b
and Alice Caplier
c
Univ. Grenoble Alpes, CNRS, Grenoble INP, GIPSA-lab, 38000 Grenoble, France
Keywords:
Semantic Similarity, Medical Language Processing, Biomedical Metric.
Abstract:
Medical language processing and deep learning techniques have emerged as critical tools for improving health-
care, particularly in the analysis of medical imaging and medical text data. These multimodal data fusion
techniques help to improve the interpretation of medical imaging and lead to increased diagnostic accuracy,
informed clinical decisions, and improved patient outcomes. The success of these models relies on the ability
to extract and consolidate semantic information from clinical text. This paper addresses the need for more
robust methods to evaluate the semantic content of medical reports. Conventional natural language process-
ing approaches and metrics are initially designed for considering the semantic context in the natural language
domain and machine translation, often failing to capture the complex semantic meanings inherent in medical
content. In this study, we introduce a novel approach designed specifically for assessing the semantic simi-
larity between generated medical reports and the ground truth. Our approach is validated, demonstrating its
efficiency in assessing domain-specific semantic similarity within medical contexts. By applying our metric to
state-of-the-art Chest X-ray report generation models, we obtain results that not only align with conventional
metrics but also provide more contextually meaningful scores in the considered medical domain.
1 INTRODUCTION
Advancements in deep learning for medical language
processing have significantly improved healthcare
clinical analysis, particularly in the domain of med-
ical imaging applications. Notably, there has been
substantial progress in generating chest X-ray reports
comparable to those written by radiologists. How-
ever, a critical challenge persists in the chest X-ray ap-
plication—assessing the semantic similarity between
generated reports and the ground truth.
Identifying semantic similarities in medical texts
is a difficult task within the language processing do-
main (Alam et al., 2020). This task necessitates a
comprehensive grasp of the entire medical text cor-
pus, the ability to recognize key content, and a pro-
found understanding of the semantic relationships
between these critical keywords at an expert level.
While existing metrics and approaches for capturing
semantic similarity in natural language are effective,
they are not designed for the complexities of medical
a
https://orcid.org/0000-0003-2675-5463
b
https://orcid.org/0000-0002-6258-6970
c
https://orcid.org/0000-0002-5937-4627
content. The need for a robust metric to assess seman-
tic similarity in medical texts has become increasingly
evident, particularly in applications like chest X-ray
report generation, and continues to be an active area
of research (Endo et al., 2021), (Miura et al., 2021),
(Yu et al., 2022).
State-of-the-art chest X-ray report generation
models (Chen et al., 2020), (Miura et al., 2021),
(Endo et al., 2021) still rely on conventional Natural
Language Processing (NLP) methods like BLEU (Pa-
pineni et al., 2002), METEOR (Banerjee and Lavie,
2005), and ROUGE (Lin, 2004) to evaluate the gen-
erated reports against ground truth references. How-
ever, these metrics produce unreliable results due to
their inability to comprehend and compare the seman-
tic similarity of key medical terms. A medical seman-
tic similarity metric would not only provide more sig-
nificant evaluation scores but could also be incorpo-
rated into the training process to improve model per-
formance, potentially leading to enhanced diagnostic
accuracy and decision-making. Additionally, as part
of our ongoing research, our goal is to focus on pro-
viding visual interpretations of chest X-ray reports us-
ing text-to-image localization. As a consequence, a
robust semantic similarity evaluation metric suitable
Gholipour Picha, S., Al Chanti, D. and Caplier, A.
Semantic Textual Similarity Assessment in Chest X-ray Reports Using a Domain-Specific Cosine-Based Metric.
DOI: 10.5220/0012429600003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 487-494
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
487

for medical content will ensure the reliability of gen-
erated reports and will enable us to achieve more ac-
curate localization and interpretation of image con-
tent.
In this context, we propose a new metric designed
to assess and assign scores about the semantic simi-
larity of medical texts. Our metric consists of two se-
quential steps: first, we identify the primary clinical
entities, and subsequently, we evaluate the similarity
between these entities using the domain-specific Co-
sine similarity score. Notably, our approach considers
the presence of negations and detailed descriptions as-
sociated with medical entities during the evaluation
process. To this end, our contributions include:
• Introduction of a novel system for clinical entity
extraction from medical texts.
• Proposition of a new scoring system for the evalu-
ation of semantic similarity that suits medical and
natural texts.
• Presentation of a validation method for scoring
verification.
This paper is structured as follows: Section 2 dis-
cusses related works; Section 3 presents the theoreti-
cal and mathematical part of the novel metric; Section
4 validates the metric; Section 5 discusses the results;
Finally, Section 6 concludes the paper.
2 RELATED WORKS
Recent studies have addressed the challenge of sim-
ilarity evaluation between generated medical reports
and the ground truth through various approaches other
than conventional NLP metrics. Researchers have of-
ten introduced innovative metrics in the process.
In the CXR-RePaiR model by Endo et al. (Endo
et al., 2021) a unique approach for automatically
evaluating chest X-ray report generation is proposed
by introducing the CheXbert vector similarity met-
ric, using the CheXbert labeler (Smit et al., 2020)
— a specialized tool for chest X-ray report labeling.
The process involves extracting labels from gener-
ated reports, comparing them with ground truth la-
bels, and presenting the final score using cosine sim-
ilarity. While this approach outperforms the BLEU
metric, its applicability is limited to the specific con-
text of chest X-ray reports and does not readily extend
to other medical applications. The limitations arise
from Chexbert being exclusively trained for chest X-
ray reports. Moreover, the Chexpert labels (Irvin
et al., 2019) (Atelectasis, Cardiomegaly, Consolida-
tion, Edema, Enlarged Cardiomediastinum, Fracture,
Lung Lesion, Lung Opacity, No Finding, Pleural Ef-
fusion, Pleural Other, Pneumonia, Pneumothorax) are
specific to the chest X-ray dataset, further limiting the
generalizability of the approach to other medical con-
texts.
In a separate study, Yu et al. (Yu et al., 2022) in-
troduced a novel metric targeting the quantification of
overlap of clinical entities between ground truth and
generated reports in chest X-ray report generation.
They use the RadGraph model (Jain et al., 2021), a
language model trained on a limited subset of reports
from the MIMIC-CXR dataset (Johnson et al., 2019).
The MIMIC-CXR dataset consists of chest X-ray im-
ages with corresponding reports, and the RadGraph
dataset includes medical entities from chest X-ray re-
ports annotated by radiologists. The approach by Yu
et al. is similar to the BLEU score, exclusively con-
sidering the exact matches among the primary entities
in generated and ground truth reports, overlooking
the semantic similarity of these entities. Furthermore,
the generalizability of this approach to other medical
applications is constrained by the RadGraph model’s
specialization in extracting only chest X-ray related
entities. Nonetheless, while the RadGraph model ac-
knowledges negations in the texts, they are treated
merely as labels to the entities, and the details of en-
tity descriptions are not factored into the evaluation
process.
In a recent study, Patricoski et al. (Patricoski et al.,
2022) conducted an evaluation of seven BERT mod-
els to assess semantic similarity in clinical trial texts.
Notably, the pre-trained BERT model known as SciB-
ERT (Beltagy et al., 2019) demonstrated better per-
formance compared to the other BERT models, even
outperforming the standard BERT model, which se-
cured the second position in this evaluation. This
study underlines the promising potential of BERT
models in semantic similarity evaluation. However,
it has a drawback associated with using BERT mod-
els without preprocessing. BERT models operate at
a token-by-token level, evaluating semantic similarity
by comparing all tokens with each other, a compu-
tationally intensive process that gives relatively low
scores. Despite this computational challenge, it is im-
portant to consider the significant potential in SciB-
ERT, particularly due to its huge clinical dictionary.
This finding underscores the need for careful consid-
eration of preprocessing strategies to maximize the
effectiveness of BERT models in semantic similarity
evaluations.
Notably, the absence of a comprehensive, general
semantic similarity evaluation metric for medical con-
tent persists. Consequently, we introduce a novel met-
ric for Medical Corpus Similarity Evaluation (MCSE)
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
488

to comprehensively address and resolve these chal-
lenges.
3 METHODOLOGY
We developed a novel metric for Medical Corpus
Similarity Evaluation (MCSE), by exclusively ex-
tracting key medical entities and employing a pre-
trained BERT model to assess the semantic similarity
of these entities within chest X-ray reports. This tar-
geted approach allows BERT to concentrate solely on
important information and reduces the computational
load during comparison. Importantly, our method-
ology goes beyond extracting main entities, we also
consider the negations and detailed descriptions asso-
ciated with the primary medical entities in chest X-ray
reports. Our MCSE metric consists of two essential
steps:
1. Clinical Entity Extraction.
2. Domain Similarity Evaluation.
3.1 Clinical Entity Extraction
The most important part of comprehending seman-
tic similarity evaluation in text relies on identifying
the key elements, often referred to as clinical entities,
within medical texts. These entities typically fall into
categories related to anatomical body parts, symp-
toms, laboratory equipment, and diagnoses. Each cat-
egory is typically signaled by certain words within a
sentence. However, there are additional words that
precede or follow these main entities, offering de-
scriptions.
To address these complexities, we employ the
Scispacy model (Neumann et al., 2019) for extract-
ing primary clinical entities from medical text us-
ing the embedded clinical dictionary in this model
(BC5CDR: a corpus comprising 1500 PubMed arti-
cles with 4409 annotated chemicals, 5818 diseases,
and 3116 chemical-disease interactions (Li et al.,
2016)). Subsequently, we automatically process the
entire text to identify associated negations and adjec-
tives related to these key entities. These elements
are then integrated to provide a comprehensive rep-
resentation of the considered text. In the context of
this research, the category of laboratory equipment is
deliberately excluded, aligning with the specific fo-
cus of our application. Table 1 presents an example
of medical text and the extracted entities using our
method and the Scispacy method without any clean-
ing process. While we employ the Scispacy model for
initial entity extraction, it is evident that this model
alone may not suffice. An additional automated post-
processing step is needed to refine and integrate re-
lated entities. The post-processing steps involve elim-
inating a single adjective or non-medical entities, ex-
cluding entities categorized as lab equipment, identi-
fying and adding the relevant adjective to the remain-
ing medical entities, including the existing negation
into these primary entities, and screening out any re-
ported diagnostic procedures terms. These processes
are essential to ensure that the final output is presented
as a cohesive set of primary medical entities, ready for
practical use.
3.2 Domain Similarity Evaluation
Having successfully extracted and shifted our focus
to the primary entities within the medical corpus, the
next step involves assessing their semantic similarities
by assigning corresponding scores.
After processing entity extraction, we calculate
a similarity score for the sequences of entities. Let
T = (t
1
, . . . , t
N
) represent the reference text entities
and
ˆ
T = (
ˆ
t
1
, . . . ,
ˆ
t
M
) represent the generated text or
candidate text entities. Initially, we identify the ex-
act same medical entities in both sequences and de-
termine the total count (|C
(i)
|). For the remaining en-
tities, we construct a similarity matrix, where each el-
ement represents the similarity score between entities,
as illustrated in table 2.
S
i
=
maxy
i, j
maxy
i, j
+ y
i, j
i = (0, 1, . . . , M) j = (0, 1, . . . , N)
(1)
y
i, j
= Similarity(r
i
, ˆr
j
) (2)
(
C
(i)
= t
i
, if t
i
=
ˆ
t
j
r
i
= t
i
& ˆr
j
=
ˆ
t
j
if t
i
̸=
ˆ
t
j
(3)
Where M is the number of total candidate entities,
r
i
and ˆr
j
are the sequence between no matched en-
tities as in equation (3), and S
i
is a normalized
similarity score between r
i
and ˆr
j
. The similarity
score Similarity(r
i
, ˆr
j
) in equation (2) is derived from
spaCy (Honnibal et al., 2020), a BERT model trained
on word2vec, to evaluate similarities using domain
cosine similarity.
To evaluate the similarity of candidate entities
with the reference entities, we compute the maximum
score for each column and normalized it with the col-
umn’s average (S
i
). We then sum these scores for each
column, adding them to |C
(i)
|. To obtain the final sim-
ilarity score between the two corpora, we divide this
sum by the total number of candidate entities. This
process is explained in Equation (4).
Semantic Textual Similarity Assessment in Chest X-ray Reports Using a Domain-Specific Cosine-Based Metric
489
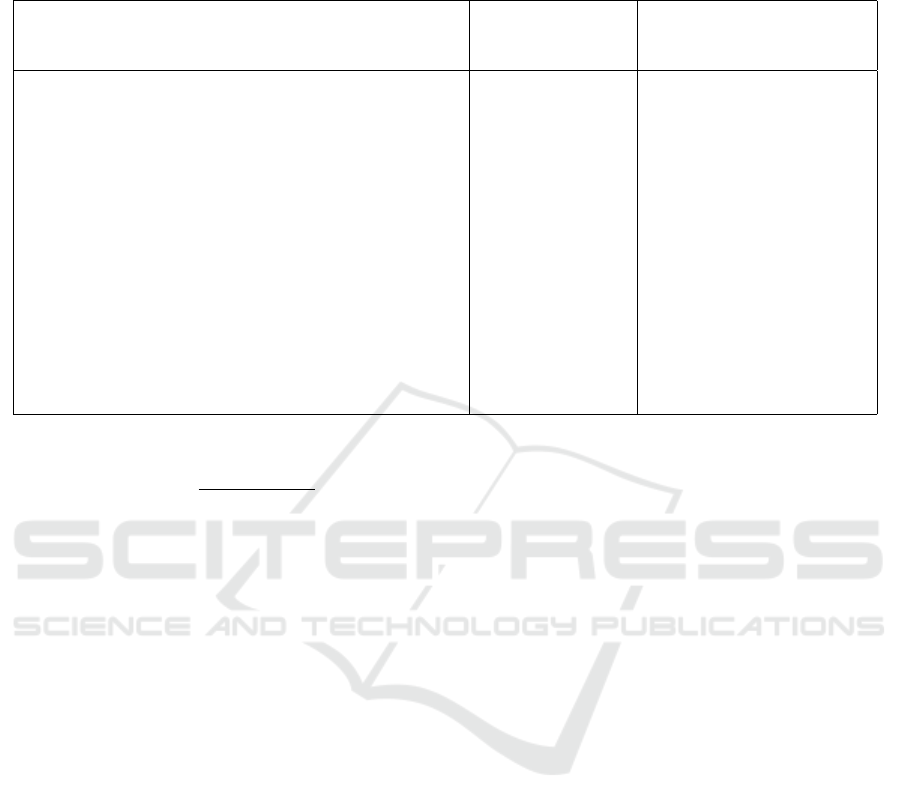
Table 1: In the right column there is an example of medical text. In the left column, there are clinical entities extracted using
the Scispacy model without any cleaning process, and In the middle column, there are clinical entities extracted using our
method.
Medical Text Extracted Entities
using our method
Extracted Entities using
Scispacy (Neumann et al.,
2019)
1. Interval clearance of left basilar consolidation. 2.
Patchy right basilar opacities, which could be seen
with minor atelectasis, but given the context clinical
correlation is suggested regarding any possibility for
recurrent or new aspiration pneumonitis at the right
lung base. 3. Increased new interstitial abnormal-
ity, suggesting recurrence of fluid overload or mild-to-
moderate pulmonary edema; aspiration could also be
considered. Inflammation associated with atypical in-
fectious process is probably less likely given the wax-
ing and waning presentation.
fluid overload,
inflammation,
aspiration pneu-
monitis, minor
atelectasis, mild
to moderate pul-
monary edema,
left basilar con-
solidation, patchy
right basilar opac-
ities, interstitial
abnormality
Interval, clearance, left
basilar, consolidation,
Patchy, right basilar,
opacities, minor, atelec-
tasis, clinical, recurrent,
aspiration, pneumonitis,
right lung base, Increased,
interstitial abnormality,
recurrence, fluid, overload,
mild-to-moderate pul-
monary edema, aspiration,
Inflammation, associated
with, atypical, infectious
process, waxing, waning,
presentation
MCSE :=
|C
(i)
| +
∑
M
i=1
S
i
M
(4)
Where |C
(i)
| is the number of exactly matched en-
tities between the two corpora of T and
ˆ
T .
For instance, Table 2 provides an example of the
probable similarity score that two sets of entities can
receive. These entities have been extracted using our
medical entity extraction procedure.
In the table, the two corpora received a score of
0.55 according to our MCSE metric. However, the
calculated BLEU score for them is approximately
zero. Upon analyzing the two medical texts, it be-
comes evident that although the candidate text does
refer to the same side of the chest as in the reference
text and that both texts indicate the presence of pul-
monary edema and pulmonary masses, their overall
similarity is relatively limited. The score of 0.55 car-
ries a more meaningful value in this context compared
to the nearly zero score generated by BLEU.
4 VALIDATION
While the underlying logic of this metric is reason-
able, it is imperative that we validate the results ro-
bustly. Given the use of chest X-ray reports for this
particular application, we have conducted an exten-
sive search within existing datasets to identify an ap-
propriate validation method. After a comprehensive
review of various datasets, we concluded that it would
be more effective to conduct separate validations for
the different steps of the proposed metric.
4.1 Clinical Entity Extraction Process
In order to rigorously validate our clinical entity ex-
traction process, we employ the RadGraph dataset
(Jain et al., 2021). This dataset is a valuable resource
in which radiologists thoroughly annotated the pri-
mary clinical entities in chest X-ray reports as either
”definitely present” within the report or ”definitely
absent”. Importantly, in cases where a negation is
associated with a particular entity, it is annotated as
”definitely absent.”
To achieve our validation objectives, we executed
our entity extraction process on the reports within this
dataset. Subsequently, we compare the number of
similar entities extracted through our method with the
annotations provided by radiologists, particularly fo-
cusing on the two categories of ”definitely present”
and ”definitely absent”. This systematic comparison
allows us to assess the accuracy and effectiveness of
our clinical entity extraction methodology in the con-
text of chest X-ray reports, aligning with radiological
standards. Throughout the validation process, cover-
ing all reports in our study, our method consistently
achieves a high level of accuracy. On average, it ac-
curately recognizes 75% of entities marked as ”defi-
nitely present” and successfully identifies 76% of en-
tities labeled as ”definitely absent”. In our entity ex-
traction process, we deliberately omit anatomical en-
tities like ”chest” or ”lung,” as they are redundant to
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
490
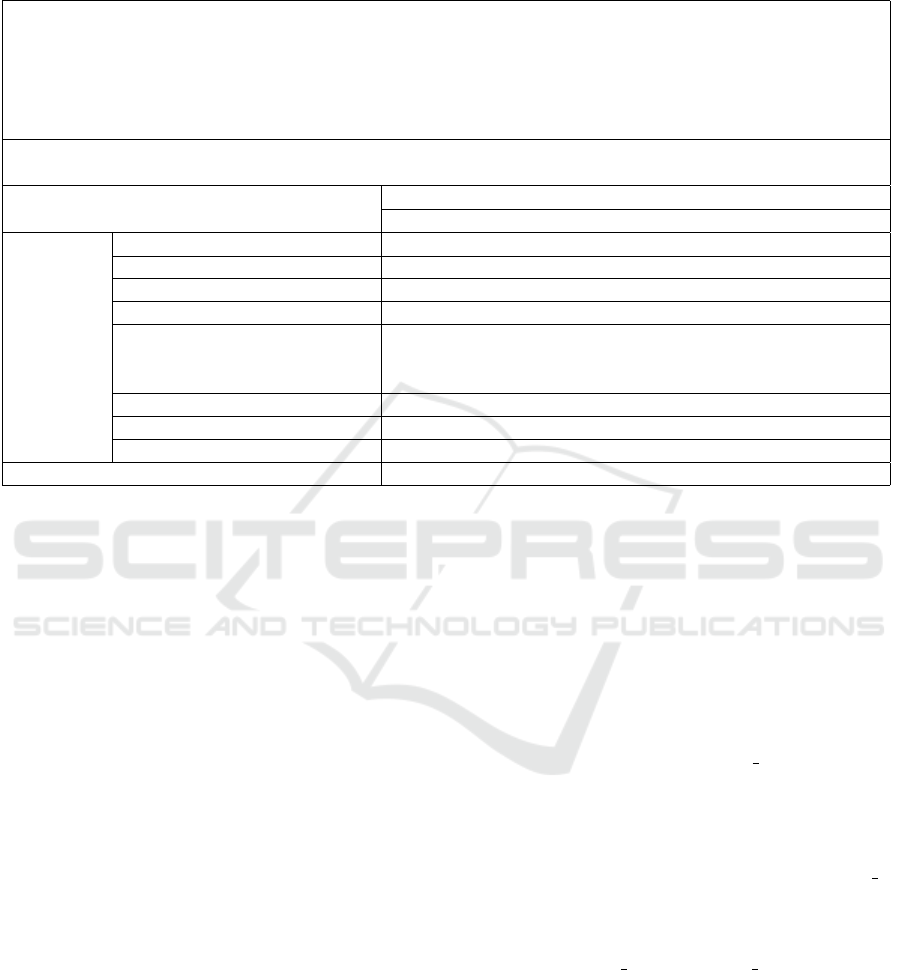
Table 2: An example of a medical similarity score between entities. Each score is calculated from equation (2), with the final
row S
i
being computed using equation (1). The scores highlighted in blue indicate the maximum value within each respective
column.
Reference: 1. Interval clearance of left basilar consolidation. 2. Patchy right basilar opacities, which
could be seen with minor atelectasis, but given the context clinical correlation is suggested regarding any
possibility for recurrent or new aspiration pneumonitis at the right lung base. 3. Increased new interstitial
abnormality, suggesting recurrence of fluid overload or mild-to-moderate pulmonary edema; aspiration could
also be considered. Inflammation associated with atypical infectious process is probably less likely given the
waxing and waning presentation.
Candidate: Stable multiple bilateral pulmonary masses and right middle lobe collapse due to hilar adenopa-
thy.
Candidate Medical Entities
pulmonary masses right middle lobe hilar adenopathy
fluid overload 0.61 0.49 0.45
inflammation 0.64 0.48 0.55
aspiration pneumonitis 0.65 0.39 0.50
minor atelectasis 0.62 0.47 0.53
Reference
Medical
Entities
mild to moderate pulmonary
edema
0.78 0.31 0.51
left basilar consolidation 0.52 0.66 0.32
patchy right basilar opacities 0.64 0.66 0.49
interstitial abnormality 0.69 0.63 0.59
S
i
0.548 0.563 0.545
the chest X-ray application and do not contribute sig-
nificantly to the process. This selective exclusion is
one of the factors contributing to the approximately
75% accuracy in our results. Nevertheless, these
results affirm the reliability and consistency of our
methodology.
4.2 Domain Similarity Score
In contrast to the initial phase of clinical entity extrac-
tion, validating the domain similarity score is more
challenging. The scoring system itself is more con-
troversial and subject to debate, and creating an au-
tomated validation method, free from reliance on ra-
diologists, necessitates a creative and innovative ap-
proach. Nevertheless, through the available tools and
databases, we establish a dedicated system for the val-
idation of this scoring method for the application of
chest X-rays.
In the chest X-ray application, the MIMIC-CXR
dataset (Johnson et al., 2019), is one of the biggest
available databases for chest X-ray images and their
corresponding reports. Notably, this dataset pro-
vides us with Chexpert labels (Medical Observation),
including Atelectasis, Cardiomegaly, Consolidation,
Edema, Enlarged Cardiomediastinum, Fracture, Lung
Lesion, Lung Opacity, No Finding, Pleural Effusion,
Pleural Other, Pneumonia, Pneumothorax, and Sup-
port Devices labels (Irvin et al., 2019). The values
of each label are 1 (definitely present), 0 (definitely
absent), -1 (ambiguous), or it carries no value at all.
Table 3 presents a sample of Chexpert labels extracted
from chest X-ray reports of five patients from the
MIMIC-CXR database. The reports corresponding to
these subjects are presented in Table 4.
Our approach involves two distinct strategies.
Firstly, we seek to identify reports sharing the same
sequence of labels and values. For instance, we
search for reports from subjects with Chexpert label
sequences similar to that of Subject 01 in Table 3. For
these reports with matching label sequences, we pro-
ceed to similarity scores computation for each pair of
reports. Simultaneously, we identify reports featur-
ing only one or two labels and with a value of ”defi-
nitely present” for these labels resembling Subject 02
in Table 3 and assess the similarity of these reports
with the reports with different label sequences. As
an example, we calculate the similarity between the
reports of Subject 02 and Subject 05 from Table 3,
given their entirely distinct label sequences. This two-
fold method allows us to analyze the semantic simi-
larity scores for both similar and contrasting reports
in terms of their labels.
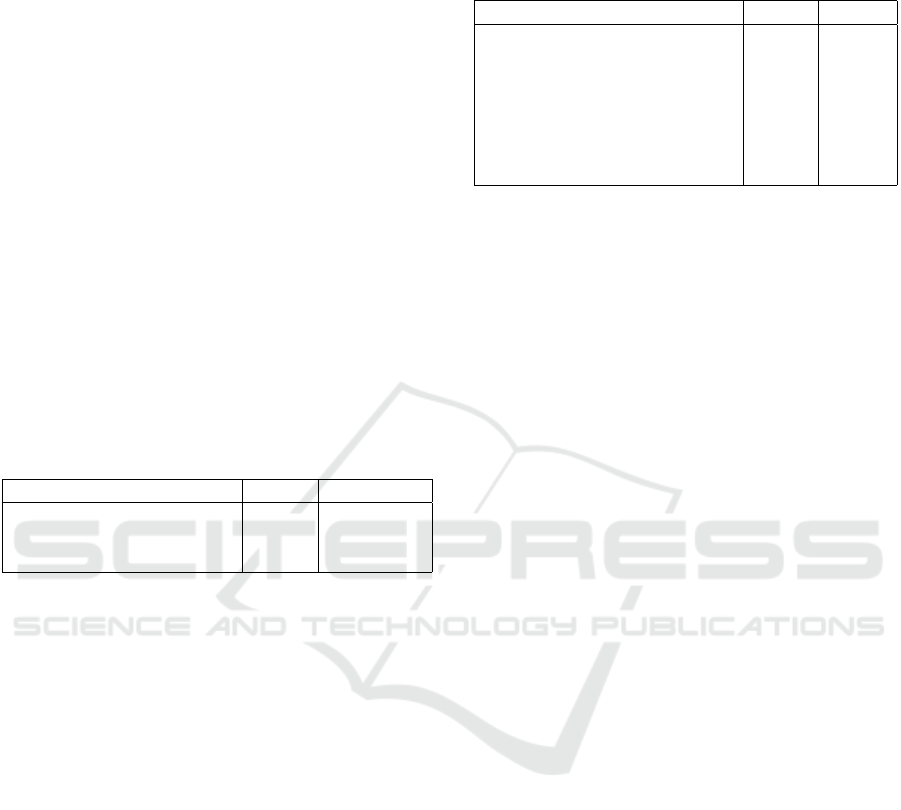
Figure 1 presents the results of the two-fold vali-
dation for our scoring method. Within the figure, blue
dots represent the average scores for semantic evalu-
ation of reports with similar label sequences, while
orange dots show the mean scores for reports with
Semantic Textual Similarity Assessment in Chest X-ray Reports Using a Domain-Specific Cosine-Based Metric
491
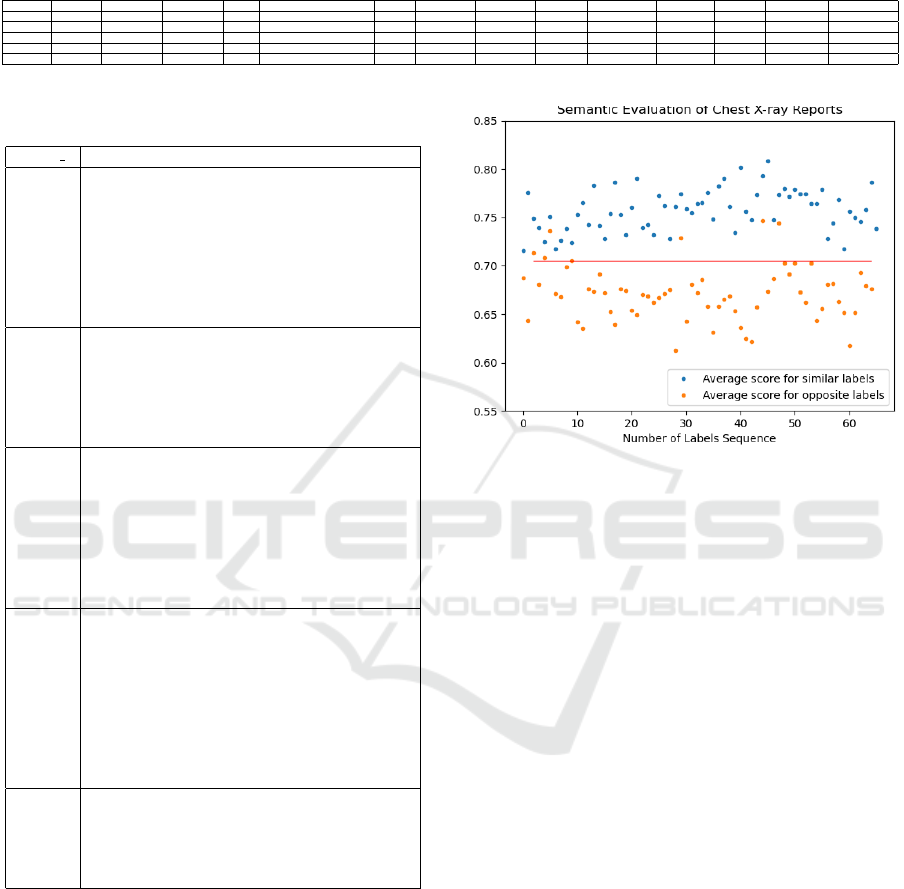
Table 3: A sample table featuring Chexpert labels (1. Atelectasis, 2. Cardiomegaly, 3. Consolidation, 4. Edema, 5. Enlarged
Cardiomediastinum, 6. Fracture, 7. Lung Lescion, 8. Lung Opacity, 9. No Finding, 10. Pleural Effusion, 11. Pleural Other,
12. Pneumonia, 13. Pneumothorax, 14. Support Devices) extracted from chest X-ray reports of five patients (Subject ##)
from the MIMIC-CXR database (Johnson et al., 2019).
Subject ## Atelectasis Cardiomegaly Consolidation Edema Enlarged Cardiomediastinum Fracture Lung Lescion Lung Opacity No Finding Pleural Effusion Pleural Other Pneumonia Pneumothorax Support Devices
01 0 1 1 -1
02 1 1
03 1 0
04 1 0 -1 0 1
05 1 1
Table 4: Reports corresponding to the subjects listed in Ta-
ble 3 from the MIMIC-CXR dataset (Johnson et al., 2019).
Subject ## Report
01 Lung volumes remain low. There are innumerable bi-
lateral scattered small pulmonary nodules which are bet-
ter demonstrated on recent CT. Mild pulmonary vascular
congestion is stable. The cardio mediastinal silhouette
and hilar contours are unchanged. Small pleural effusion
in the right middle fissure is new. There is no new focal
opacity to suggest pneumonia. There is no pneumotho-
rax.
02 A triangular opacity in the right lung apex is new from
prior examination. There is also fullness of the right
hilum which is new. The remainder of the lungs are clear.
Blunting of bilateral costophrenic angles, right greater
than left, may be secondary to small effusions. The heart
size is top normal.
03 Mild to moderate enlargement of the cardiac silhouette
is unchanged. The aorta is calcified and diffusely tor-
tuous. The mediastinal and hilar contours are otherwise
similar in appearance. There is minimal upper zone vas-
cular redistribution without overt pulmonary edema. No
focal consolidation, pleural effusion or pneumothorax is
present. The osseous structures are diffusely demineral-
ized.
04 The endotracheal tube tip is 6 cm above the carina. Na-
sogastric tube tip is beyond the GE junction and off the
edge of the film. A left central line is present in the tip
is in the mid SVC. A pacemaker is noted on the right in
the lead projects over the right ventricle. There is prob-
able scarring in both lung apices. There are no new ar-
eas of consolidation. There is upper zone redistribution
and cardiomegaly suggesting pulmonary venous hyper-
tension. There is no pneumothorax.
05 A moderate left pleural effusion is new. Associated
left basilar opacity likely reflect compressive atelectasis.
There is no pneumothorax. There are no new abnormal
cardiac or mediastinal contour. Median sternotomy wires
and mediastinal clips are in expected positions.
contrasting labels. The red horizontal line within the
figure serves as the dividing line distinguishing be-
tween similar and opposite evaluations. Upon review-
ing these results, it becomes evident that a distinct
boundary exists between reports sharing the same
clinical diagnoses and those with entirely dissimilar
diagnoses. Notably, there are no blue dots below
a 70% similarity threshold, whereas six orange dots
have scores above 70% across 70 label sequences,
which is certainly not very high. Nevertheless, de-
Figure 1: Semantic Evaluation of Chest X-ray reports. Each
blue dot represents the mean score of semantic evaluation
for reports with similar label sequences, while each orange
dot signifies the mean score of semantic evaluation for re-
ports with opposing labels. The red horizontal line repre-
sents the classification boundary.
spite this differentiation between similar and opposite
evaluations, some level of similarity, exceeding 50%,
persists within the opposing category. This can be at-
tributed to the implemented cosine similarity within
the medical domain, which introduces a certain bias
towards tokens in the same medical domain. Unfor-
tunately, this bias cannot be entirely eliminated, as
it plays a substantial role in the evaluation process.
However, a clear boundary remains between similar
and contrasting reports.
5 RESULTS AND DISCUSSION
In our original application of chest X-ray report gen-
eration, we incorporate our metric to assess the out-
puts of various models. We compare our results with
the BLEU scores evaluated by these models, specifi-
cally, the CXR-RePaiR (Endo et al., 2021) and R2Gen
(Chen et al., 2020) models, both being state-of-the-art
models for generating chest X-ray reports. Our eval-
uation focuses on measuring the semantic similarity
between the generated reports and the ground truth.
Table 5 presents the BLEU scores obtained from these
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
492
models and our metric’s semantic evaluation. As an-
ticipated, the BLEU scores are relatively low, signi-
fying a substantial dissimilarity between the gener-
ated results and the ground truth for both the CXR-
RePaiR and R2Gen models despite being regarded as
state-of-the-art models for chest X-ray report gener-
ation. These models still employ the BLEU metric
for evaluation, primarily due to the scarcity of more
suitable metrics and the need for a standardized eval-
uation process for comparative purposes. Conversely,
our metric produces more promising results for both
of these models. While our metric’s scores align with
the BLEU scores, indicating higher scores for both
BLEU and our MCSE metric in the case of the R2Gen
model compared to the CXR-RePaiR, our metric pro-
vides a deeper evaluation. It suggests a degree of sim-
ilarity to the ground truth rather than outright dissim-
ilarity in BLEU, thus making the generated reports
more reliable and trustworthy, which is a crucial ad-
vancement in the field.
Table 5: The result of BLEU score of 2-gram for state-of-
the-art models and the result of our novel metric on these
models outcomes.
Models BLEU Our MCSE
R2Gen (Chen et al., 2020) 0.212 0.71
CXR-RePair (Endo et al.,
2021)
0.069 0.64
Table 6 provides an example of medical text gen-
erated and evaluated using both a BLEU score and
our MCSE metric. It’s evident that, according to the
BLEU score, these two texts appear vastly different,
even though they share the same primary medical en-
tities. However, when we delve into the context, we
can notice that ”moderately severe” serves as a de-
scription for the main entity, ”pulmonary edema”, in
the generated text. Similarly, in the second part of the
text, the main medical entity is ”pleural effusions”,
and terms like ”likely” and ”no large” are used to de-
scribe this entity, which may not be identical but share
semantic similarities. This subtle context evaluation
is precisely what our metric considers, yielding a sim-
ilarity score of 0.64 for these texts, which we argue
is a more accurate reflection compared to the BLEU
score.
Lastly, the significant benefit of employing this
metric lies in its capacity for comparative analysis
alongside other evaluation measures. For instance,
when examining the outcomes of the BLEU score,
with its word-by-word analysis, situations may arise
where the results are totally inaccurate, casting doubt
on their reliability, despite the models performing
well overall. Integrating the results of our novel
Table 6: A comparative example of using the BLEU score
and our adapted metric with medical reference and gener-
ated text.
BLEU MCSE
Reference Sentence: ”Pul-
monary edema, cardiomegaly,
likely pleural effusions.”
Generated Sentence: ”Mod-
erately severe bilateral pul-
monary edema with no large
pleural effusion.”
0.047 0.64
MCSE metric into the evaluation process allows us
to semantically analyze and ascertain the dependabil-
ity of the models’ textual outputs within the context
of medical content.
6 CONCLUSION
In our research, we tackle the challenge of semantic
similarity scoring in medical corpora, driven by the
inadequacy of existing metrics that, while suitable for
machine translation evaluation, fall short in the field
of medical semantic assessment. Our innovative met-
ric draws inspiration from how humans comprehend
text, centering on the extraction of key terms and their
relational context. It introduces a novel approach for
extracting clinical entities from medical text, consid-
ering not only the entities themselves but also the as-
sociated descriptions and negations. Additionally, we
created a new method for scoring the semantic rela-
tionships between these entities by using the domain
cosine similarity. The validation process allowed us to
analyze and validate each of these steps individually,
unraveling a clear distinction between reports sharing
the same diagnosis and those diverging in this regard.
For our research, we focused on the application of
chest X-rays, a critical domain where a robust seman-
tic evaluation metric is highly valuable. We applied
our metric to some of the latest state-of-the-art mod-
els, and the results harmonized with other evaluation
metrics, affirming their reliability.
While our validation process and implementa-
tion yielded successful outcomes, we encountered the
challenge of an inherent bias in domain cosine simi-
larity. This challenge has illuminated a promising di-
rection for our future research, as we explore ways
to mitigate this bias and advance the field of medical
semantic evaluation.
Material, Codes, and Acknowledgement: Results
can be reproduced using the code available in the
GitHub repository https://git hub.com/sayeh19
Semantic Textual Similarity Assessment in Chest X-ray Reports Using a Domain-Specific Cosine-Based Metric
493

94/Medical-Corpus-Semantic-Similar ity-Eva
luation.git. All the computations presented in this
paper were performed using the (Gricad, ) infrastruc-
ture (https://gri cad.univ-grenoble-alpes.f
r), which is supported by Grenoble research commu-
nities.
REFERENCES
Alam, F., Afzal, M., and Malik, K. M. (2020). Comparative
analysis of semantic similarity techniques for medical
text. In 2020 International Conference on Information
Networking (ICOIN), pages 106–109.
Banerjee, S. and Lavie, A. (2005). METEOR: An automatic
metric for MT evaluation with improved correlation
with human judgments. In Proceedings of the ACL
Workshop on Intrinsic and Extrinsic Evaluation Mea-
sures for Machine Translation and/or Summarization,
pages 65–72, Ann Arbor, Michigan. Association for
Computational Linguistics.
Beltagy, I., Lo, K., and Cohan, A. (2019). SciBERT: A
pretrained language model for scientific text. In Inui,
K., Jiang, J., Ng, V., and Wan, X., editors, Proceed-
ings of the 2019 Conference on Empirical Methods
in Natural Language Processing and the 9th Inter-
national Joint Conference on Natural Language Pro-
cessing (EMNLP-IJCNLP), pages 3615–3620, Hong
Kong, China. Association for Computational Linguis-
tics.
Chen, Z., Song, Y., Chang, T.-H., and Wan, X. (2020). Gen-
erating radiology reports via memory-driven trans-
former. In Proceedings of the 2020 Conference on
Empirical Methods in Natural Language Processing
(EMNLP), pages 1439–1449, Online. Association for
Computational Linguistics.
Endo, M., Krishnan, R., Krishna, V., Ng, A. Y., and Ra-
jpurkar, P. (2021). Retrieval-based chest x-ray report
generation using a pre-trained contrastive language-
image model. In Proceedings of Machine Learning
for Health, volume 158 of Proceedings of Machine
Learning Research, pages 209–219.
Gricad. infrastructure supported by grenoble research com-
munities.
Honnibal, M., Montani, I., Van Landeghem, S., and Boyd,
A. (2020). spaCy: Industrial-strength Natural Lan-
guage Processing in Python.
Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S.,
Chute, C., Marklund, H., Haghgoo, B., Ball, R., Sh-
panskaya, K., Seekins, J., Mong, D. A., Halabi, S. S.,
Sandberg, J. K., Jones, R., Larson, D. B., Langlotz,
C. P., Patel, B. N., Lungren, M. P., and Ng, A. Y.
(2019). Chexpert: A large chest radiograph dataset
with uncertainty labels and expert comparison. Pro-
ceedings of the AAAI Conference on Artificial Intelli-
gence, 33(01):590–597.
Jain, S., Agrawal, A., Saporta, A., Truong, S., Duong,
D. N., Bui, T., Chambon, P., Zhang, Y., Lungren,
M. P., Ng, A. Y., Langlotz, C., and Rajpurkar, P.
(2021). Radgraph: Extracting clinical entities and re-
lations from radiology reports. In Thirty-fifth Con-
ference on Neural Information Processing Systems
Datasets and Benchmarks Track (Round 1).
Johnson, A. E. W., Pollard, T. J., Berkowitz, S. J., Green-
baum, N. R., Lungren, M. P., Deng, C.-y., Mark, R. G.,
and Horng, S. (2019). Mimic-cxr, a de-identified pub-
licly available database of chest radiographs with free-
text reports. Scientific Data, 6(1):317.
Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H., Lea-
man, R., Davis, A. P., Mattingly, C. J., Wiegers, T. C.,
and Lu, Z. (2016). BioCreative V CDR task cor-
pus: a resource for chemical disease relation extrac-
tion. Database, 2016:baw068.
Lin, C.-Y. (2004). ROUGE: A package for automatic evalu-
ation of summaries. In Text Summarization Branches
Out, pages 74–81, Barcelona, Spain. Association for
Computational Linguistics.
Miura, Y., Zhang, Y., Tsai, E., Langlotz, C., and Jurafsky,
D. (2021). Improving factual completeness and con-
sistency of image-to-text radiology report generation.
In Toutanova, K., Rumshisky, A., Zettlemoyer, L.,
Hakkani-Tur, D., Beltagy, I., Bethard, S., Cotterell, R.,
Chakraborty, T., and Zhou, Y., editors, Proceedings of
the 2021 Conference of the North American Chapter
of the Association for Computational Linguistics: Hu-
man Language Technologies, pages 5288–5304, On-
line. Association for Computational Linguistics.
Neumann, M., King, D., Beltagy, I., and Ammar, W. (2019).
ScispaCy: Fast and Robust Models for Biomedical
Natural Language Processing. In Proceedings of the
18th BioNLP Workshop and Shared Task, pages 319–
327, Florence, Italy. Association for Computational
Linguistics.
Papineni, K., Roukos, S., Ward, T., and Zhu, W.-J. (2002).
Bleu: a method for automatic evaluation of machine
translation. In Proceedings of the 40th Annual Meet-
ing of the Association for Computational Linguistics,
pages 311–318, Philadelphia, Pennsylvania, USA.
Association for Computational Linguistics.
Patricoski, J., Kreimeyer, K., Balan, A., Hardart, K., Tao,
J., Anagnostou, V., Botsis, T., Investigators, J. H. M.
T. B., et al. (2022). An evaluation of pretrained bert
models for comparing semantic similarity across un-
structured clinical trial texts. Stud Health Technol In-
form, 289:18–21.
Smit, A., Jain, S., Rajpurkar, P., Pareek, A., Ng, A., and
Lungren, M. P. (2020). Chexbert: Combining auto-
matic labelers and expert annotations for accurate ra-
diology report labeling using bert. In Conference on
Empirical Methods in Natural Language Processing.
Yu, F., Endo, M., Krishnan, R., Pan, I., Tsai, A., Reis, E. P.,
Fonseca, E. K. U. N., Ho Lee, H. M., Abad, Z. S. H.,
Ng, A. Y., Langlotz, C. P., Venugopal, V. K., and Ra-
jpurkar, P. (2022). Evaluating Progress in Automatic
Chest X-Ray Radiology Report Generation. preprint,
Radiology and Imaging.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
494
