
Approach and Method for Bayesian Network Modelling:
The Case for Pregnancy Outcomes in England and Wales
Scott McLachlan
1a
, Bridget J. Daley
2
, Sam Saidi
3
, Evangelia Kyrimi
4
, Kudakwashe Dube
1
,
Crina Grossan
1
, Martin Neil
4
, Louise Rose
1
and Norman E. Fenton
4
1
Nursing, Midwifery and Palliative Care, Kings College London, London, U.K.
2
Maternity Unit, Liverpool Women’s Hospital NHS Trust, Liverpool, U.K.
3
School of Medicine, University of Sydney, Sydney, Australia
4
Electronic Engineering and Computer Science, Queen Mary University of London, London, U.K.
Keywords: Clinical Decision-Support Systems, Bayesian Networks, Predictive Models, Pregnancy Outcomes.
Abstract: For predicting and reasoning about outcomes of specific medical condition Bayesian Networks (BNs) can
provide significant benefits over traditional statistical prediction models. However, developing appropriate
and accurate BNs that incorporate key causal aspects of the condition is challenging and time-consuming.
This work introduces a novel development approach, merging expert elicitation, literature knowledge, and
national health statistics that enables such BNs to be developed efficiently. The approach is applied to build
a BN for pregnancy complications and outcomes in England and Wales using 2021 data. The BN showed
comparable predictive performance against logistic regression and nomograms, but additionally provides
powerful support for decision-making and risk assessment across diverse pregnancy-related conditions and
outcomes.
1 INTRODUCTION
Traditional pregnancy prediction models focus on
singular health issues such as gestational diabetes
mellitus (GDM) or preeclampsia (PE) without
considering the broader context of the pregnancy.
Typically, these models are statistical, relying on a
limited set of risk factors which leads to several
limitations (a full set of references for this and other
imputations in this paper can be found in the
expanded preprint version: McLachlan et al, 2024).
These limitations include: (i) a focus on predicting the
presence of a condition without considering the
absence of that condition; (ii) overfitting to available
data, leading to poor performance in the presence of
uncertain or missing data; (iii) lack of transparency
and interpretability, making it difficult to understand
how the model makes its predictions; and (iv) limited
ability to generalize to new populations or settings.
To address these limitations we propose a new
approach to pregnancy prediction based on Bayesian
networks (BNs). BNs are a type of probabilistic
a
https://orcid.org/0000-0002-2528-8050
model that can represent complex relationships
between variables and have been shown effective in a
wide range of medical applications.
Our proposed approach involves using BNs to
model the entire pregnancy rather than focusing on
singular health issues. This allows us to draw on a
wider range of information including symptoms, risk
factors, and medical history and to simultaneously
make predictions about multiple health issues.
We have evaluated our proposed approach using
the domain of pregnancy outcomes and found that it
can outperform traditional methods in terms of
accuracy, generalisability, and interpretability. We
believe our approach has potential to transform how
prediction models, and particularly pregnancy
outcome prediction models, are developed and used.
The resulting model is extensively validated using
vignettes and concurrency analysis.
The rest of the paper is structured as follows:
Section 2 covers theoretical and application domain
backgrounds and reviews literature related to the
research problem. Section 3 outlines approach and
method for knowledge, data, and expert-driven
604
McLachlan, S., Daley, B., Saidi, S., Kyrimi, E., Dube, K., Grossan, C., Neil, M., Rose, L. and Fenton, N.
Approach and Method for Bayesian Network Modelling: The Case for Pregnancy Outcomes in England and Wales.
DOI: 10.5220/0012428600003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 604-612
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

modelling using causal BNs. Section 4 presents the
results of applying this approach to develop a BN for
pregnancy complications and outcomes. Section 5
discusses the experience of utilizing the approach and
method, highlighting potential limitations in
application to other problem domain spaces.
2 BACKGROUND AND RELATED
WORKS
In our contemporaneous work, we screened a
collection of 100 works published between 2000 and
2023 that proposed predictive risk screening models
for pregnancy complications (Dube, Kyrimi &
McLachlan, 2023).
2.1 Risk Factors and Symptoms
Risk screening typically occurs during the initial
maternal clinic visit known as the booking visit
(Tandu-Umba et al, 2014). While risk screening
scores may be updated throughout antenatal care as
new clinical and non-clinical information emerges,
the specific signs, symptoms or clinical tests used
vary depending on the adopted guideline or scoring
model (Tandu-Umba et al, 2014; Stott et al, 2016).
Some models rely on common factors like maternal
age, BMI, and pregnancy history collected during the
booking visit (Tandu-Umba et al, 2014), while others
incorporate antenatal care records, pregnancy
outcomes, or even novel variables like paternal DNA
or vaginal swab results (Stott et al, 2016).
2.2 Common Issues
Developing healthcare risk, probability and decision
support models can be challenging because: (i)
obtaining a sufficiently large and high-quality dataset
remains a hurdle and data may only be available for
small patient groups, with demographic or clinical
risk factors significantly reducing subgroup sample
sizes (North et al, 2011; Pitchforth & Mengersen,
2013); (ii) traditional model evaluation relies on
internal statistical methods (Dube et al, 2023) such as
ROC curves and CIs that have limitations in assessing
BNs (Pitchforth & Mengersen, 2013); (iii) prediction
accuracy varies when some observations are missing
or the patient lacks the predicted health condition,
often due to overfitting that occurs when models are
trained solely on data identifying the medical
condition of interest, excluding information about
what isn't that condition (Kumar et al, 2022); and (iv)
most are not presented with clear examples like risk-
scored vignettes, hindering clinical comprehension
and adoption (North et al, 2011; Mehta-Lee et al,
2017).
3 METHOD
Our research initially focused on constructing causal
models for singular health issues affecting patients
with rheumatoid arthritis, angina, acute traumatic
coagulopathy, and GDM (McLachlan et al, 2020).
However, we too overlooked the broader perspectives
of general health, the accumulated effect of
comorbidity, and healthcare access and experience
within an entire population. We now stress the
importance of adopting a holistic approach to model
not only the patient, but also the community and
disease; in essence, a complete digital twin that can
be used to establish the credibility of our models in:
(a) identifying or explaining risk; and (b) providing
computer-based clinical decision support using
machine learning (ML) or artificial intelligence (AI).
Creating a community-wide baseline is crucial to
fully evaluate causal relationships among known and
novel symptoms, and modelling treatment and
prognostic counterfactuals.
3.1 Hypothesis
This work proposes a broadly accurate BN model for
diagnosis and treatment outcomes can be constructed
using expert clinical knowledge, privacy-preserving
datasets and population-wide statistics. By analysing
commonly recorded medical observations, the model
can predict health outcomes, incorporating causal
interactions among clinical data, patient information
and publicly available clinical data. Notably, this
approach is unprecedented in using large-scale
health and outcome statistics.
3.2 Bayesian Networks
BNs, also termed probabilistic graphical models,
offer a graphical framework for probabilistic
reasoning under uncertainty through a directed
acyclic graph (DAG) consisting of structure and
parameters. The structure includes nodes representing
variables and edges indicating causal relationships.
Parameters consist of conditional probability
functions for each node, representing its strength
given its parents. Bayesian probabilistic reasoning
involves updating prior beliefs (prior probability)
based on new evidence, resulting in posterior
Approach and Method for Bayesian Network Modelling: The Case for Pregnancy Outcomes in England and Wales
605
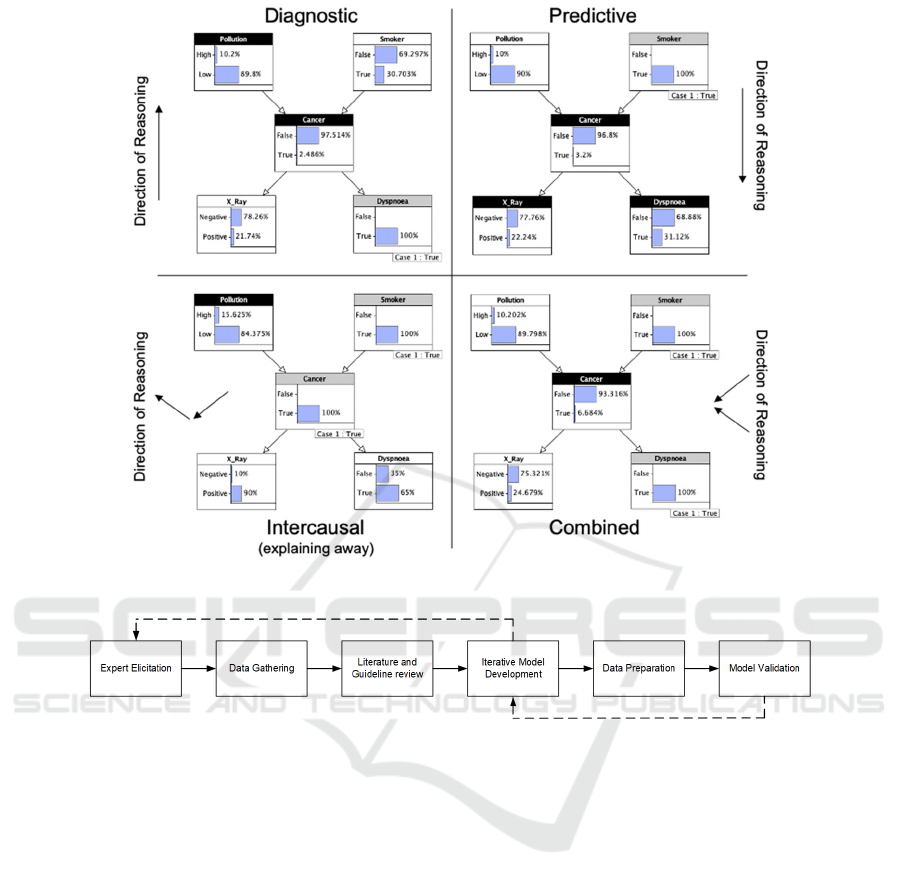
Figure 1: Types of reasoning.
Figure 2: BN Development Process Flow Diagram.
probability. In Figure 1, various scenarios illustrate
reasoning from evidence using a lung cancer model.
A node head in grey indicates observed evidence,
while black determines the question being reasoned
about. Forward reasoning - following the arc
direction, and backward reasoning - counter to the arc,
represent causal or predictive and diagnostic
reasoning, respectively. Combining forward,
intercausal and backward reasoning produces
intercausal and combined reasoning. The approach
used in this work ensures the model's capacity for all
four modes of reasoning.
3.3 Study Population
The model in this study used publicly available
privacy-preserving aggregate statistics from various
sources. The data covered 624,828 pregnancies in
England and Wales during 2021 encompassing live
births, stillbirths, and neonatal deaths. Additionally,
evidence for risk factors and causal relationships was
drawn from guidelines and academic studies
published between 2019 and 2022, focusing on UK
populations in 2021.
3.4 BN Development
Our main design objective was to create a model that
credibly encapsulates current clinical knowledge on
pivotal risk factors and interacting signs and
symptoms affecting pregnancy outcomes. This
objective is pursued through a six-phase development
process outlined in Figure 2 and detailed in the
subsequent section.
3.4.1 Expert Elicitation
The BN's structure and parameters can be derived
entirely from data with an extensive dataset. However,
BNs exhibit flexibility, capable of seamlessly
integrating less comprehensive datasets, multiple
expert’s knowledge, and diverse information sources
HEALTHINF 2024 - 17th International Conference on Health Informatics
606
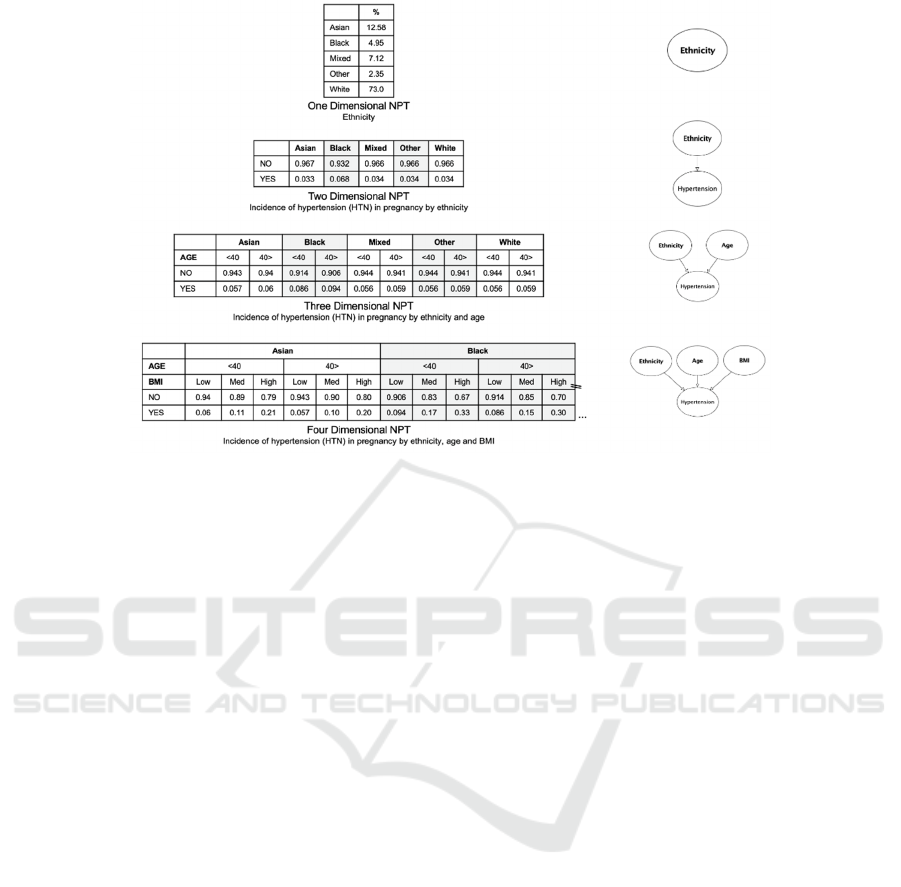
Figure 3: Examples of dimensionality in NPTs.
(Christophersen et al, 2018). Expert input enriches
BN's design, ensuring up-to-date domain knowledge
(Sanford & Moosa, 2015) Expert elicitation yielded
caremaps; visualisations of disease progression,
diagnosis, treatment processes and potential patient
outcomes through flow diagrams or process maps.
The caremap development process, has been detailed
previously (McLachlan et al; 2020a).
3.4.2 Data Gathering
We sought national datasets that described the
incidence of pregnancy complications and outcomes
for an entire population. A key focus was publicly
available privacy-preserving datasets whose use
would not require, or violate, institutional ethics
policies. This limited us to secondary or aggregate
statistical sources such as those of national health
services, health departments or statistics agencies.
We collected datasets for the year 2021 as these were
the most recent complete and published statistical
datasets available for the UK.
3.4.3 Literature and Clinical Guideline
Review
We performed a search to locate literature, clinical
practice guidelines and protocols relevant to the
medical condition(s) being modelled. The literature
included was aligned to the data gathered in Phase
3.4.2. Priority was given to articles published during
the same time period that described incidence of the
medical condition(s) in like populations.
3.4.4 Model Development
The iterative model development process was: (1)
medical idioms identifying key structural fragments
were identified from the combination of caremap and
knowledge derived from the clinical experts; (2) data
was identified from statistical and literary sources to
populate node probability tables (NPTs), describing
incidence of the variable described by the node and
incidence of interaction across arcs between that node
and parent or child nodes; (3) structural fragments
were brought together to form a single contiguous
BN; and (4) the resulting model structure was
reviewed with clinical experts and where changes
were identified, the process returned to the first step.
The process for identifying medical idioms and using
these to support expert elicitation was previously
described in (Kyrimi et al, 2020).
3.4.5 Data Preparation
Each node within a BN has a NPT. Absolute parent
nodes (such as the pollution node in the example in
Figure 1) have a single dimensional NPT. Where a
node has a single parent (such as the dyspnoea node
in the example in Figure 1) it will have a two-
dimensional NPT. Where a node has two parents
(such as the cancer node in the example in Figure 1)
it will have a three-dimensional NPT etc. Nodes with
six parents or greater are generally avoided due to
complexity of elicitation and computation. Examples
of data dimensionality in NPT are provided in Figure
3.
Approach and Method for Bayesian Network Modelling: The Case for Pregnancy Outcomes in England and Wales
607
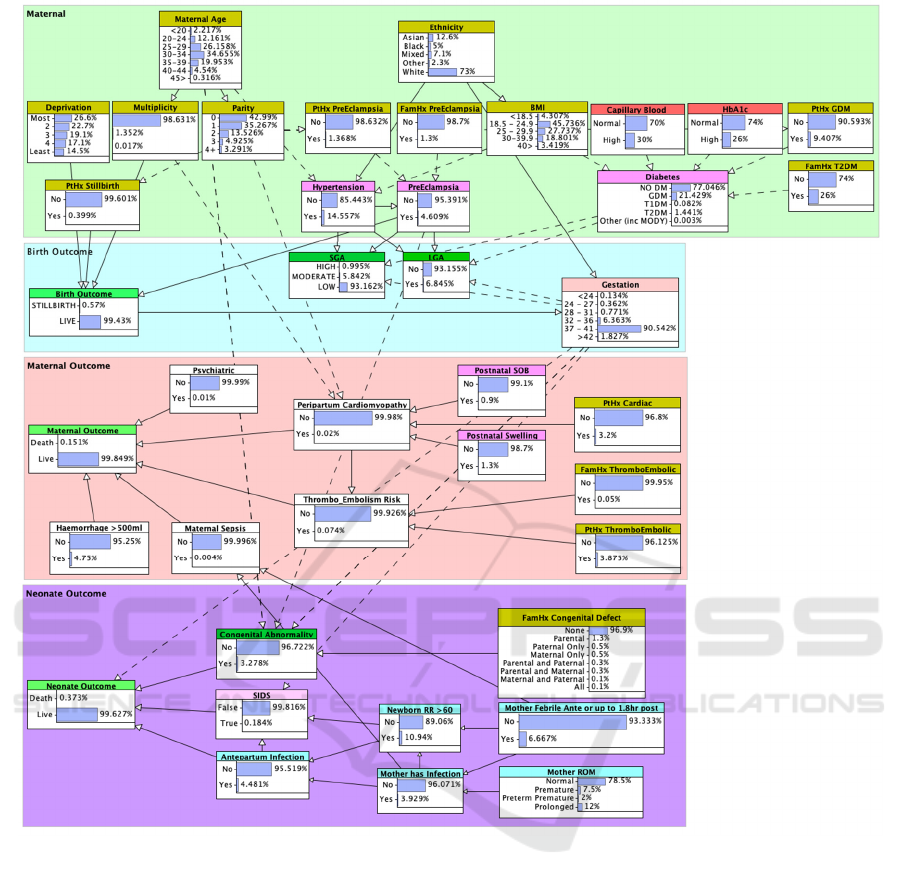
Figure 4: BN model showing background priors.
The content of each column in an NPT should sum
to 1.0 (100%). Discretisation allows the modeller to
convert continuous variable factors like BMI and
capillary blood glucose (CBG) by assigning them
clinically relevant intervals, ordinal states or
categories (for example: low, medium, high). Some
variables such as BMI in the example shown in Figure
3 were discretised in this way. Tables using
population-level continuous variable data were
prepared in Microsoft Excel and converted on
ingestion by the Agenarisk BN modelling tool.
3.4.6 Model Validation
The validation process for our BN models followed a
multi-step methodology recommended by various
authors (Pitchforth & Mengersen, 2013). We initially
undertook face validity with clinical experts
(Pitchforth & Mengersen, 2013). However, we
recognised the potential weakness in situations
wherein experts involved in design are unlikely to
disagree with their own judgment as reflected in the
resulting model. To mitigate this we also used: (i)
content validity to assess the BN structure against
identified literature and clinical practice guidelines,
evaluating the relationships between crucial risk
factors and symptoms (Pitchforth & Mengersen,
2013); and (ii) concurrent validity to compare BN
predictions against published models using clinical
vignettes (Pitchforth & Mengersen, 2013). Due to the
extreme rarity of the primary model outcomes;
HEALTHINF 2024 - 17th International Conference on Health Informatics
608
“Stillbirth” as Birth Outcome and “Death”
as Maternal Outcome, classical validation tests for
model’s accuracy, discrimination and calibration
were not performed.
4 RESULTS
Figure 4 presents the Maternal Outcomes BN model,
comprising four fragments that are used to group
nodes relevant to: (1) maternal risk factors and
common health conditions that may affect the
pregnancy; (2) the immediate pregnancy outcome for
the neonate and (3) mother; and (4) survival of the
neonate.
The model structure allows the relevant impact of
observations on factors in the primary maternal zone
to carry over onto maternal and neonate outcomes.
Solid lines indicate direct relationships, while dashed
lines indicate the presence of hidden nodes used for
alternate discretisation of variables. Model priors are
also shown in Figure 4.
4.1 BN Validation
This section provides an overview of the processes
used to validate the maternal outcomes model.
4.1.1 Face Validity
Throughout development of our BN, a collaborative
effort with a small group of clinical experts ensured
validation through comparisons with literature and
clinical guidelines. Face validity ensures the model's
visual representation aligns with expectations. The
iterative development process incorporated clinical
insights and weighed variables and causal pathways
against evidence from clinical texts, medical journals,
and available data. For parameterisation, clinicians
played a crucial role in providing initial estimates for
BN parameters. We updated these in the final model
using national statistics for the entire England &
Wales population.
The model holistically addresses primary
pregnancy outcomes: (1) Birth Outcome: This
fragment encompasses live birth or stillbirth,
including considerations for late-term miscarriage
based on nuanced definitions (NHSInform, 2022); (2)
Maternal Outcome: Predicting maternal death
aligned to 2021 mortality statistics (ONS, 2023); (3)
Neonate Outcome: Predicting death in live-born
babies aligned to 2021 ONS and MBRRACE-UK
birth outcome statistics.
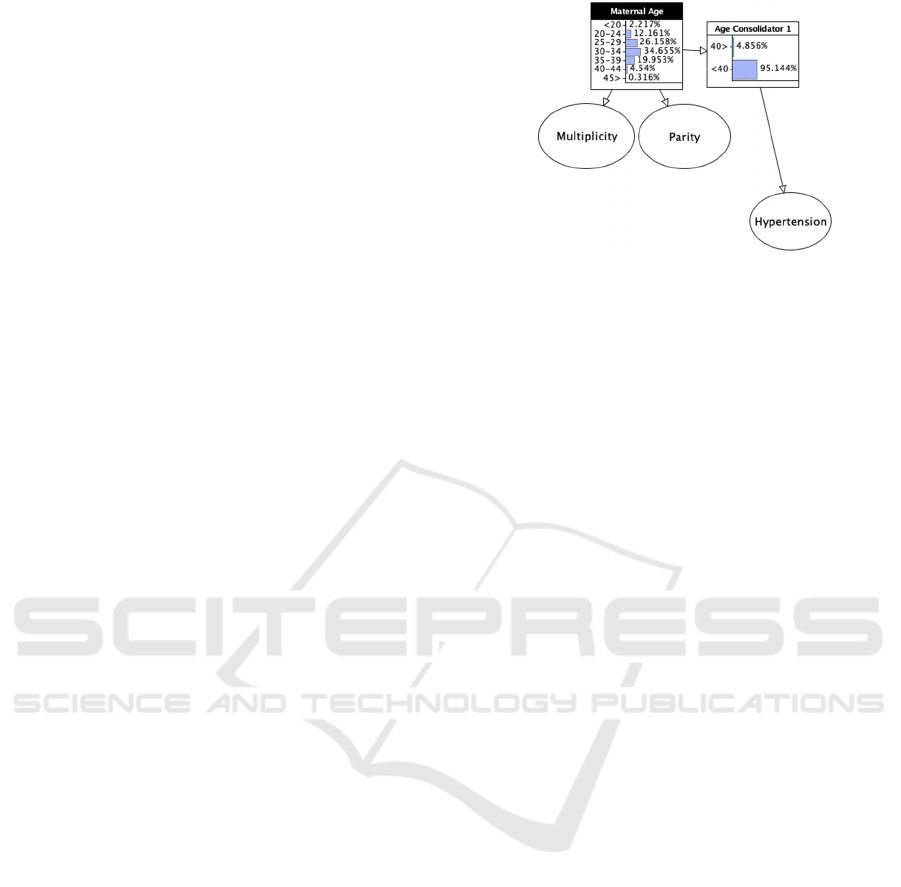
Figure 5: Maternal Age substructure showing hidden Age
Consolidator node.
Secondary outcomes linked to the baby are also
identified, including: (1) Small for Gestational Age
(SGA): Stratifying risk into three categories, the
model offers a nuanced perspective on this outcome,
grounded in a total incidence for 2021 derived from
substantial data (NMPA Project Team, 2021); (2)
Large for Gestational Age (LGA): Incorporating the
impact of gestational diabetes mellitus (GDM), the
model aligns with 2021 datasets, capturing the
nuanced nature of this outcome; (3) Congenital
Abnormality: Grounded in probability and informed
by UK national statistical data and research on
increasing prevalence rates due to inheritance, the
model projects a prior probability of 3.278% for this
outcomes.
The collaborative and iterative approach, coupled
with reliance on expert input and robust statistical
grounding, ensures a model's robustness and
relevance in reasoning complex outcomes.
4.1.2 Content Validity
The key demographic risk factors identified in
predictive models included: (1) Maternal Age (74%):
Models varied in representing maternal age,
reflecting it either as a continuous variable or
discretizing it into intervals. Our model employs five-
year increments that align with national maternity
statistics (ONS, 2023). To simplify situations where
a binary identifying advanced maternal age was
required, a hidden boundary age node was included
as shown in Figure 5; (2) BMI (59%): BMI statistics,
categorized into five groups, were derived from
Public Health England’s report (PHE, 2019). BMI is
strategically placed between ethnicity and child nodes
representing diabetes, hypertension, and pregnancy
outcomes; (3) Parity (42%): Nulliparity and grand
parity were categorized into five groups to aid
consideration of their impact on conditions and
outcomes. Maternal age is linked, especially in
Approach and Method for Bayesian Network Modelling: The Case for Pregnancy Outcomes in England and Wales
609

extreme cases, updating probabilities for nodes like
diabetes, hypertension, and birth outcomes (Ananth et
al, 1996); (4) Ethnicity (36%): Ethnicity's influence is
integrated across the model, impacting hypertension,
gestation and BMI, with connections to various
pregnancy outcomes; (5) Gestation (32%): Gestation
can be both an outcome and a risk factor, and affects
various pregnancy outcomes including prematurity
and post-term deliveries. The gestation node is
informed by the 2021 dataset (ONS, 2023) to ensure
accuracy in predicting maternal and neonate risks and
outcomes.
4.1.3 Concurrent Validity
We re-examined papers included in our screening
review (Dube et al, 2023) to locate any works that
included a vignette with prediction suitable for use in
concurrent validity testing, identifying only two.
North et al (2011) propose a model using statistical
methods to predict incidence of pre-eclampsia. Their
model is based on demographic and risk factors of the
first-time mother along with observable signs and
symptoms routinely collected by the midwife during
the initial (booking) patient appointment. They used
their model to compute the following vignette:
A 28 year old nulliparous woman whose birth
weight was 2400 g, with a mean arterial
pressure of 96 mmHg, BMI 30, a family
history of pre-eclampsia, and no protective
factors, her probability of pre-eclampsia is
39%.
Making the same observations (maternal age, BMI,
parity, family history of pre-eclampsia, and maternal
hypertension) our model indicates a 43% probability
for pre-eclampsia. The 4% difference can be
attributed to: (i) diverse country origins in their
dataset (England, Wales, Ireland, New Zealand, and
Australia) versus our England and Wales focus; (ii)
their 2004-2008 data versus our 2021 dataset; (iii)
their 4961 pregnancies versus our larger 624,828; and
(iv) reported increasing incidence of pregnancy-
related conditions globally between 2010-2020
(Cameron et al, 2022).
Mehta-Lee et al (2017) used statistical methods on
pregnancy data collected between 2004-2009 to
develop a nomogram for predicting preterm delivery.
They began with a larger number of potential factors,
but the resulting nomogram includes only the nine
factors they identified as most predictive from a
cohort of 192,208 pregnancies. Their vignette
describes:
A 35 year old (13 points) African American
(41 points) woman planning to get pregnant
for the first time (46 points) who has no history
of diabetes (0 points) but who smokes (12
points) would have a total of 112 points. This
approximates to a baseline probability of
preterm birth of 12-13% prior to conception.
Using the same observations, our model predicted an
8.7% preterm birth rate - which is the sum of
predictions for all gestations prior to 37 weeks with
the exclusion of smoking as a factor (Smith et al,
2023). Discrepancies observed with how other
models incorporated smoking may have arisen due to
underreported smoking rates influenced by social
stigma (Smith et al, 2023). Global variations in
reporting, and confounded outcomes in UK studies,
only contribute to the confusion (Smith et al, 2023).
UK studies generally report lower smoking rates
compared to the USA, with some incredibly reporting
no smoking at all (Stott et al, 2016). Vaping, a
smoking alternative, poses uncertainties in long-term
pregnancy outcomes. Omitting smoking from our
model considers these issues and ensures robust
predictions unaffected by potential smoking-related
biases (Smith et al, 2023).
Finally, Du & Li (2021) developed a nomogram
for prediction of the baby’s survivability in
pregnancies complicated by GDM using data
collected from 626 Chinese mothers receiving
outpatient antenatal care between 2016 and 2019. We
used their nomogram to evaluate the following
scenario:
A 35 year old (22 points) Asian woman at 31
weeks or 217 days gestation (10 points) with a
BMI of 35 (25 points), first degree family
history of T2DM (10 points), history of GDM
in a previous pregnancy (4 points) and a
mildly high fasting plasma glucose (FPG) of
6.0 mmol (47.5 points). This gave a total of
118.5 points which their nomogram
approximated to 82-83% survivability for the
baby.
Our BN model with these observations predicted a
live birth, birth outcome of 84.7%. The dataset of
pregnancies used in Du & Li (2021) were temporally
the closest to those used to develop our model, and
the resulting predictions are not significantly different.
HEALTHINF 2024 - 17th International Conference on Health Informatics
610

5 DISCUSSION
During our model validation, two unaddressed issues
in reviewed models emerged. First, some works
included potentially unquantifiable elements: self-
assessed, unmeasurable, or flexible factors (Mehta-
Lee et al, 2017). Second, some models included
potentially unknowable elements: data challenging to
reliably procure (North et al, 2011).
While resource-intensive, our model's design
proves efficient. It eliminates redundant data entry
across different predictive models, streamlining the
process for clinicians. Rather than inputting the same
variables multiple times for various conditions, our
model allows one-time entry, computing probabilities
for both primary and subsequent conditions along the
disease pathway (Angeli et al, 2011). Traditional
models often focus on prediction of a singular health
condition, neglecting a holistic view of health. Our
model considers the patient comprehensively,
capturing interactions between risk factors,
symptoms, and various health issues. Unlike
condition-limited models, our approach models the
patient as an entire organism, preserving information
about the overall impact of common symptoms or
concomitant diseases on health outcomes.
While adapting the model for New Zealand, future
work includes exploring treatment selection and
outcome counterfactuals. This involves testing
alternate hypotheses, such as the potential outcomes
with or without specific interventions. Limitations
include the need for granular national health statistics
and access to expert support for model development
and face validity assessment. Resources and time are
substantial in constructing complex models like ours,
contrasting with the preference for simpler, single-
condition statistical models.
6 CONCLUSIONS
This work has introduced a novel pregnancy risk
prediction model addressing limitations in existing
approaches. Our holistic model considers not only the
condition of interest but also related conditions and
outcomes. Unlike models relying on limited local
data, we utilise publicly available national health
statistics, allowing versatile model development.
Employing a causal Bayesian probabilistic approach,
we navigate uncertain or missing data. Validation
involves ongoing face, content, and concurrent
methods, revealing an accurate description of
pregnancies nationally and individually. Three case
vignettes provide exemplar predictions for future
model comparisons. The model's reliability and
clinical holism, achieved at low cost, can instil
confidence in both clinicians and patients.
REFERENCES
Ananth, C. V., Wilcox, A. J., Savitz, D. A., Bowes Jr, W.
A., & Luther, E. R. (1996). Effect of maternal age and
parity on the risk of uteroplacental bleeding disorders
in pregnancy. Obstetrics & Gynecology, 88(4), 511-
516.
Angeli, F., Angeli, E., Reboldi, G., & Verdecchia, P.
(2011). Hypertensive disorders during pregnancy:
clinical applicability of risk prediction models. Journal
of hypertension, 29(12), 2320-2323.
Belbasis, L., & Panagiotou, O. A. (2022). Reproducibility
of prediction models in health services research. BMC
Research Notes, 15(1), 1-5.
Cameron, N. A., Everitt, I., Seegmiller, L. E., Yee, L. M.,
Grobman, W. A., & Khan, S. S. (2022). Trends in the
incidence of new‐onset hypertensive disorders of
pregnancy among rural and urban areas in the United
States, 2007 to 2019. Journal of the American Heart
Association, 11(2), e023791.
Christophersen, A., Deligne, N. I., Hanea, A. M., Chardot,
L., Fournier, N., & Aspinall, W. P. (2018). Bayesian
network modeling and expert elicitation for
probabilistic eruption forecasting: Pilot study for
Whakaari/White Island, New Zealand. Frontiers in
Earth Science, 6, 211.
Daley, B., Hitman, G.A., Fenton, N., & McLachlan, S.
(2019) Assessment of the quality and content of
national and international guidelines on the
identification and management of Diabetes in
Pregnancy: An AGREE II Study. BMJ Open, e:027285.
Du, R., & Li, L. (2021). Estimating the risk of insulin
requirement in women complicated by gestational
diabetes mellitus: a clinical nomogram. Diabetes,
Metabolic Syndrome and Obesity: Targets and
Therapy, 2473-2482.
Dube, K., Kyrimi, E. & McLachlan, S. (2023). Predictive
Models for Health Conditions: A Review of Pregnancy
Models, Validation Methods, Risk Factors and
Symptoms Used. Manuscript in preparation.
Kumar, M., Ang, L. T., Png, H., Ng, M., Tan, K., Loy, S.
L., ... & Karnani, N. (2022). Automated machine
learning (AutoML)-derived preconception predictive
risk model to guide early intervention for gestational
diabetes mellitus. International Journal of
Environmental Research and Public Health, 19(11),
6792.
Kyrimi, E., Neves, M., McLachlan, S., Neil, M., Marsh, W.,
& Fenton, N. (2020). Medical Idioms for clinical
Bayesian Network Development. Journal of
Biomedical Informatics, 108. https://doi.org/10.1016/
j.jbi.2020.103495
Approach and Method for Bayesian Network Modelling: The Case for Pregnancy Outcomes in England and Wales
611

McLachlan, S. (2023). When is a new study NOT a new
study? Law, Health and Technology. Last accessed:
13th June, 2023. Sourced from:
https://lawhealthandtech.substack.com/p/when-is-a-
new-study-not-a-new-study
McLachlan, S., Kyrimi, E., Dube, K., & Fenton, N. (2020a).
Standardising Clinical Caremaps: Model, Method and
Graphical Notation for Caremap Specification.
Communications in Computer and Information
Science, 1212, pp 429-452. Springer Nature, DE.
McLachlan, S., Paterson, H., Dube, K., Kyrimi, E.,
Dementiev, E., Neil, M., ... & Fenton, N. E. (2020,
February). Real-time online probabilistic medical
computation using Bayesian networks. In 2020 IEEE
International Conference on Healthcare Informatics
(ICHI) (pp. 1-8). IEEE.
McLachlan, S., Daley, B., Saidi, S., ... & Fenton, N. (2024).
Approach and method for Bayesian Network
modelling: A case study in pregnancy outcomes for
England and Wales. Preprint.
http://dx.doi.org/10.13140/RG.2.2.20057.29281
Mehta-Lee, S. S., Palma, A., Bernstein, P. S., Lounsbury,
D., & Schlecht, N. F. (2017). A preconception
nomogram to predict preterm delivery. Maternal and
child health journal, 21, 118-127.
NCARDRS. (2022). NCARDRS Congenital Abnormality
Official Statistics Report, 2020. Last accesed: 26th
May, 2023. Sourced from: https://digital.nhs.uk/data-
and-information/publications/statistical/ncardrs-
congenital-anomaly-statistics-annual-data/ncardrs-
congenital-anomaly-statistics-report-2020
NHSInform. (2022). Miscarriage. Last accessed: 25th May,
2022. Sourced from: https://www.nhsinform.scot/
illnesses-and-conditions/pregnancy-and-childbirth/mis
carriage
NMPA Project Team. (2021) National Maternity and
Perinatal Audit: Clinical Report 2021. Based on births
in NHS maternity services in England, Scotland and
Wales between 1 April 2017 and 31 March 2018.
London: RCOG. Last accessed: 26th May, 2023.
Sourced from: https://maternityaudit.org.uk/FilesUplo
aded/Ref315%20NMPA%20clinical%20report%2020
21_v1.1.pdf
North, R. A., McCowan, L. M., Dekker, G. A., Poston, L.,
Chan, E. H., Stewart, A. W., ... & Kenny, L. C. (2011).
Clinical risk prediction for pre-eclampsia in nulliparous
women: development of model in international
prospective cohort. Bmj, 342.
ONS (2023). Dataset: Birth Characteristics. Last accessed:
10th June, 2023. Sourced from: https://www.ons.gov.
uk/peoplepopulationandcommunity/birthsdeathsandma
rriages/livebirths/datasets/birthcharacteristicsinengland
andwales
PHE (2019). Health of women before and during
pregnancy: health behaviours, risk factors and
inequalities. An updated analysis of the maternity
services dataset antenatal booking data. Last accessed:
10th June, 2023. Sourced from:
https://assets.publishing.service.gov.uk/government/up
loads/system/uploads/attachment_data/file/844210/He
alth_of_women_before_and_during_pregnancy_2019.
pdf
Pitchforth, J., & Mengersen, K. (2013). A proposed
validation framework for expert elicited Bayesian
Networks. Expert Systems with Applications, 40(1),
162-167.
Sanford, A., & Moosa, I. (2015). Operational risk
modelling and organizational learning in structured
finance operations: A Bayesian network approach.
Journal of the Operational Research Society, 66, 86-
115.
Smith, M. S. R., Saberi, S., Ajaykumar, A., Zhu, M. M.,
Gadawski, I., Sattha, B., ... & Côté, H. C. (2023).
Robust tobacco smoking self-report in two cohorts:
pregnant women or men and women living with or
without HIV. Scientific Reports, 13(1), 7711.
Stott, D., Bolten, M., Salman, M., Paraschiv, D., Clark, K.,
& Kametas, N. A. (2016). Maternal demographics and
hemodynamics for the prediction of fetal growth
restriction at booking, in pregnancies at high risk for
placental insufficiency. Acta Obstetricia et
Gynecologica Scandinavica, 95(3), 329-338.
Tandu-Umba, B., Mbangama, M. A., Kamongola, K. M. B.,
Kamgang Tchawou, A. G., Kivuidi, M. P., Kasonga
Munene, S., ... & Kasikila Kuzungu, S. (2014). Pre-
pregnancy high-risk factors at first antenatal visit: how
predictive are these of pregnancy outcomes?.
International Journal of Women's Health, 1011-1018.
HEALTHINF 2024 - 17th International Conference on Health Informatics
612
