
CT to MRI Image Translation Using CycleGAN: A Deep Learning
Approach for Cross-Modality Medical Imaging
Anamika Jha
1
and Hitoshi Iima
2
1
Department of Information Science, Kyoto Institute of Technology, Matsugasaki, Kyoto, Japan
2
Department of Information and Human Sciences, Kyoto Institute of Technology, Matsugasaki, Kyoto, Japan
Keywords: MRI, CT, Deep Learning, CycleGAN, Unpaired Dataset, Image Translation.
Abstract: Medical imaging plays a crucial role in healthcare, with Magnetic Resonance Imaging (MRI) and Computed
tomography (CT) as key modalities, each having unique strengths and weaknesses. MRI offers exceptional
soft tissue contrast, but it is slow and costly, while CT is faster but involves ionizing radiation. To address
this paradox, we leverage deep learning, employing CycleGAN to translate CT scans into MRI-like images.
This approach eliminates the need for additional radiation exposure or costs. Our results, which show the
effectiveness of our image translation method with an MAE of 0.5309, MSE of 0.37901, and PSNR of 52.344,
demonstrate the promise of this invention in lowering healthcare costs, expanding diagnostic capabilities, and
improving patient outcomes. The model was trained for 500 epochs with a batch size of 500 on an Nvidia
GPU, RTX A6OOO.
1 INTRODUCTION
A key component of contemporary healthcare is
medical imaging, which gives medical personnel a
visual representation and comprehension of the
human body's interior architecture. Computed
tomography (CT) and magnetic resonance imaging
(MRI) are two of the most widely utilized medical
imaging techniques. These technological
advancements offer unique yet complementary
perspectives on the human anatomy.
MRI is a non-invasive medical imaging method
that creates finely detailed images of the body's
internal structures by utilizing radio waves, strong
magnets, and a computer. A well-known feature of
MRI is its remarkable soft tissue contrast. It is a vital
tool for many medical applications, such as
neuroimaging, cancer, and musculoskeletal imaging,
due to its exceptional ability to visualize organs,
muscles, nerves, and other soft tissues.
Contrarily, CT is an alternative imaging technique
that makes use of X-ray technology. It produces
"slices," or cross-sectional, images of the body that
can be assembled into three-dimensional
representations. CT scans are renowned for their
effectiveness and speed, which enables quick picture
capture. They are very helpful for seeing blood
arteries, identifying fractures, and imaging bone
structures.
There are many CT scanners, but a few MRI ones.
Therefore, the idea of image translation from a CT
scan to an MRI image is extremely important in the
realm of medical imaging. The goals of this study
project are to realize this image translation and to
greatly improve diagnostic capacities. The image
translation enables medical practitioners to take
advantages of both methods, using MRI's soft tissue
contrast and CT scans' comprehensive information.
Thus, this development is promising for more
thorough and precise diagnoses, which eventually
enhance patient care and treatment results. Both
patients and healthcare providers stand to gain from
this substantial reduction in medical expenses and
waiting times.
To provide context and insight into the
significance of our work, we begin by taking up
methodologies of prior research studies that have
paved the way for our contributions. We have not
found any previous work on translating a CT scan to
an MRI image, but previous work in other medical
image translation has introduced the concept of using
Generative Adversarial Networks (GANs) for image-
to-image translation (Denck et al.,2021). Pix2pix (Li
et al.,2021), UNIT (Welander et al.,2018),
CycleGAN (Zhu et al.,2017) and UNET
Jha, A. and Iima, H.
CT to MRI Image Translation Using CycleGAN: A Deep Learning Approach for Cross-Modality Medical Imaging.
DOI: 10.5220/0012422900003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 3, pages 951-957
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
951

(Ronneberger et al.,2015) models have been used in
previous research. Training images used in our work
are not paired. CycleGAN can seamlessly handle
such unpaired data (Wolterink et al.,2017). Therefore,
our method to translate a CT scan into an MRI image
leverages CycleGAN’s capacity. By embracing cycle
consistency, the CycleGAN model learns to map CT
and MRI images in both directions. It generates
synthetic MRI images from CT and can revert these
generated MRI images to their original CT-like
representations. The effectiveness of our model is
examined through experiments.
2 DATASET
The dataset used in this research was obtained from
an open-source repository on Kaggle. The dataset was
meticulously aggregated to serve as the foundation
for training the CycleGAN model, specifically
designed for image-to-image translation.
This dataset is essential to our work because it
allows us to develop and assess our methodology for
translating CT to MRI images. It supplies the basis
for the CycleGAN model's training and testing,
ultimately leading to improvements in cross-modality
medical imaging.
2.1 Dataset Content
The dataset comprises a collection of CT and MRI
scans, focusing on brain cross sections. These images
were sourced from various listed repositories and
were subsequently organized into separate directories
for both training and testing purposes. The dataset is
divided into two primary domains: Domain A, which
contains CT scans, and Domain B, which comprises
MRI scans. This clear separation enables the effective
utilization of the dataset for CycleGAN-based image
translation, ensuring that the model can learn and map
the distinct features and characteristics of CT scans to
their MRI counterparts.
The dataset is available under the Creative
Commons Attribution-Non-Commercial-Share Alike
4.0 International License (CC BY-NC-SA 4.0). This
licensing arrangement governs the usage,
redistribution, and modification of the dataset,
emphasizing the importance of proper attribution,
non-commercial usage, and the continuity of the
open-source spirit.
3 METHODLOGY
Deep learning has become a viable approach to bridge
the image gap. Specifically, image-to-image
translation challenges have demonstrated the
potential of GANs.
GANs could be a great option in the field of
medical imaging, as CT and MRI scans offer many
forms of information. The contrast, texture, and
anatomical characteristics of these modalities differ,
hence a model that can capture complex data
distributions is required. GANs are highly effective in
simulating intricate transformations.
3.1 GAN Model Selection
One significant obstacle in the field of medical
imaging is the dearth of paired data, or sets of
comparable CT and MRI pictures of the same
individuals. CycleGAN is a great option for the CT to
MRI translation challenge because of its ability to
handle unpaired data. In order to guarantee the
model's efficacy even when the amount of paired data
is restricted, it incorporates a cycle consistency loss
that compels translated images to return to their
original domains.
3.2 CycleGAN
The core of this research's image-to-image translation
lies in the innovative architecture of CycleGAN.
CycleGAN is a type of GAN that is particularly well-
suited for unpaired image translation tasks, making it
a powerful choice for transforming CT scans into
MRI-like images. CycleGAN comprises two key
components: the generator and the discriminator. The
generator is responsible for creating the translated
images, in this case, generating synthetic MRI scans
from CT scans. The discriminator, on the other hand,
is tasked with distinguishing between real MRI
images and those generated by the generator.
3.2.1 Generator Architecture
Figure 1 shows the architecture of CycleGAN
generator where s is the stride. The CycleGAN
generator has 3 sections: Encoder, Transformer and
Decoder (Zhu et al.,2017).
The encoder receives the input CT image. The
encoder uses convolutions to extract features from the
input image and compresses the image representation
while increasing the number of channels. Three
convolutions make up the encoder, which shrinks the
representation to one-fourth the size of the original
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
952
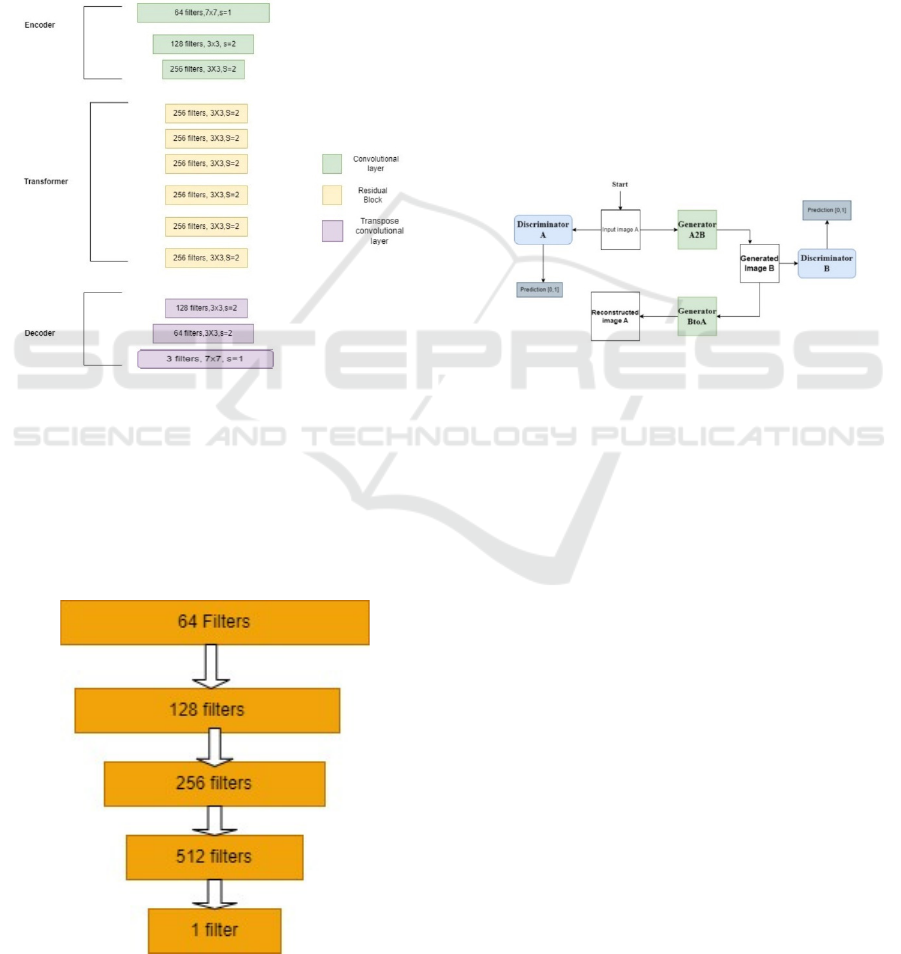
image. When we feed an image into the encoder with
dimensions of (256, 256, 3), the result is (64, 64, 256).
Following the application of the activation
function, the encoder's output is then fed into the
transformer. General transformers contain six or nine
residuals blocks, depending on the magnitude of the
input. We adopt six residual blocks for medical image
translation. The transformer's output is then fed into
the decoder, which increases the representation's size
to its initial size by using a 2-deconvolution block of
fractional strides.
Figure 1: CycleGAN Generator.
3.2.2 Discriminator Architecture
The CycleGAN discriminator uses PatchGAN [12].
The Patch GAN differs from a regular GAN
discriminator in that the regular GAN maps a
256x256 image to a single scalar output that indicates
whether the image is real or fake. In contrast the Patch
Figure 2: CycleGAN Discriminator.
GAN maps a 256x256 image to an NxN array of
outputs X, where each element Xij indicates whether
the patch ij in the image is real or fake. Figure 2 shows
the architecture of the discriminator.
3.2.3 CycleGAN Architecture
The strength of CycleGAN lies in its cycle
consistency constraint, a defining feature of ensuring
the model translates an input image from one domain
to the other and back to the original input image. In
the context of this study, this means that if we
translate a CT scan into an MRI-like image and then
revert it to the original domain, it should closely
resemble the original CT scan. This cycle consistency
is integral to achieving high-quality and anatomically
accurate translations. Figure 3 shows the architecture
of CycleGAN. In this study, image A is a CT scan,
and image B is an MRI-like image.
Figure 3: CycleGAN.
CycleGAN architecture also incorporates
adversarial losses, which compel the generator to
produce images that are indistinguishable from real
MRI scans, as judged by the discriminator. This
adversarial training encourages the generator to
create highly realistic images.
The architecture's ability to work with unpaired
datasets is a significant advantage. In traditional
supervised learning, paired data (where each input
has a corresponding output) is required
(Armanious,2019), which can be challenging to
obtain in medical imaging. CycleGAN's ability to
handle unpaired data makes it a valuable tool for this
CT-to-MRI image translation task.
3.3 CycleGAN Losses
The effectiveness of CycleGAN in image-to-image
translation tasks is attributed to a collection of
carefully designed loss functions (Armanious et
al.,,2019), each serving a specific purpose to guide
the training process and ensure the desired results.
The key losses employed in CycleGAN architecture
are explained in this subsection.
CT to MRI Image Translation Using CycleGAN: A Deep Learning Approach for Cross-Modality Medical Imaging
953

3.3.1 Adversarial Loss
Adversarial loss is fundamental in GAN-based
models and aims to make the generated images
indistinguishable from real images. The discriminator
and generator networks are trained to compete against
one another using the adversarial loss. The
discriminator network seeks to discern between real
and generated images, while the generator network
attempts to produce realistic images enough to trick
it. The adversarial loss is given by:
𝐿𝑜𝑠𝑠
= (1−𝐷
(𝐺(
𝐴
)))
(1)
𝐿𝑜𝑠𝑠
= (1−𝐷
(𝐹(𝐵)))
(2)
where
𝐺: Generator transforming input image A to B.
𝐹: Generator transforming image B to A.
𝐷
: Discriminator for B.
𝐷
: Discriminator for A.
In the context of CT to MRI translation, the
generator is pitted against the discriminator, which
learns to differentiate between genuine MRI scans
and translated MRI-like images. The generator's
objective is to minimize this loss by creating
convincing images enough to fool the discriminator.
3.3.2 Cycle Consistency Loss
Cycle consistency loss is the defining characteristic
of CycleGAN. It enforces the model to maintain
consistency when translating images in both
directions.
To make the generator network learn the proper
mapping between the two domains, the cycle
consistency loss is employed. An image is translated
from one domain to the other, and then back to the
original domain to calculate cycle consistency loss.
When the translated image is as similar to the original
image as possible, the cycle consistency loss is light.
The cycle consistency is given by
𝐿𝑜𝑠𝑠
=(F(G
(
A
)
− A + (GF
(
B
)
−B).
(3)
In the case of this research, it ensures that when a
CT scan is transformed into an MRI-like image and
then reverted to the CT domain, the resulting image
closely resembles the original CT scan. This loss
plays a critical role in ensuring anatomical accuracy
and image fidelity.
The overall CycleGAN loss function is a weighted
sum of the adversarial loss and the cycle consistency
loss.
4 EXPERIMENTS
This section provides a detailed description of the
experimental setup that was used to evaluate the
suggested approach.
4.1 Experimental Setup
Python 3.8.10 was used throughout the development
of the complete framework, with TensorFlow 2.6.5
serving as the neural network computing backend and
Keras serving as the deep learning framework. The
integrated programming environment Visual Studio
Code was used for both the framework's development
and implementation.
To facilitate efficient model training and
accelerate the image translation process, we
leveraged the computational power of a dedicated
GPU. Specifically, the experiment was conducted on
an Nvidia GPU, RTX A6OOO, equipped with CUDA
Version 11.3. This GPU configuration allowed for the
expedited execution of deep learning operations,
significantly reducing the training time. The choice of
such hardware specifications was instrumental in
achieving the low time complexity of the proposed
method, making it more time-efficient compared to
other complex deep learning models. The utilization
of this GPU configuration, combined with the
streamlined deep learning framework, enables a
seamless and efficient image translation process from
CT to MRI scans.
4.2 Dataset Pre-Processing
The primary objective of data pre-processing is to
load and standardize the dimensions of the CT and
MRI images. Each image is loaded, and its
dimensions are resized to a uniform scale of 256x256
pixels. This resizing ensures consistency across all
images, which is vital for neural network training. In
order to expedite the training process, a subset of the
data is selected. For the CT scans, 500 images out of
1742 are chosen, and for the MRI images, a subset of
500 out of 1744 images are selected. This
subsampling facilitates a more efficient training
process, especially for demonstration purposes. Note
that CT and MRI scans are an unpaired dataset.
To make the data compatible with the neural
network architecture, an essential pre-processing step
is applied. The pixel values of the images are scaled
to fit within the range of [-1, 1]. This scaling is
imperative because the generator in the CycleGAN
model employs the tanh activation function in its
output layer, producing values within this range.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
954

Scaling the data accordingly, ensures that the
generator can produce realistic and meaningful
images.
These meticulous pre-processing steps result in a
well-structured and appropriately scaled dataset. The
dimensions of the data, after pre-processing, are as
follows: The dataset consists of 1000 images, each
with dimensions of 256x256x3 (width, height, and
channels).
Data augmentation is done to compensate the
limited dataset. Effective deep learning model
training requires the diversification of datasets, which
is facilitated by data augmentation. Our goal in using
augmentations is to reduce the likelihood of
overfitting by simulating variables found in the real
world.
4.3 Evaluation
Several metrics are used to assess the suggested CT
to MRI image translation model based on the
CycleGAN architecture in order to determine the
model's performance. When comparing the translated
images to actual MRI scans, these metrics objectively
evaluate the translated images' fidelity and accuracy.
The main assessment metrics include the Mean
Absolute Error (MAE), Mean Squared Error (MSE),
and Peak Signal-to-Noise Ratio (PSNR).
4.3.1 MAE
MAE quantifies the average absolute difference
between the pixel values of the translated MRI-like
images and the corresponding real MRI scans. It is a
valuable indicator of the overall dissimilarity between
the generated and ground truth images. A lower MAE
suggests a closer resemblance between the translated
and real MRI images. MAE is given by
𝑀𝐴𝐸=
1
𝑛
|
𝑦
−𝑦
|
(4)
where
𝑛: no of samples or data,
𝑦
: actual (observed) value for the ith sample,
𝑦
: predicted value for the ith sample.
4.3.2 MSE
MSE computes the mean of the squared differences
between the pixel values of the generated MRI-like
images and the true MRI scans. This metric provides
insights into the magnitude of errors of the generated
MRI-like images, with smaller MSE values
indicating reduced image dissimilarity. MSE is given
by
𝑀𝑆𝐸=
1
𝑛
(
𝑦
−𝑦
)
.
(5)
4.3.3 PSNR
PSNR is a standardized measure to evaluate the
quality of the generated images. It calculates the ratio
of the peak intensity of an image to the root mean
square error. Higher PSNR values signify a closer
match to the real MRI scans, with increased image
fidelity and reduced noise
.
PSNR is given by
𝑃𝑆𝑁𝑅=10.log
𝑀𝐴𝑋
𝑀𝑆𝐸
(6)
where MAX is the maximum possible pixel value of
the image.
5 RESULTS
5.1 Generated Images and Their
Evaluation
Figures 4 and 5 show the visual representation of the
generated MRI-like images as the result of evaluating
the effectiveness of the CycleGAN model for CT to
MRI image translation.
After more than 50,000 iterations of rigorous
training, the model was able to generate artificial MRI
scans from CT data, as seen in these images. The
presented pictures demonstrate how well the model
can create MRI-like images from CT scans.
Figure 4: MRI images generated after training the
CycleGAN model for 100 epochs.
CT to MRI Image Translation Using CycleGAN: A Deep Learning Approach for Cross-Modality Medical Imaging
955
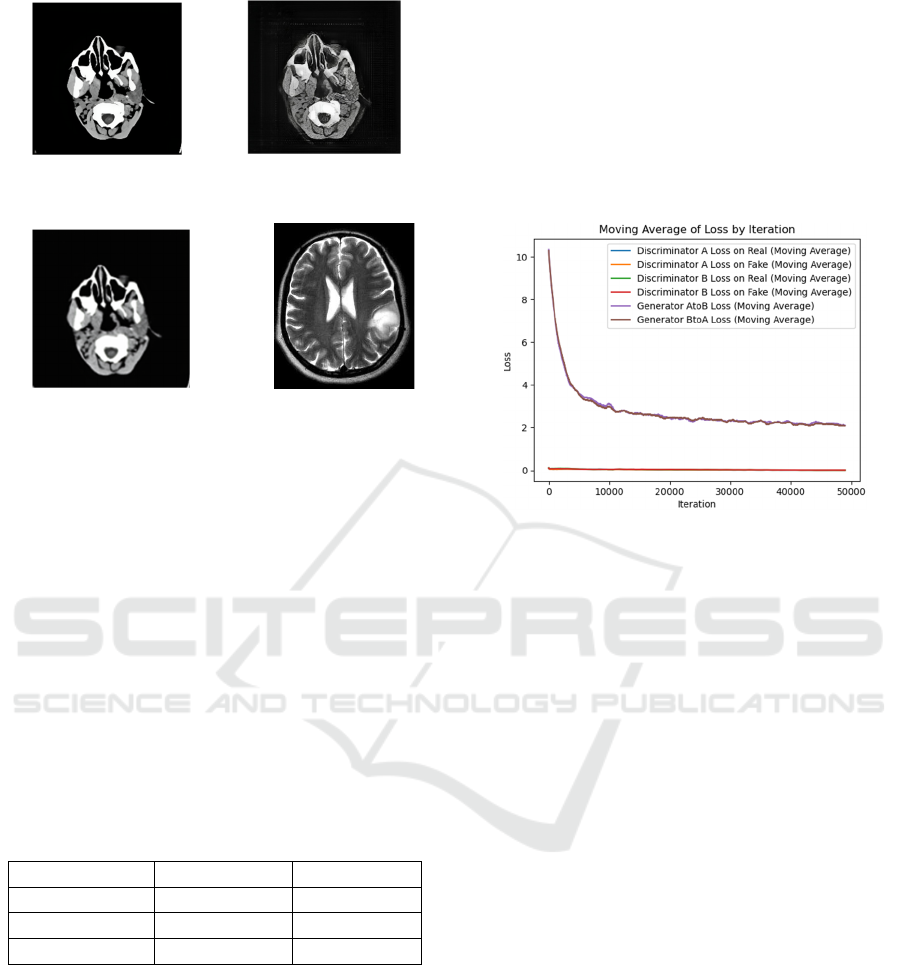
(a) (b)
(c) (d)
Figure 5: Output after using the CycleGAN model for test
dataset: (a) ground truth of CT, (b) translated MRI ,(c)
reconstructed CT scan ,(d) MRI image from unpaired test
dataset for reference.
The result shown in Figure 5(b) is a T1-weighted
MRI image of the brain generated from a CT scan
using a CycleGAN model. The image shows a decent
overall representation of the brain anatomy, with
clear visualization of the gray matter, white matter,
cerebrospinal fluid, and major blood vessels.
However, it is important to note that this is a
synthetic image and should not be used thoughtlessly
for clinical diagnosis. Some subtle details may be lost
in the generation process, and the image may not be
as accurate as a real MRI scan.
Table 1: The evaluation metrics with CNN and CycleGAN
for CT to MRI translation.
CNN CycleGAN
MAE 70.44 0.5309
MSE 60.867 0.37901
PSNR 9.457 52.344
Table 1 shows the metrics when the test dataset of
CT scan images is passed through the model and
translated as the MRI images and the real MRI images
as well as translated MRI are compared. The CNN in
this table is adopted as a baseline method that cannont
be learned using unpaired dataset. It is trained using
a dataset in which each CT scan is paired with an MRI
scan randomly. The results of CNN were not
satisfying enough as it is not capable of handling the
unpaired dataset. In contrast, CycleGAN
demonstrates high performance.
5.2 Loss Plot
The training progression is depicted through loss
graphs shown in Figure 6, illustrating the evolution of
these loss components over time. Notably, the graphs
showcase a consistent and substantial decrease in the
loss values for all six components throughout the
training process. This trend signifies the model's
remarkable capacity to learn and adapt.
Figure 6: Loss of 100 epochs.
6 CONCLUSIONS
This work represents a significant advancement in the
field of cross-modality medical imaging, especially
with regard to the complex process of translating CT to
MRI images. The fact that the CycleGAN model was
able to be implemented successfully shows how well it
can bridge the gap between these modalities and
convert CT scans into high-fidelity MRI-like images
pix2pix (Cao et al.,2021). This study has far-reaching
implications, particularly in the field of healthcare,
where the synthesis of radiation-free and economically
viable MRI-like data has the potential to transform
diagnostic capabilities, save costs associated with
healthcare, and shorten patient wait times.
The comprehensive evaluation of the model's
performance, quantified by pivotal metrics such as
Mean Absolute Error (MAE), Mean Squared Error
(MSE), and Peak Signal-to-Noise Ratio (PSNR),
solidifies the model's efficacy. Exhibiting low MAE
and MSE alongside a notably high PSNR, the
translated MRI-like images manifest an exceptional
resemblance and fidelity to actual MRI scans. This
not only underscores the model's adeptness in
generating top-tier images but also bolsters its
diagnostic prowess, paving the way for more accurate
medical assessments.
Furthermore, to fortify the significance of this
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
956

study, a comparative analysis was conducted between
the CycleGAN and a fundamental CNN model,
showcasing the former's superiority in image
translation capabilities.
7 FUTURE WORK
With the CycleGAN model, this work has established
a solid basis for practical CT-to-MRI image
translation, which could lead to major breakthroughs
in cross-modality medical imaging (Kazeminia et
al.,,2020). Looking ahead, several interesting
directions for more study and advancement become
apparent.
The next step in the research is incorporation of
Super Resolution GAN(SRGAN) into the image
enhancement process offers substantial benefits to
this research (Ledig et al.,,2017). With the goal of
creating high-resolution images from lower-
resolution inputs, SRGAN is an expert in super-
resolution tasks. SRGAN has the potential to improve
the overall quality and fine details of the MRI images
that are generated in the context of CT-to-MRI image
translation. It enhances the current CycleGAN
framework by improving the resolution and fidelity
of the translated MRI-like images, which could lead
to sharper, more realistic representations that closely
resemble actual MRI scans.
Moreover, an exciting prospect involves the
creation of a hybrid model merging SRGAN with
CycleGAN, aiming to capitalize on the strengths of
both architectures. This hybrid approach intends to
leverage the super-resolution capabilities of SRGAN
to enhance fine details and resolution in the MRI-like
images generated by CycleGAN. By integrating these
models, the goal is to produce sharper, high-
resolution MRI-like images with enriched visual
quality, closely resembling authentic MRI scans.
Furthermore, the results will be compared with other
models like UNET, CycleGAN etc.
ACKNOWLEDGEMENTS
This work was partly supported by JSPS KAKENHI
Grant Number JP23K11263.
REFERENCES
Anaya, E. and Levin, C. (2021) Evaluation of a Generative
Adversarial Network for MR-Based PET Attenuation
Correction in PET/MR, In 2021 IEEE Nuclear Science
Symposium and Medical Imaging Conference
(NSS/MIC), pp. 1-3. https://doi.org/10.1109/NSS/
MIC44867.2021.9875556
Armanious, K., Jiang, C., Abdulatif, S., Küstner, T.,
Gatidis, S.,and Yang, B., (2019). Unsupervised Medical
Image Translation Using Cycle-MedGAN,27
th
European Signal Processing Conference.
Cao, G., Liu, S., Mao, H., and Zhang, S. (2021) Improved
CyeleGAN for MR to CT synthesis, In 2021 6th
International Conference on Intelligent Informatics and
Biomedical Sciences (ICIIBMS), pp. 205-208.
https://doi.org/10.1109/ICIIBMS52876.2021.9651571.
Denck, J., Guehring, J., Maier, A., and Rothgang, E.
(2021).MR-contrast-aware image-to-image translations
with generative adversarial networks, International
Journal of Computer Assisted Radiology and
Surgery,Vol.16,pp.2069-2078.
Isola, P., & Zhu, J.,-Y., & Zhou, T., & Efros, A. (2017).
Image-to-Image Translation with Conditional
Adversarial Networks. , In 2017 IEEE Conference on
Computer Vision and Pattern Recognition (CVPR), pp.
5967-5976. https://doi.org/10.1109/CVPR.2017.632.
Kazeminia, S., Baur, C., & Kuijper, A., Ginneken, B.,
Navab, N., Albarqouni, S.,and Mukhopadhyay, A.
(2020). GANs for Medical Image Analysis. Artificial
Intelligence in Medicine. vol.109. https://doi.org/
10.1016/j.artmed.2020.101938.
Ledig, C., Theis, L.,Huszar, F.,Caballero, J.,Cunningham,
A.,Acosta, A., Aitken, A., Tejani, A., Totz, J.,Wang,
Z.,and Shi, W. (2017). Photo-Realistic Single Image
Super-Resolution Using a Generative Adversarial
Network. 105-114. 10.1109/CVPR.2017.19
Li, M., Zhang, T. and Li, S. (2021) An innovative image
segmentation approach for brain tumor based on 3D-
Pix2Pix, In 2021 6th International Symposium on
Computer and Information Processing Technology
(ISCIPT),pp.542-545. https://doi.org/10.1109/ISCIPT
53667.2021.00115.
Ronneberger, O., Fischer, P.,and Brox, Thomas. (2015). U-
Net: Convolutional Networks for Biomedical Image
Segmentation. Medical Image Computing and
Computer-Assisted Intervention, MICCAI 2015,
Lecture Notes in Computer Science(), vol. 9351.
https://doi.org/10.1007/978-3-319-24574-4_28.
Welander, P., Karlsson, S.,Eklund A. (2018). Generative
Adversarial Networks for Image-to-Image Translation
on Multi-Contrast MR Images - A Comparison of
CycleGAN and UNIT, arXiv:1806.07777.
Wolterink, J.M., Dinkla, A.M., Savenije, M.H.F., Seevinck,
P.R., van den Berg, C.A.T., and Išgum, I. (2017). Deep
MR to CT Synthesis Using Unpaired Data Simulation
and Synthesis in Medical Imaging. SASHIMI 2017.
Lecture Notes in Computer Science(), vol, 10557.
Zhu, J.-Y., Park, T., Isola, P., and Efros A.A. (2017).
Unpaired Image-to-Image Translation Using Cycle-
Consistent Adversarial Networks, In 2017 IEEE
International Conference on Computer Vision, pp.
2242-2251. https://doi.org/10.1109/ICCV.2017.244.
CT to MRI Image Translation Using CycleGAN: A Deep Learning Approach for Cross-Modality Medical Imaging
957
